Abstract
We have developed a novel chromatography for the rapid isolation of active ribosomes from bacteria without the use of harsh conditions or lengthy procedures that damage ribosomes. Ribosomes interact with an alkyl linker attached to the resin, apparently through their RNA component. Examples are given with ribosomes from Escherichia coli, Deinococcus radiodurans, and with clinical isolates of Streptococcus pneumoniae and methicillin-resistant Staphylococcus aureus (MRSA). The ribosomes obtained by this method are unusually intact, so that highly active ribosomes can now be isolated from the clinical isolates, enabling significantly improved in vitro functional assays that will greatly assist the discovery and development of new ribosomally targeted antibiotics.
Keywords: ribosomes, chromatography, purification, pathogens
INTRODUCTION
Almost half of the known antibiotics target the bacterial ribosome, an attractive and proven target for antibacterial drug discovery. However, increasing levels of bacterial drug resistance threaten to erode the medical advances of recent decades. The spread of multiply resistant strains, both in hospitals and now in the community, has further added to the threat.
To efficiently develop new generations of antibiotics to overcome this resistance, clinical isolates, especially multi-drug-resistant strains, must be studied. However, the biochemical and structural biology tools used in ribosome-targeted antibiotic research usually employ “laboratory” strains. This is because isolation of fully functional ribosomes from clinical isolates such as methicillin-resistant Staphylococcus aureus (MRSA) is prevented by very high levels of protease and nuclease activity that degrade their ribosomes during purification. This, in turn, limits our ability to precisely understand resistance mechanisms and discover effective new therapeutic reagents.
Purification of ribosomes by ultracentrifugation has remained the standard method for ribosome isolation since the field began. However, it is lengthy and laborious. In the 1970’s and 1980’s, three new methods were advanced. These were affinity purification using ribosomally targeted antibiotics as bait (Le Goffic et al. 1974, 1980), size exclusion chromatography (SEC) (Jelenc 1980), and hydrophobic interaction chromatography (HIC) (Kirillov et al. 1978; Fabry et al. 1981; Saruyama 1986). Initial reports of antibiotic affinity chromatography were promising, but have not led to further publications. SEC has been successfully used by a couple of laboratories, but was not widely adopted. Meanwhile, HIC has only recently gained popularity as a method for isolating ribosomes from Thermus thermophilus for use in X-ray crystallography (Clemons et al. 2001; Petry et al. 2005; Korostelev et al. 2006; Selmer et al. 2006). HIC requires high salt concentrations (typically, 1.5–2 M) to effect resin binding. Such high concentrations of salt appear well tolerated by the more robust thermophilic ribosomes, but can damage ribosomes from nonextremophiles like Escherichia coli. Some early methods also employed buffers containing detergents or solvents. Thus, purification of ribosomes from E. coli by HIC can result in dissociation of ribosomal proteins from the ribosome (Kirillov et al. 1978; Fabry et al. 1981) and/or loss of ribosomal activity (Saruyama 1986).
The advent of molecular biology methods for affinity tagging has enabled expression of tagged ribosomal RNA, which, once assembled into ribosomes, can be exploited for their purification (for review, see Youngman and Green 2005). While tagging each of the (usually) multiple copies of rRNA genes is not practicable, this technique is especially suited to isolation of a tagged subpopulation of mutant ribosomes from a pool of wild-type ribosomes. Tagging of ribosomal proteins has also been used (Inada et al. 2002). However, modification of ribosome components requires investment in strain construction and is not available for all species.
Therefore, a chromatographic method for isolating ribosomes from any organism that does not require prior strain construction or employ conditions that lead to ribosome damage, would be highly advantageous. We describe here a novel chromatography that satisfies all of these requirements, allowing efficient and scaleable isolation of ribosomes from bacterial lysates without harsh conditions and rapidly enough to limit degradation, resulting in highly active ribosomes even from Streptococcus pneumoniae and MRSA.
RESULTS
Sulfolink resin (Pierce, Inc.) is a cross-linked agarose resin (CL-6B) derivatized with 18-atom alkyl chains carrying a reactive iodoacetyl group at their terminus. The resin is designed for covalent immobilization of proteins and other ligands by reaction of the iodoacetyl group with free sulfhydryls in the protein. Reaction with free cysteine is then used to modify any remaining iodoacetyl groups. For this study, all of the alkyl chains were reacted with cysteine to produce the resin shown in Figure 1 (R group number 4).
FIGURE 1.
Chemical structures of resins used in this study. (A) Sulfolink resin was reacted with (1) ethanethiol, (2) cysteamine, (3) 3-mercaptopropionic acid, or (4) l-cysteine, to yield the derivatives terminated by the corresponding R-groups shown. (B) Structure of Biotin-PEO-LC amine resin. The biotin moiety on the left of the linker is bound to the streptavidin of the streptavidin-agarose resin.
Binding of ribosomes to the resin was first demonstrated using crude ribosomes from a laboratory strain of E. coli. These bound to the cysteine-Sulfolink resin in binding buffer (see Materials and Methods), and over 90% of the input ribosomes were recovered by increasing the concentration of NH4Cl to 0.5 M to elute the bound material. With all resins tested, either >90% of the ribosomes were recovered in the elution or >90% in the flowthrough and washes, so that the distinction between success and failure was clear. The agarose resin alone did not bind ribosomes. A biotin-PEO-LC-amine linker was also tested (Fig. 1B). (The biotin on one end of the linker is to allow it to be bound to a streptavidin-agarose support). This less hydrophobic linker failed to give any binding, suggesting that hydrophobic interactions may play a part. Elution in high salt suggests ion exchange, perhaps by the zwitterionic cysteine head group on the resin. To test this hypothesis, we made resins with the charged and uncharged headgroups shown in Figure 1A (R groups 1–3), but none of them prevented binding or elution under these conditions. A slight preference for cysteamine (R=2), or less so, ethanethiol (R=1), was detected, but only in a buffer of higher ionic strength and with limiting resin. Since preparation of cysteine-Sulfolink is straightforward, this resin was used in subsequent experiments.
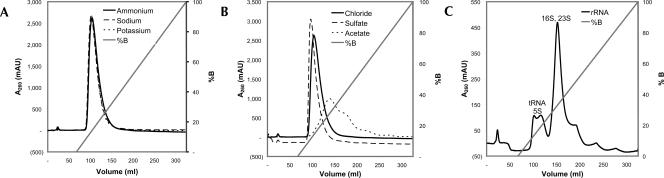
We next tested the effectiveness of different salts in elution of ribosomes from the resin. For convenience, the ribosomes were first purified from cells using the resin and aliquots of this pure sample were reapplied to the column so that the ribosomes could be tracked by their absorbance alone. Changing the cation had no effect on elution (Fig. 2), while changing the anion (Fig. 2B) altered elution in the order—sulfate first, then chloride, followed by acetate. This is similar to the “activity series” seen for elution of anion exchangers with different ions (Roe 1989). The multiple peaks obtained with the acetate salt were investigated further by sucrose-density gradient to determine whether different components were resolving, but all contained 30S, 50S, and 70S. When RNA was extracted from the ribosomes and chromatographed on its own with NH4Cl as the eluting salt (Fig. 2C), it too chromatographed, with early eluting peaks of mainly 5S and tRNA and a later peak comprising 16S and 23S rRNA.
FIGURE 2.
Chromatograms of E. coli ribosomes separated on cysteine-Sulfolink. (A) Elution by different chloride salts. (B) Elution by different ammonium salts. (C) E. coli RNA separated on cysteine-Sulfolink, with elution by ammonium chloride.
The utility of the resin was demonstrated by loading cell lysates from Deinococcus radiodurans, E. coli, S. pneumoniae, or methicillin-resistant S. aureus (MRSA) directly onto a preparative scale column. An example with Deinococcus is shown in the chromatography profile of Figure 3. Analysis of fractions by sucrose-density gradient centrifugation shows that the ribosomes contained in the cell lysate (Fig. 3B) are retained by the column (Fig. 3D), while the low molecular weight contaminants, such as soluble proteins, flow straight through the column (Fig. 3C). Elution releases the ribosomes, which show a similar gradient profile to those in the original lysate, including a peak of disomes (two 70S ribosomes on the same mRNA) that migrate at about 100S.
FIGURE 3.
Chromatography of D. radiodurans cell lysate on cysteine-Sulfolink. (A) Absorbance trace with fractions containing flowthrough, wash, and elution indicated on the plot. Note that absorbance exceeds detector range for the two highest peaks. (B–D) Absorbance profiles of samples of the cell lysate, flowthrough, and elution, respectively, after centrifugation on sucrose-density gradients. Peaks of 30S and 50S subunits, as well as 70S ribosomes and 100S disomes are indicated.
Based on absorbance at 260 nm, 51% of the material was in the flowthrough, 3% in the wash, and 42% was recovered in the elution, leaving only 4% unaccounted for. Forty-five A260 units of Deinococcus ribosomes were bound per milliliter of resin. Binding in this case was at a salt concentration of 0.15 NH4Cl, with elution at about 0.3 M NH4Cl, so binding and release are finely balanced. E. coli lysates gave similar results, except that more free tRNA was recovered in the elution. It may be that binding of tRNA to the resin is less favored in the milieu of the Deinococcus lysate.
The ribosome mixture eluted from the column can be pelleted and resuspended in appropriate buffers for storage or subjected to preparative scale sucrose-density gradient centrifugation to isolate 30S and 50S subunits or 70S ribosomes, if desired. Since the separation of 30S and 50S subunits by sucrose-density gradient is laborious, attempts were made to separately elute the dissociated subunits of E.coli or D. radiodurans (in 0.25 mM Mg2+) from the column with a very gradual salt gradient (0.28–0.64 M over 50 column volumes), but these were unsuccessful (data not shown).
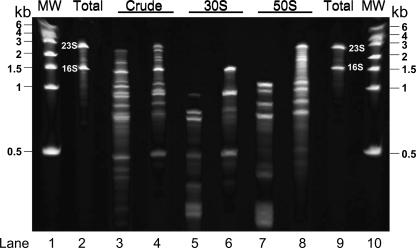
To compare the purity of the preparations, proteins were extracted and subjected to SDS-PAGE (Fig. 4). Lanes 2 and 3 in Figure 4 compare the proteins in a lysate of D. radiodurans with those in a sample of pure ribosomes. While the lysate contains many proteins that migrate above the 36 kD molecular weight marker, S1 (69 kD) is the only ribosomal protein with a molecular weight higher than 36 kD. The lysate also contains nonribosomal proteins below 36 kD, but these do not resolve from the ribosomal proteins. (Two-dimensional ribosomal protein gels give excellent resolution of individual ribosomal proteins, but most contaminants are insufficiently small or basic to be detected, so that purity was more easily visualized in this one-dimensional system). Crude ribosome preparations show some contaminants, whether prepared by centrifuge (Fig. 4, lane 4) or by chromatography (Fig. 4, lane 5), although the latter shows less contamination (and a different pattern of contaminants). The same is true with crude ribosomes from E. coli in lanes 8 and 9 (Fig. 4). Again, the only ribosomal protein that migrates above the 36 kD marker is S1 (61 kD). No differences were seen between 50S subunits purified from Deinococcus by centrifuge alone (Fig. 4, lane 6) or by chromatography and centrifuge (Fig. 4, lane 7), either on this gel, on two-dimensional gels, or by LC-MS analysis (data not shown).
FIGURE 4.
PAGE of proteins extracted from ribosome samples. (Lanes 1,10) Molecular weight standards. D. radiodurans proteins extracted from a cell lysate (lane 2) or from a 1:1 mixture of 30S and 50S subunits purified separately by centrifuge (lane 3). Proteins extracted from crude Deinococcus ribosomes that were purified by centrifuge (lane 4) or by chromatography (lane 5). (Lanes 8,9) The same from E. coli. Proteins from 50S subunits from Deinococcus by centrifuge alone (lane 6) or by chromatography and centrifuge (lane 7).
Preparation of Deinococcus ribosomes by centrifuge alone can lead to loss or degradation of several proteins (data not shown) and degradation of the ribosomal RNA, resulting in decreased activity. While ribosomes purified from Deinococcus by this resin showed comparable protein integrity to preparations made by centrifuge (LC-MS; data not shown), their rRNA was much more intact (Fig. 5). Comparison of RNA extracted from centrifuge-purified (Fig. 5, lane 3) and resin-purified ribosomes (Fig. 5, lane 4) shows that the latter contain much more intact 23S and 16S RNA (Fig. 5, lanes 2,9). When 30S and 50S subunits are derived from these ribosomes, they still contain intact 16S (Fig. 5, lane 6) and 23S rRNA (Fig. 5, lane 8), respectively. However, with subunits derived from centrifuge-purified ribosomes (Fig. 5, lanes 5,7), no intact rRNA can be seen.
FIGURE 5.
PAGE of RNA extracted from samples of D. radiodurans ribosomes. (Lanes 1,10) Molecular weight standards; (lanes 2,9) total RNA directly extracted from D. radiodurans cells (intact 16S and 23S as indicated). RNA extracted from centrifuge-purified (lanes 3,5,7) or resin-purified (lanes 4,6,8) ribosomes: crude ribosomes, 30S subunits, and 50S subunits as indicated above the gel.
To evaluate the biological activity of the ribosomes, we first compared binding of resin-purified and centrifuge-purified Deinococcus ribosomes to antibiotics such as erythromycin using a filter-binding assay (Fig. 6A). While centrifuge-purified ribosomes showed little binding to 14C-erythromycin, the resin-purified ribosomes demonstrated a stoichiometric binding profile typical of active ribosomes. This difference was reproducible, but assay specific, as it was not found by using the gentler method of equilibrium dialysis, suggesting that binding is more robust for subunits isolated by chromatography.
FIGURE 6.
Effect of the purification method on in vitro activities of ribosomes. (A) Binding of erythromycin to ribosomes purified from D. radiodurans by resin (▪) or by centrifuge alone (□). (B) Protein synthetic activity of ribosomes purified from MRSA (squares) or S. pneumoniae (diamonds) by resin (filled) or centrifuge (open) in bacterial transcription and translation assay.
No differences were seen for the Deinococcus ribosomes in a coupled transcription/translation assay (TNT). However, when we isolated ribosomes from clinical pathogens and reconstituted them with required soluble factors in the TNT assay, resin-purified ribosomes from clinical isolates of S. pneumoniae and MRSA had very high activity in protein synthesis (at levels comparable to standard E. coli ribosomes), while ribosomes isolated by centrifugation had very little (Fig. 6B).
DISCUSSION
We reported here a novel chromatography system for rapid isolation of active ribosomes from a broad spectrum of bacteria including clinical pathogenic isolates such as multidrug-resistant S. aureus. The system employs cysteine-Sulfolink resin and shows unique features compared with other chromatography methods used for ribosomes or RNA. Of the many methods available for chromatography of RNA (McLaughlin 1989; Tanner 1989; Alpert 1990), only anion exchange or mixed-bed chromatography adsorb at low salt and elute with high salt, as does cysteine-Sulfolink. The importance of the anion is another common feature. However, while both anion exchange and mixed bed require an exchanging group, our tests with different resins suggest that such a group is not required in our system. While hydrophobic interactions seem to play a part, the mechanism matches neither the solvent elution of reversed phase chromatography nor the reverse salt gradient of HIC, suggesting a novel mechanism.
As detailed in the introduction, other chromatographic purifications of ribosomes have not achieved widespread use, perhaps due to difficulties in easily reproducing them, in scaling them up, or because they are not applicable to all organisms. The utility of cysteine-Sulfolink chromatography has been clearly demonstrated with ribosomes from four very different species. Both 30S and 50S subunits, as well as 70S monosomes and polysomes all bind and elute from the resin. The ability to reduce degradation of ribosomes (Fig. 5) is likely a matter of speed, where the proteases and nucleases, which are present at very high levels in clinical isolates, start to separate from the ribosomes as soon as the latter bind to the resin. Other chromatography methods, no doubt, also benefit from this advantage, but do not share the extremely mild conditions employed with cysteine-Sulfolink. The resin is commercially available and easy to prepare for use. Large-scale separations can be conveniently run using automatable chromatography platforms requiring less hands-on time than the multiple centrifugations that would otherwise be required. Such convenient features make the method advantageous for standard laboratory strains, as well as their less tractable pathogenic relatives. Additional applications can also be envisaged, such as parallel ribosome purification from multiple strains and purification of other ribonucleoprotein complexes or nucleic acids.
Although “laboratory strains” can serve as a model organism in antibacterial research, there are significant differences between ribosomes from different species, so that instances inevitably arise where the model diverges from the target. Furthermore, where a model organism must be used, the ability to test its validity as a model by in vitro comparisons with pathogens will be invaluable.
Even the minimal activity seen in Figure 6B for ribosomes purified from clinical isolates by conventional methods could in theory be used to generate IC50 curves with inhibitors, but the use of damaged ribosomes is not ideal and may give misleading results. Such low activities are typical of those obtained by us in repeated efforts over many years with different clinical isolates (a single report of TNT with a clinical S. pneumoniae did not record activities) (Capobianco et al. 2000). When partial reactions of protein synthesis, such as peptide transfer, are studied, residual activity can still be detected even after drastic treatments (Noller et al. 1992). However, complete systems such as the TNT assay are more physiologically relevant than partial reactions and are especially important if the exact mechanism of inhibition is unclear. The new chromatography described here has allowed us to isolate highly active ribosomes from clinical pathogens such as MRSA, and thus, is a significant advance for antibacterial research and antibiotic drug discovery.
MATERIALS AND METHODS
Reagents
Biotin-PEO-LC-amine, Sulfolink coupling gel, and Streptavidin-agarose gel were purchased from Pierce, Inc. Reagents and buffers for resin preparation were purchased from Sigma Aldrich. Yeast extract was from Difco, and tryptone from Oxoid Ltd. 14C-erythromycin was from Perkin Elmer. All other chemicals were from JT Baker.
Preparation of resins
A total of 3 mL of a 50% slurry of Sulfolink coupling gel was placed in a 5-mL plastic vial, centrifuged, and the storage buffer carefully decanted. The gel was washed three times with Sulfolink coupling buffer (3 mL) using the centrifuge/decant method. Next, 3 mL of a 50 mM solution of ethanethiol in coupling buffer was added and the slurry gently shaken for 1 h at 25°C. Residual ethanethiol was removed by centrifuge/decant to afford reagent (1) in Figure 1. Using Sulfolink coupling gel along with cysteamine, 3-mercaptopropionic acid and l-cysteine, respectively, products (2), (3), and (4) shown in Figure 1 were similarly prepared.
Biotin-PEO-LC-amine (2.1 mg) was dissolved in 150 μL N,N-dimethylformamide (DMF) and added to a solution of 400 μL of acetonitrile with 50 μL of N,N-diisopropylethylamine and 1 mg (1 eq.) of bromoacetyl bromide. The reaction was followed by liquid chromatography mass spectral analysis (LCMS) to completion. (Expected mass: 539.49 Da.; Found: 539.9 Da.). A solution of 1.0 mg of l-cysteine in 50 uL of DMF was added, and the reaction followed by LCMS to completion. (Expected mass: 565.71 Da.; Found: 566.1 Da.). The reaction mixture was stripped to a solid, dissolved in coupling buffer, and added to 1 mL of prewashed streptavidin-agarose gel. After sitting 1 h at 25°C, the gel was washed with coupling buffer by centrifuge/decant, to afford product (5) in Figure 1.
Growth of cells
E. coli strain MRE600 was grown in 10 L of L-broth (10 g of tryptone, 5 g of yeast extract, 5 g of NaCl) supplemented with 10 mM MgSO4 in a BF4 fermentor (New Brunswick Scientific Co., Inc.) from A600 0.05–0.5 at 37°C. Cells were harvested by centrifugation, washed in THM buffer (10 mM Tris-HCl at pH 7.4, 10 mM Mg acetate), repelleted, and stored at −80°C.
D. radiodurans (DSM#20539) was similarly grown, but at 30°C from A600 0.1–3, with no added MgSO4 but with 5 g of glucose added to the broth. Cells were washed in DR lysis buffer (10 mM HEPES-NaOH at pH 7.8, 30 mM MgCl2, 150 mM NH4Cl, 6 mM β–mercaptoethanol) during harvest.
Methicillin-resistant S. aureus (MRSA) was grown overnight in 100 mL of brain heart infusion (BHI) medium at 37°C. This was then used to inoculate 2 L of fresh BHI medium, which was then grown to A600 of 2–4. Cells were pelleted and washed successively with 200 mL of cold S30 buffer A (10 mM Tris-acetate at pH 8.0, 14 mM Mg-acetate, 1 mM DTT, 0.1 mM Pefabloc SC, and 1 M KCl). The pellet was then washed with 100 mL of buffer A containing 50 mM KCl and stored at −80°C.
S. pneumoniae was grown in BHI medium buffered with 50 mM MOPS (pH 7.4), at 37°C, with 5% CO2 under static conditions until a final OD600 of 0.3–0.5. Cells were harvested, washed, and stored as described for MRSA above.
Preparation of ribosomes by centrifugation
E. coli ribosomes were prepared by centrifugation as described by Spedding (1990). E. coli lysis buffer was 20 mM Tris-HCl (pH 7.5), 10.5 mM Mg Acetate, 100 mM NH4Cl, 0.5 mM EDTA, and 3 mM β–mercaptoethanol. After removal of unbroken cells by centrifugation, the supernatant was centrifuged at 45 K rpm for 2.5 h in a Beckman Ty50.2 rotor to pellet the ribosomes. Ribosome pellets were resuspended in buffer and centrifuged overnight at 45 K in the Ty50.2 rotor with an underlay of 10 mL of the same buffer supplemented to 500 mM NH4Cl and 1.1 M sucrose. Pellets were resuspended in lysis buffer and flash frozen in liquid nitrogen. D. radiodurans ribosomes were prepared similarly, but using DR lysis buffer and only pelleting the ribosomes once (through a 1.1-M sucrose underlay without extra salt).
The procedure for making S. aureus extracts was described previously (Murray et al. 2001). S. pneumoniae was microfluidized at a cell suspension of 0.5 g/mL, using sufficient cycles at 12 K psi to produce at least 50% cell disruption. The lysed cells were spun at 4°C, followed by treatment as described (Murray et al. 2001).
Batch binding to resin
A 1.5-mL bed of resin was packed in Polyprep columns (Bio-Rad) and pre-equilibrated with binding buffer (10 mM Tris-HCl at pH 7.5, 10 mM MgCl2, 60 mM NH4Cl). Ten A260 units of crude E. coli ribosomes were diluted in 1 mL of binding buffer, loaded onto the column, and allowed to flow through by gravity at 4°C. The columns were washed with five 1-mL portions of the same buffer and then eluted with five 1-mL portions of the same buffer, but with 0.5 M NH4Cl. Each 1-mL fraction was collected and its A260 determined.
Standard chromatography method
Cell are thawed in either the E. coli or the D. radiodurans lysis buffer (2 mL/g wet cells), French pressed at 10,000 psi and incubated on ice for 30 min with 2 μL/mL of RNAse-free DNase (Roche Diagnostics Corp.). Unbroken cells are then removed by centrifugation at 18.5 K rpm for 30 min in a Sorvall SS34 rotor at 4°C and the supernatant filtered through a 0.22 μ filter (GP Express plus Stericup; Millipore, Inc.) before loading on a column of cysteine Sulfolink resin, using about 1 mL of resin per 20 absorbance units of sample and a flow rate of 0.5–1 mL per minute. The conductivity of the E. coli buffer is 17 mS/cm, while that of the Deinococcus buffer is 25 mS/cm. This latter value is close to that at which elution occurs, but cell lysates typically have a lower conductivity (20 mS/cm) that allows robust binding. Other buffers can be used provided that conductivity is kept below 20–25 mS/cm; the resin is unaffected by reducing agents.
The column is washed with lysis buffer (10 column volumes, or until the baseline returns to zero) and then eluted with the same buffer but containing 0.3 M NH4Cl, either by isocratic elution (10 CV) or with a gradient from 0%–100% elution buffer (10 CV). Flow rates for washing and elution are set by the maximum pressure limit for the resin (0.15 MPa). Ribosomes are pelleted from pooled fractions (e.g., 35 K in a Beckman Ty45 rotor for 15 h at 4°C) and resuspended in buffer for snap freezing in liquid nitrogen and storage at −80C. The resin is rinsed with water after use and stored in 20% ethanol at 4°C. We have successfully reused the same resin over several years.
Sucrose-density gradient centrifugation serves well as a secondary purification to separate ribosomal subunits, monosomes, and polysomes, but gel filtration could also be considered.
Testing effects of anions/cations on elution from resin
A lysate (3 mL) prepared from 1.5 g of MRE600 cells in DR lysis buffer was loaded onto a 25-ml column of cysteine-Sulfolink resin equilibrated in DR lysis buffer, washed with 125 mL of lysis buffer and then eluted with a linear 0%–100% gradient where buffer B was lysis buffer containing 0.5 M NH4Cl. Fractions containing the ribosomes were pooled and placed in an Amicon stirred cell (Millipore, Inc.) at 15 psi nitrogen pressure with a YM100 membrane for concentration and buffer exchange with lysis buffer to restore the NH4Cl concentration to 0.15 M. Aliquots were loaded onto the column (pre-equilibrated in lysis buffer), washed with 70 mL of lysis buffer, and eluted with a linear 0%–100% gradient where buffer B was lysis buffer containing either 1 M NH4Cl, 1 M NaCl, KCl, (NH4)2SO4, or NH4Acetate. RNA was extracted from one aliquot of ribosomes and subjected to chromatography on the column with 1 M NH4Cl in the elution buffer. To identify the RNA species in different peaks, samples were subjected to PAGE.
Large-scale chromatography
A lysate was prepared from 50 g of D. radiodurans cells as described above and loaded from a 150 mL Superloop onto a 6.5 cm long column of 125 mL of cysteine-Sulfolink resin on an ÄKTA basic chromatography platform (GE Healthcare). Once loaded, the column was washed with 625 mL of lysis buffer at 5 mL/minute and then eluted with a linear 625 mL 0%–100% gradient, where buffer B is the lysis buffer containing 1 M NH4Cl.
Analytical sucrose-density gradient centrifugation
Linear 15%–30% (w/w) sucrose gradients were made in lysis buffer and centrifuged at 4°C for 2.3 h in a Beckman SW55 rotor at 50 K rpm. Gradients were fractionated using an ISCO Type 11 optical cell and model UA6 absorbance monitor.
Protein extraction and analysis
Proteins were extracted from ribosomes or cell lysate using 66% acetic acid/10 mM MgCl2, the RNA was pelleted, and proteins in the supernatant precipitated with 5 vol of acetone at −20°C. Pellets were dried and resuspended in 8 M urea containing 1% (v/v) β-mercaptoethanol. Samples were loaded onto a Novex 12% Bis-tris gel (Invitrogen, Inc.) buffered with MOPS-SDS buffer and run at 200 V for 45 min before staining with Coomassie G-250.
RNA extraction and analysis
RNA was phenol/chloroform extracted from samples and electrophoresed on a Novex 6% acrylamide TBE/urea gel according to the manufacturer's instructions (Invitrogen, Inc.), but for 100 min at 180 V. RNA molecular weight standards were High Range RNA Marker from United States Biological. Electrophoresed gels were stained with ethidium bromide for detection of RNA by fluorescence.
Ribosome binding assay
Two A260 units of ribosomes were mixed with 2.5 μM 14C-erythromycin in 50 μL of final reaction mixture containing 10 mM Tris-HCl (pH 7.5), 60 mM KCl, 10 mM NH4Cl, and 6 mM MgCl2. The reaction was incubated at 35°C for 30 min, stopped, and precipitated with 200 μL of 75% cold ethanol, and placed at −20°C for 20 min. Bound radiolabel was harvested onto GF/B filter plates using a Packard Unifilter 96 harvester with a 75% ethanol wash. Filters were dried and bound radioactivity counted in a Packard Topcount Scintillation counter.
Bacterial transcription and translation (TNT) assay
Bacterial transcription and translation assay was performed as described (Murray et al. 2001). To study the activity of MRSA and S. pneumoniae ribosomes, they were supplemented with S150 from S. aureus RN4220 and S. pneumoniae R6, respectively.
ACKNOWLEDGMENTS
We are grateful to Tim Subashi for skilled fermentations of the nonpathogenic strains and to Drs. Jim Boyd, Kieran Geoghegan, Andrew Seddon, Jeff Culp, and John Mueller for helpful discussions.
Footnotes
Abbreviations: rRNA, ribosomal RNA; TNT, transcription and translation; PAGE, polyacrylamide gel electrophoresis; IC50, inhibitor concentration that gives 50% inhibition; CPM, counts per minute; HIC, hydrophobic interaction chromatography; SEC, size exclusion chromatography; LC-MS, liquid chromatography mass spectrometry; CV, column volumes.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.692408.
REFERENCES
- Alpert, A.J. Hydrophilic-interaction chromatography for the separation of peptides, nucleic acids, and other polar compounds. J. Chromatogr. 1990;499:177–196. doi: 10.1016/s0021-9673(00)96972-3. [DOI] [PubMed] [Google Scholar]
- Capobianco, J.O., Cao, Z., Shortridge, V.D., Ma, Z., Flamm, R.K., Zhong, P. Studies of the novel ketolide ABT-773: Transport, binding to ribosomes, and inhibition of protein synthesis in Streptococcus pneumoniae . Antimicrob. Agents Chemother. 2000;44:1562–1567. doi: 10.1128/aac.44.6.1562-1567.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemons W.M., Jr, Brodersen, D.E., McCutcheon, J.P., May, J.L.C., Carter, A.P., Morgan-Warren, R.J., Wimberly, B.T., Ramakrishnan, V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: Purification, crystallization and structure determination. J. Mol. Biol. 2001;310:827–843. doi: 10.1006/jmbi.2001.4778. [DOI] [PubMed] [Google Scholar]
- Fabry, M., Kalvoda, L., Rychlik, I. Hydrophobic interactions of Escherichia coli ribosomes. Biochim. Biophys. Acta. 1981;652:139–150. doi: 10.1016/0005-2787(81)90217-3. [DOI] [PubMed] [Google Scholar]
- Inada, T., Winstall, E., Tarun S.Z., Jr, Yates J.R., III, Schieltz, D., Sachs, A.B. One-step affinity purification of the yeast ribosome and its associated proteins and mRNAs. RNA. 2002;8:948–958. doi: 10.1017/s1355838202026018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelenc, P.C. Rapid purification of highly active ribosomes from Escherichia coli . Anal. Biochem. 1980;105:369–374. doi: 10.1016/0003-2697(80)90472-8. [DOI] [PubMed] [Google Scholar]
- Kirillov, S.V., Makhno, V.I., Peshin, N.N., Semenkov, Y.P. Separation of ribosomal subunits of Escherichia coli by Sepharose chromatography using reverse salt gradient. Nucleic Acids Res. 1978;5:4305–4315. doi: 10.1093/nar/5.11.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev, A., Trakhanov, S., Laurberg, M., Noller, H.F. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- Le Goffic, F., Baca, B., Moreau, N. New rapid and powerful technique to obtain purified ribosomes. FEBS Lett. 1974;41:69–72. doi: 10.1016/0014-5793(74)80956-7. [DOI] [PubMed] [Google Scholar]
- Le Goffic, F., Moreau, N., Chevelot, L., Langrene, S., Siegrist, S. Purification of bacterial ribosomes using chloramphenicol and erythromycin columns. Biochimie. 1980;62:69–77. doi: 10.1016/s0300-9084(80)80372-5. [DOI] [PubMed] [Google Scholar]
- McLaughlin, L.W. Mixed-mode chromatography of nucleic acids. Chem. Rev. 1989;89:309–319. [Google Scholar]
- Murray, R.W., Melchior, E.P., Hagadorn, J.C., Marotti, K.R. Staphylococcus aureus cell extract transcription-translation assay: Firefly luciferase reporter system for evaluating protein translation inhibitors. Antimicrob. Agents Chemother. 2001;45:1900–1904. doi: 10.1128/AAC.45.6.1900-1904.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller, H.F., Hoffarth, V., Zimniak, L. Unusual resistance of peptidyl transferase to protein extraction procedures. Science. 1992;256:1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- Petry, S., Brodersen, D.E., Murphy, F.V.I.V., Dunham, C.M., Selmer, M., Tarry, M.J., Kelley, A.C., Ramakrishnan, V. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell. 2005;123:1255–1266. doi: 10.1016/j.cell.2005.09.039. [DOI] [PubMed] [Google Scholar]
- Roe, S. Separation based on structure. In: Harris E.L.V., Angal S., editors. Protein purification methods: A practical approach. IRL press; Oxford, UK: 1989. pp. 175–242. [Google Scholar]
- Saruyama, H. Isolation of ribosomal subunits from an extremely halophilic archaebacterium Halobacterium halobium by hydrophobic interaction chromatography. Anal. Biochem. 1986;159:12–16. doi: 10.1016/0003-2697(86)90300-3. [DOI] [PubMed] [Google Scholar]
- Selmer, M., Dunham, C.M., Murphy, F.V.I.V., Weixlbaumer, A., Petry, S., Kelley, A.C., Weir, J.R., Ramakrishnan, V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Spedding, G. Isolation and analysis of ribosomes from prokaryotes, eukaryotes, and organelles. In: Spedding G., editor. Ribosomes and protein synthesis. IRL press; Oxford, UK: 1990. pp. 1–29. [Google Scholar]
- Tanner, N.K. Purifying RNA by column chromatography. Methods Enzymol. 1989;180:25–41. doi: 10.1016/0076-6879(89)80089-8. [DOI] [PubMed] [Google Scholar]
- Youngman, E.M., Green, R. Affinity purification of in vivo-assembled ribosomes for in vitro biochemical analysis. Methods. 2005;36:305–312. doi: 10.1016/j.ymeth.2005.04.007. [DOI] [PubMed] [Google Scholar]