Abstract
Post-transcriptional ribonucleotide modifications are widespread and abundant processes that have not been analyzed adequately due to the lack of appropriate detection methods. Here, two methods for the analysis of modified nucleotides in RNA are presented that are based on the quantitative and site-specific DNAzyme-mediated cleavage of the target RNA at or near the site of modification. Quantitative RNA cleavage is achieved by cycling the DNAzyme and its RNA substrate through repeated periods of heating and cooling. In a first approach, DNAzyme-directed cleavage directly 5′ of the residue in question allows radioactive labeling of the newly freed 5′-OH. After complete enzymatic hydrolysis, the modification status can be assessed by two-dimensional thin layer chromatography. In a second approach, oligoribonucleotide fragments comprising the modification site are excised from the full-length RNA in an endonucleolytic fashion, using a tandem DNAzyme. The excised fragment is isolated by electrophoresis and submitted to further conventional analysis. These results establish DNAzymes as valuable tools for the site-specific and highly sensitive detection of ribonucleotide modifications.
Keywords: DNAzyme, tandem DNAzyme, modification, modified nucleotide, modification enzymes
INTRODUCTION
Modified ribonucleotides occur in great chemical variety and are found in all classes of known natural RNAs. Analysis of their function is largely impeded by the lack of efficient methods for their detection and quantification. Modification levels and sites are difficult to identify with techniques based on RT-PCR, and in most cases the information must therefore be derived directly from the RNA sample without amplification. As a consequence, preparation of sufficient quantities of modified RNAs is usually the limiting factor in modification analyses, and it follows that a major criterion for method development is sensitivity.
A large number of methods are based on separation of modified nucleotides, nucleosides, or bases by column chromatography, thin-layer chromatography (TLC), capillary electrophoresis, or related techniques. In these cases, the sample is usually degraded to the nucleotide level or further, thereby causing the loss of sequence information. The physicochemical properties of the single nucleotides or nucleosides permit their separation and also serve for their identification and characterization by retention values. Coupling of the separation step, in particular of HPLC, with mass spectroscopy is the method of choice to obtain more direct data concerning the chemical nature of the modification, because it is suited for relatively small amounts of sample (Matsuyama et al. 1998).
When references are available, the use of retention values alone permits application of radioactive labeling techniques to increase sensitivity. Under favorable circumstances, the combination of 32P-labeling and two-dimensional TLC separation on cellulose enables the detection of femtomole quantities of modified nucleotides. Rf values on cellulose plates in three solvent systems have been mapped for more than 70 modifications, constituting the commonly accepted reference system in the field (Silberklang et al. 1977; Kuchino et al. 1987; Keith 1995; Grosjean et al. 1998, 2004).
Radioactive labels, in particular 32P, may be conjugated to the nucleotide in question before or after the actual modification event. While prelabeling of native modified RNA has become uncommon because it requires high amounts of radioactivity in living cells (Helm et al. 1999), prelabeling with [α-32P]-NTPs in T7 run-off transcription reactions is frequently employed for the measurement of modification activities in vitro (Grosjean et al. 1998; Motorin and Grosjean 1999). Degradation of the modified RNA to single nucleotides may be performed by either of two types of RNases. While nuclease P1 leaves the label on the 5′-OH of the nucleotide by which it was originally introduced, RNase T2 puts the label on the 3′-OH of the 5′-neighboring nucleotide. Hence, this nearest neighbor analysis offers limited access to sequence information of the modification site (Grosjean et al. 1998). More sequence information can be obtained from an RNase T1 fragment catalog. To this end, the substrate is submitted to complete digestion by RNase T1, and, after size fractionation, the resulting fragments are separately investigated by TLC. In general, analysis of such prelabeled fragments in combination with nearest neighbor analysis allows unambiguous assignment of modification sites (Brule et al. 1998; Grosjean et al. 1998). The T1-catalog technique is also suitable for combination with mass spectroscopy as a label-free detection method providing somewhat more detailed sequence information (Pomerantz and McCloskey 2005; Goll et al. 2006). More advanced methods rely on the use of sequence information contained in oligonucleotides to direct the analysis of modifications with high specificity. In particular, the ability of modified nucleotides to impede polymerase reactions can be detected by primer elongation assays. Yu et al. (1997) have reported an RT-based assay in which the polymerase is arrested by 2′-O-Me modifications at low dNTP concentrations. In the case of pseudouridine, which normally has template properties indistinguishable from uridine, treatment with CMCT produces a covalent adduct that arrests primer extension (Bakin and Ofengand 1993, 1998). Discrimination of modified nucleotides by T4 DNA ligase has recently been exploited in the development of a high-throughput method with low detection thresholds (Saikia et al. 2006).
The advent of modern nucleic acid technologies has provided new avenues to harness the sequence information contained in oligonucleotides. Targeting of site-specific RNas H cleavage by 2′-O-Me modified oligonucleotides, originally developed as a therapeutic antisense approach, has been applied by Hou et al. (2006) to degrade RNAs that did not contain base-pair-disrupting nucleotide modifications. In a most elegant method, Zhao and Yu (2004) have combined the site-specific cleavage by RNase H with post-labeling at the cleavage site and subsequent analysis of the labeled nucleotide by TLC. This latter approach allows targeting of virtually any nucleotide in an RNA sequence for quantitative modification analysis, provided the modification itself does not interfere with the cleavage reaction. Adjustment of the proper stoichiometry of enzyme, oligonucleotide, and substrate must be performed to guarantee quantitative cleavage, which is a prerequisite for accurate quantification. Aiming to overcome the major drawback of this method, the high cost of 2′-O-Me modified oligonucleotides, we promote here the use of 10–23 DNAzymes (Santoro and Joyce 1997; Cairns et al. 2003) as a convenient alternative. Such inexpensive oligodeoxyribonucleic acids display RNA hydrolyzing activity, which can be site directed by the choice of proper targeting sequences flanking the catalytic site. Moreover, tandem DNAzymes can be used to excise fragments of defined length, which can then be submitted to further analysis, e.g., by TLC. Using these two methods, the vast majority of RNA modifications become amenable to sensitive and site-specific analysis.
RESULTS
Site-specific labeling of nucleotides at modification sites using DNAzymes
We have devised two approaches to utilize the sequence-specific RNA cleavage ability of DNAzymes for the analysis of base modifications. The first approach is a site-specific post-labeling approach of the nucleotide under investigation (Fig. 1). More specifically, we have analyzed the formation of pseudouridine (Ψ) at position 27 of an in vitro transcript corresponding to the genetic sequence of human mitochondrial tRNALys mediated by mouse pseudouridine synthase 1 (mPus1p) (Chen and Patton 1999; Behm-Ansmant et al. 2006). Instead of the canonical cloverleaf structure, this RNA displays an extended hairpin conformation (Fig. 1B; Helm et al. 1998) and its recognition by tRNA modifying enzymes is thus of particular interest (Helm and Attardi 2004).
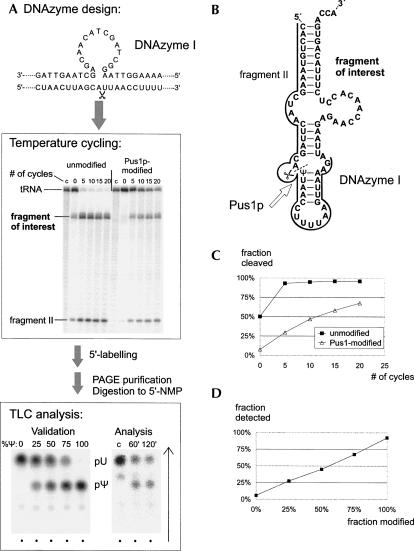
FIGURE 1.
Post-labeling after site-specific DNAzyme mediated cleavage. (A) Overview of the experimental procedure, showing annealing of the DNAzyme to the target RNA, separation by PAGE of the fragments resulting from cleavage, and TLC analysis after digestion to mononucleotides. The middle panel shows a phosphorimager scan of a denaturing PAGE of fragmented tRNALys after temperature cycling for the indicated number of times. C denotes a control lane with RNA incubated without DNAzyme. Lanes labeled “unmodified” contain untreated tRNALys transcript, lanes labeled “Pus1p-modified” contain tRNALys transcript treated with Pus1p enzyme prior to analysis. The downstream fragment resulting from DNAzyme cleavage, indicated by “fragment of interest” in the figure, was excised for further analysis by 5′ labeling with 32P, digestion to mononucleotides, and thin layer chromatography. The RNA in the gel is internally labeled with [α-32P]-UTP to facilitate cleavage detection. Note that the actual analysis is performed on RNA, which is nonradioactive until the point of post-labeling. (Lower left) TLC of a series of calibration samples with known U/Ψ composition at position 27 after application of the analytic procedure. The spots of U and Ψ are indicated. (Lower right) Application to the analysis of tRNALys incubated with Pus1p. Times of incubation with Pus1p are indicated in minutes; C designates a nonmodified control. (B) DNAzyme design for targeting residue Ψ/U 27 in human mitochondrial tRNALys. The target tRNA is shown in its unusual extended hairpin form (Helm et al. 1998). The hybridization sequences of DNAzyme I are indicated by a bold line. The loop in the bold line indicates the catalytic center near the targeted cleavage site. Pseudouridine synthase 1 and its respective target site for post-transcriptional modification of uridine to pseudouridine (Ψ) are indicated. (C) Cleavage efficiency is affected by modifications. Target cleavage yields of unmodified tRNA (solid squares) and a tRNA pre-incubated with Pus1p (open triangles) are compared as a function of the number of temperature cycles. (D) Validation of detection efficiency. The composition of a mixture of tRNAs containing defined ratios of Ψ/U 27 is plotted versus the ratio detected after site-specific labeling.
As outlined in Figure 1A, the analytical approach consisted of the sequence-specific cleavage directly 5′ to the nucleotide to be investigated, followed by 5′ labeling of the resulting downstream fragment, complete digestion to 5′-monophosphate-nucleotides, and separation of the latter by TLC. This approach is conceptually similar to that of Zhao and Yu (2004), except that a DNAzyme is used instead of RNase H and a 2′-O-Me Oligo. Conveniently, the resulting downstream target fragment has a 5′-OH, making a dephosphorylation step unnecessary. A 10–23 DNAzyme with the catalytic core sequence as published by Santoro and Joyce (1997) and flanking targeting sequences corresponding to nucleotides 1–25 and 27–45 of the tRNA (Fig. 1B) cleaves 40%–70% of the target within 30 min, but the reaction does not proceed to completion. However, when temperature cycling is applied to create repeated annealing and refolding conditions for the DNAzyme, cleavage yields increase (Fig. 1A), and eventually, i.e., after 60 cycles, the reaction can be driven to completion (not shown). The DNAzyme is supplied in 10-fold excess of the substrate, but at least at a final concentration of 0.5 μM. Temperatures for cleavage and denaturation (provided in Materials and Methods) may be optimized for a specific pair of substrate RNA and corresponding DNAzyme if initial attempts do not yield quantitative cleavage. As evident in Figure 1, previous incubation of tRNALys with Pus1p clearly decreased cleavage efficiency, suggesting that the presence of Ψ negatively affects RNA substrate properties. This is an explicit illustration of why quantitative cleavage of the RNA substrate is required to produce modification results that are representative of the RNA sample. For the same reason, it is important to achieve quantitative labeling of the 5′-OH of the resulting 3′-fragment with [γ-32P]-ATP in the subsequent phosphorylation step. In our experience, at low ATP concentrations uridines are labeled more efficiently by T4-PNK than are pseudouridines, but labeling is quantitative at 10 μM ATP.
To verify that varying proportions of Ψ at position 27 can be accurately quantified, we have synthesized derivatives of tRNALys containing either exclusively U27 or exclusively Ψ27. To this end, we have ligated RNA fragments into which the respective nucleotides had been incorporated by chemical solid-phase synthesis, using a splint ligation approach (Kurschat et al. 2005). These derivatives were then mixed in known proportions, and their cleavage and labeling efficiencies were studied. Figure 1 shows an autoradiography of such calibration standards after separation of the labeled nucleotide 27 by TLC. Analysis by densitometry shows a linear correlation between Ψ/U content and detected Ψ/U ratio (Fig. 1D), thus validating this method for quantifying samples of unknown Ψ content. Application to tRNALys incubated with Pus1p does, as expected, reveal U27 of this RNA to be modified to Ψ27 (Fig. 1).
Site-directed cleavage of modified RNAs using DNAzymes
A second application of DNAzymes consists of the site-directed scission of the target RNA molecule into length-defined fragments, which are then isolated and submitted to further analysis. Here, we have used this approach to characterize the in vitro activity of affinity-purified Dnmt2, an enzyme that belongs to the family of DNA-methyltransferases, but has recently been shown to also exhibit tRNA methylating activity (Goll et al. 2006; Jeltsch et al. 2006). This has suggested that Dnmt2 enzymes might utilize novel catalytic mechanisms and has rendered the analysis of Dnmt2-mediated tRNA modifications particularly important. Native tRNAAsp from DNMT2 knock-outs was reported to function as a Dnmt2 substrate in tritium incorporation assays with methyl-3H-labeled S-adenosylmethionine (SAM), while in vitro transcripts of tRNAAsp were reported to be inactive (Goll et al. 2006). We found that immunopurified Dnmt2 from Drosophila melanogaster cells catalyzed the incorporation of 0.05 mol 3H-methyl groups per mole RNA, using a full-length in vitro transcript of tRNAAsp from D. melanogaster or a 3′-truncated derivative lacking the terminal 4 nucleotides (nt) as substrates (data not shown). For comparison, recombinant Trm4 from Saccharomyces cerevisiae (Motorin and Grosjean 1999) catalyzed incorporation of 0.8 mol 3H-methyl groups per mole tRNA (data not shown). Trm4 is known to catalyze m5C-formation of cytosine residues 48 and 49 in the variable loop of tRNAs. Being a multi-site-specific enzyme, Trm4 also catalyzes the formation of m5C40 in yeast tRNAPhe by methylating an intron-containing precursor (Motorin and Grosjean 1999).
To demonstrate that the observed signal in tritium incorporation assays with tRNAAsp was caused by site-specific, Dnmt2-mediated methylation of C38, we initially used DNAzymes for site-specific cleavage and subsequent post-labeling, as described above. However, these experiments were unsuccessful, because a DNAzyme with the same catalytic loop as in Figure 1 and targeting sequences corresponding to cleavage between nucleotides 37 and 38 produced only limited cleavage yields, even when excessive temperature cycling was applied. In addition, neither target site for Trm4 displayed a dinucleotide sequence favorable for efficient DNAzyme cleavage. We therefore designed a new approach to experimentally analyze the tRNAAsp modifications mediated by Dnmt2 and Trm4. The truncated transcript of tRNAAsp, internally labeled by transcription in the presence of [α-32P]-CTP, was incubated in vitro with either Dnmt2 or Trm4. DNAzymes II and III were designed to cleave the tRNA at positions 30, 39, and 57, to yield defined fragments of 9, 14, 18, and 30 nt upon complete cleavage of the target RNA at all sites (Fig. 2A). We used the truncated, 71-nt RNA rather than the full-length tRNA, because the DNAzyme cleavage generated 14-mer and 18-mer fragments from the 3′ end, rather than two 18-mers, which could not have been separated by PAGE (Fig. 2). Of particular interest, the 9-mer fragment originating from cleavages at positions 30 and 39 was obtained with the tandem DNAzyme II, containing a sequence complementary to the 9mer target in between two catalytic loops, as well as 13-nt-long complementary targeting sequences on each side (Fig. 2A,B). Figure 2C shows a separation of cleavage fragments by denaturing PAGE after cycling with both DNAzymes. Treatment with either DNAzyme for 25 cycles results in additional fragments 27, 32, 39, 41, and 57 nt in length. Near complete cleavage is achieved by sequential cycling with DNAzyme II (25 times) and DNAzyme III (additional 25 cycles). The gel-purified fragments of interest were digested by nuclease P1. The resulting mononucleotide mixtures were separated by two-dimensional TLC to determine their m5C/C ratios. In fragments from Trm4-modified RNA, the 18-mer contained m5C (0.8 mol m5C/mol RNA; Fig. 2D), but none of the other fragments contained any modification (data not shown). In particular, TLC analysis of the 9-mer fragment containing C38 does not show any evidence of modification (Fig. 2D), which is consistent with the notion that Trm4 does not methylate C38. The distinct spot corresponding to pGp is caused by a cytosine at position 40. This residue carries a 32P-label on its 5′, which remains 3′ to guanosine 39 (the 3′ end of the 9mer) upon cleavage by the DNAzyme, which creates a 2′-3′-cyclic phosphate. Digestion with nuclease P1 then creates diphosphate nucleotide 5′-pG-2′-3′-cyclic-[32P]-p, visible by TLC.
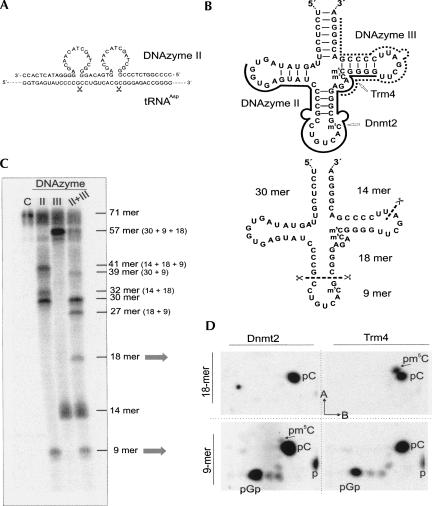
FIGURE 2.
Tandem DNAzyme-mediated excision of target oligoribonucleotides for modification analysis. (A) Design of a tandem DNAzyme for double cleavage of RNA. Scissors indicate cleavage sites on the RNA. The catalytic centers of the DNAzyme are displayed as loops. (B, top) Secondary structure of 3′-truncated tRNAAsp. The hybridization sequences of DNAzymes II (bold line) and III (dotted line) are indicated. Loops indicate the catalytic centers near the targeted cleavage site. Methyltransferases Dnmt2 and Trm4 and their respective target sites for post-transcriptional modification to 5-methylcytosine are indicated. (Bottom) RNA fragments resulting from the combined action of DNAzymes II and III. Scissors indicate cleavage sites on the RNA. (C) PhosphorImager scan of fragments generated by DNAzyme-directed cleavage. Lanes designated II, III, and II+III show incubations in the presence of the respective DNAzymes, and C designates the control incubation without DNAzyme. The size of the RNA fragments is indicated on the right. Arrows indicate fragments that have been isolated from a reaction after cycling to near-complete hydrolysis. (D) 2D-TLC of mononucleotide mixtures resulting from the excised 9-mer and 18-mer fragments of [α-32P]-CTP labeled RNA after incubation with Dnmt2 and Trm4, respectively. pC and pm5C identify spots corresponding to the 5′-phosphates of cytosine and 5-methylcytosine, p stands for free phosphate, and pGp for 5′-pG-cyclic-2′-3′-[32P]-p diphosphate, generated from the 3′ end of the 9-mer by P1-digestion. Dimensions of chromatography in solvents A and B are indicated by arrows.
Interestingly, while Dnmt2-treated full-length RNA did not give a detectable m5C signal, the 9-mer excised from RNA modified with Dnmt2 shows a weak m5C spot (corresponding to 0.05 mol m5C/mol tRNA), but no m5C is visible in the 18 mer. This shows that our approach improves the detection threshold in addition to facilitating the localization of a modification.
DISCUSSION
Two approaches using sequence-specific RNA cleavage directed by DNAzymes for the analysis of post-transcriptional modifications are demonstrated in this article. In both cases, the flanking sequence of the 10–23 DNAzyme contains the information that directs the RNA cleavage. Proper hybridization of these nucleotide stretches is crucial for sequence specificity, and stability of the resulting complex is important for efficient cleavage.
The first approach directs DNAzyme-mediated cleavage to the 5′ end of the to-be-analyzed nucleotide. The properties of a modified nucleotide at this position may affect its detection and the accuracy of its quantification. For example, in our hands, pseudouridine negatively affected both the cleavage yield and the labeling reaction, which is consistent with the findings of Buchhaupt et al. (2007), who have used differential cleavage activities of DNAzymes to assess the pseudouridine content of rRNAs. In our case, incomplete cleavage would lead to the overrepresentation of cleaved and labeled 3′ fragments. Hence, accurate analysis requires quantitative cleavage of the substrate RNA. We have so far successfully applied this technique to the analysis of pseudouridine and 5′-fluorouracil, which do not interfere with Watson–Crick base pairing. For an extension to other nucleotide modifications and to other sequences, the following aspects must be considered: (1) the 10–23 DNAzyme preferably cleaves purine–pyrimidine sequences, some of which require the incorporation of inosine into the DNAzyme (Cairns et al. 2003); (2) the lengths of the targeting sequences influence hybridization properties to the target; and (3) the temperature cycling parameters may have to be optimized for quantitative cleavage. Other modified nucleotides may completely prevent cleavage, in which case modification yield can be obtained simply from the cleavage yield, provided the nature of the nucleotide modification is known (compare with Hou et al. 2006).
This approach is aimed at the quantification of a single modification site. While the inherent properties of DNAzymes make the approach unsuitable for sequencing an unknown RNA, it shows increased sensitivity, e.g., for the quantification of incompletely modified sites, because the signal stems only from the target site and thus produces low background.
Quantification of selected modification sites in an unknown RNA is only possible if an efficient DNAzyme can be designed based on knowledge of the primary sequence, at least on the genomic DNA level. However, extra precautions have to be taken to verify that hybridization or cleavage efficiency of the DNAzyme is not impeded by unknown modifications. For selected applications, this approach presents a suitable alternative to LC-MS or LC-MS-MS methods. Typical LC-MS methods for quantification of modifications in tRNA samples rely on complete digestion to nucleosides, which are then separated on HPLC, identified by MS or MS-MS, and quantified based on their UV absorption. This analysis, which does not address sequence information, typically employs sample sizes from 80 to 200 pmol (Matsuyama et al. 1998; Tomita et al. 1999; G. Björk, pers. comm.), while TLC signals shown in Figure 1 were obtained from 2-pmol samples.
The most severe drawback of this approach is the requirement for efficient DNAzymes (Silverman 2005), because otherwise extensive thermocycling is needed for quantitative cleavage, resulting in increased unspecific hydrolysis. The specific use of DNAzymes of the 10–23 type restricts the choice of cleavage sites to R-Y sites; however, other catalytic nucleic acids may conceivably be used to target additional sites. For example, the bipartite DNAzyme efficiently cleaves at A-A sites (Feldman and Sen 2001)and various DNAzyme variants of the 8–17 type (Cruz et al. 2004) show appreciable activity toward N-G and R-A sites.
The second approach presented here makes extensive use of the flanking sequences to cleave an RNA substrate into fragments of defined length, which are then isolated after size fractionation by PAGE and submitted to analysis. This approach, previously described for RNase T1 as “T1-catalogue,” is demonstrated here as a prelabeling approach using RNA labeled by transcription in the presence of [α-32P]-NTPs. In the present case, the m5C modification in the C38 position of the anticodon could be detected and quantified in a 9-mer fragment that was excised from the full-length RNA by a tandem DNAzyme. Because this fragment contains only 4 [α-32P]-labeled cytosine residues, as opposed to 19 in the 3′-truncated tRNAAsp, this method not only allows clarifying the location of the m5C residue under investigation, but it also enhances detection efficiency and quantification accuracy.
The use of DNAzymes as an alternative to a T1 catalogue is not restricted to the pre-labeling approach employed here; rather, analysis of the resulting fragments by MS or HPLC-MS is equally suitable, especially with native material that is not easily amenable to pre-labeling (e.g., Goll et al. 2006). The minimal amount required for quantitative analysis of pre-labeled RNA is 40–120 pmol for modification levels >40% and somewhat higher for modification levels as low as the 5% investigated here. Sample sizes employed for the investigation of T1-catalogues by LC-MS-MS vary from 5 to 120 pmol (Pomerantz and McCloskey 2005; Guymon et al. 2007). Thus, the quantities required are roughly similar for both approaches. The choice between HPLC-MS and TLC analysis might indeed be governed by availability of the respective technique; while very few laboratories have established the expertise for highly sensitive HPLC-MS analysis, use of DNAzymes and TLC represents an inexpensive low-tech approach available to most laboratories.
One clear advantage of DNAzyme cleavage over T1 digestion is that the number and size of the resulting fragments can be well controlled. For this DNAzyme approach, too, the disadvantages include the requirement for sequence information and the possible interference of unknown modifications with DNAzyme hybridization and cleavage activity. Compared with the DNAzyme approach discussed above, the problem to find a suitably active DNAzyme is somewhat relieved by a certain flexibility in the choice of target sites.
With particular respect to the tRNA methyltransferase activity of Dnmt2, the approach has allowed us to provide confirmation of the enzymatic formation of m5C in tRNAAsp and to show that in vitro transcripts of tRNAAsp are accepted as substrates by Dnmt2. In conclusion, our results establish DNAzymes as valuable tools for the site-specific and highly sensitive detection of ribonucleotide modifications.
MATERIALS AND METHODS
Oligonucleotides used in this study were from IBA except for oligoribonucleotide II, which was from Dharmacon, and DNAzymes II and III, which were from MWG Biotech.
In vitro transcription with T7-RNA polymerase
Unmodified tRNAs were obtained by run-off transcription from a DNA template containing a hammerhead ribozyme adjacent to the tRNA sequence. The human mitochondrial tRNALys sequence was transcribed from a corresponding pTFMA-vector derivative (Helm et al. 1998). Drosophila melanogaster tRNAAsp was transcribed from a double-stranded template obtained through a PCR-like polymerase reaction from the synthetic single-stranded template TCCCCGTCGGGGAATCGAACCCCGGTCTCCCGCGTGACAGGCGGGGATACTCACCACTATACTAACGAGGAGACGGTACCGGGTACCGTTTCGTCCTCACGGACTCATCAGTCCTCGTTATCTCCCTATAGTGAGTCGTATT, using the conditions described by Helm et al. (2005). Transcription was performed in a buffer containing 80 mM Tris-HCl (pH 8.1), 30 mM MgCl2, 0.2 mM spermidine, 0.002% TritonX, 10 mM DTT, 5 μg/mL BSA, and 4 mM of each NTP, except if 1 nt was supplemented as radioactive [α-32P]-XTP (0.5–1 μCi/μL, Hartmann Analytics), in which case its concentration was reduced to 1 mM. Transcription was started by the addition of 4 u/μL of T7 RNA polymerase (Fermentas). The mixture was incubated at 37°C for 3 h. The reaction mixture was submitted to denaturing PAGE and the desired RNAs excised, eluted, and precipitated.
Synthesis of chemically modified Ψ27-containing tRNAs
Oligonucleotides comprising full-length tRNALys were joined by splinted ligation (Kurschat et al. 2005) of appropriate combinations of oligoribonucleotides I (CACUGUAAAGCUAACUUAGC), II (pAΨUAACCUUUUAA), III (AUUAACCUUUUAA), and IV (GU dtCy5AAAGAUUAAGAGAACCAACACCUCUUUACAGUGACCA) using the DNA splint V (GGGTGGGTCTGCTTGGTCACTGTAAAGAGGTGTTGGTTCTCTTAATCTTTAACTTAAAAGGTTAATGCTAAGTTAGCTTTACAGTGATTTTGGGTACC). At the site denoted dt, 5′-Dimethoxytrityl-5-[N-(trifluoroacetylaminohexyl)-3-acrylimido]-2′-deoxyUridine,3′-[(2-cyanoethyl)-(N,N-diisopropyl)]-phosphoramidite was coupled during solid phase synthesis instead of uridine phosphoramidites, replacing uridine in the sequence with a 2′-deoxythymidine that carries a linear spacer of an 11-atom chain ending in a primary amino group, to which an N-hydroxysuccinimide derivative of Cy5 dyes was conjugated post-synthetically. The respective conjugation site is hence indicated as dtCy5. Stoichiometric amounts of (I, II, IV, and V) or (I, III, IV, and V) were hybridized to stoichiometric amounts of DNA splint by heating to 75°C and slow cooling to room temperature over 10 min in KL buffer supplemented with 5 mM DTT and 2 mM ATP. T4 DNA ligase (2 u/μL; Fermentas) was added and ligation was carried out over night at 16°C. Template DNA was removed by addition of 0.02–0.1 u/μL DNase I (Fermentas), followed by 15 min of incubation at 37°C. RNAs were purified by denaturing PAGE, eluted from the gel, and precipitated with ethanol. Concentrations were calculated from absorption at 254 nm, as determined on a Nanodrop ND-1000 spectrometer.
Protein purification
Recombinant Trm4p from Saccharomyces cerevisiae was expressed and purified as described by Motorin and Grosjean (1999). Pus1p from mouse was expressed and purified as described by Chen and Patton (1999). Protein concentration was assessed by Bradford reagent using the NanoDrop spectrometer. Flag-tagged Dnmt2 was purified from S2R+ cells that had been stably transfected with pRmt/A-3-Dnmt2-Flag and pCoBlasticidin (kind gift of Dr. Matthias Schaefer, German Cancer Research Center [DKFZ], Heidelberg, Germany).
Modification of tRNA in vitro
The m5C methyltransferase activity was tested by monitoring the [3H]-methyl group incorporation from [3H]SAM (488 GBq/mmol; Hartmann) into nonradioactive RNA. Two hundred picomoles of in vitro transcribed tRNAAsp were incubated in modification buffer (100 mM Tris-HCl at pH 8.0, 100 mM NH4OAc, 1 mM DTT, 0.1 mM EDTA) supplemented with 9 μM SAM (1.375 μCi) and 0.8 μM Flag-Dnmt2 in a total volume of 50 μL for 2 h at 37°C. The respective negative controls contained either no RNA or no enzyme. Fifteen-microliter aliquots were spotted on Whatman filters, and the tRNA was precipitated in ice-cold 5% TCA. The filters were washed two times in 5% TCA for 10 min and once in 100% ethanol for 5 min. After drying, the filters were submerged in Ultima Gold scintillation fluid (PerkinElmer), and radioactivity was measured by liquid scintillation counting in a multipurpose scintillation counter LS 6500 (Beckmann Coulter).
For DNAzyme analysis of the modification of tRNAAsp with Dnmt2 or Trm4, 350–500 pmol of [α-32P]-CTP-labeled tRNAAsp were incubated with enzyme (0.5 μM end concentration) for 2–3 h at 37°C in modification buffer containing 70 μM SAM; for Flag-Dnmt2 modification, the buffer was supplemented with 10 mM MgCl2. The reaction was stopped by addition of NH4OAc to 0.5 M and 2 volumes of 100% ethanol, subsequent storage at −20°C, and precipitation. The pellet was resuspended in 20 μL H2O, and the RNA was submitted to DNAzyme cleavage.
Enzymatic modification with mPus1 was performed by incubation of 0.8 pmol of RNA with 0.16 pmol recombinant pseudouridine synthase 1 (Pus1p) from mouse (Behm-Ansmant et al. 2006) (plasmid kindly provided by Jeff Patton, University of South Carolina School of Medicine, Columbia, SC, USA) in modification buffer (100 mM Tris-HCl at pH 8.0, 10 mM MgCl2, 0.1 M NH4OAc, 0.1 mM EDTA, 0.5 mM DTT) at 37°C in a final volume of 120 μL. tRNA was purified by two phenol extractions with 1 volume of saturated phenol, followed by extraction with 3 volumes of saturated diethylether three times. The RNA was then ethanol precipitated and redissolved in water.
DNAzyme-mediated cleavage and site-specific detection of modified bases
DNAzyme cleavage of tRNALys was performed by up to 60 iterations of incubation at 37°C for 3 min, preceded by a denaturing step at 85°C for 30 sec, with DNAzyme I (CTTTAACTTAAAAGGTTAAGGCTAGCTACAACGAGCTAAGTTAGCTTTACAGTG). DNAzyme cleavage was performed in 50 mM Tris-HCl (pH 7.5), 150 mM KCl, 10 mM MgCl2 by submitting the reaction mixtures in a PCR tube. For the analysis of cleavage yields, [α-32P]-UTP-labeled tRNALys was used, and aliquots from the reaction mixture were taken after 5, 10, 15, and 20 cycles, supplemented with 1 volume of loading buffer (90% formamide, 1× TBE), and the fragments were separated on a 12% denaturing PAGE. Cleavage yields were determined from a phosphorimager scan (Imagequant 5.2, GE).
Mixtures of the ligation-derived constructs of tRNALys containing amounts of 0, 25, 50, 75, and 100 mol pseudouridine per mole tRNA at position 27 were digested with the corresponding DNAzyme in 15 μL with 60 temperature cycles. The mixture was supplemented with 15 μL of labeling buffer (50 mM Tris-HCl at pH 7.6, 10 mM MgCl2, 5 mM DTT, 1 mM spermidine, 1 mM EDTA), and the fragmented RNA was 5′ labeled with [γ-32P] ATP (40 μCi, 10 μM final) by incubation for 1 h at 37°C in the presence of 10 u of T4-PNK. Enzyme inactivation was performed by incubation at 75°C for 10 min, and the DNAzyme was then digested by addition of 2 u DNase I (Fermentas) and incubation at 37°C for at least 30 min. The labeled RNA was purified on a 12% denaturing PAGE, excised, eluted, precipitated in the presence of carrier tRNA, and submitted to TLC analysis.
For TLC analysis, radiolabeled RNAs were digested to mononucleotides by incubation in 20 mM NH4OAc (pH 5.3), 0.5 mM ZnCl2 in the presence of 0.3 u nuclease P1 (Roche) for 3 h at 50°C–55°C. The resulting 5′-monophosphate nucleotides were separated by TLC on a 20-cm cellulose plate (Merck). For detection of pseudouridine in tRNALys, single dimension TLC in buffer C (70% isopropanol, 15% conc. HCl, 15% H2O, v/v/v) was performed.
For modification analysis of tRNAAsp, in vitro modified RNA was cleaved by 25 iterations of incubation at 37°C for 3 min, preceded by a denaturing step at 80°C for 30 sec, with tandem-DNAzyme II (CCCCGGTCTCCCGGGCTAGCTACAACGAGTGACAGGGGCTA GCTACAACGAGGGGATACTCACC) and DNAzyme III (CCCCGTCGGGGAAGGCTAGCTACAACGACGAACCCCGGTCT). Fragments were purified by denaturing PAGE, excised, eluted, precipitated in the presence of carrier tRNA, digested to mononucleotides as described above, and submitted to TLC analysis. Analysis of m5C in tRNAAsp was carried out by TLC in two dimensions in buffers A (70% isobutyric acid, 28.9% H2O, 1.1% ammonia [25%] v/v/v) and B (60% w/v (NH4)2SO4, 2% v/v 1-propanol, 0.1 M Na2HPO4/NaH2PO4 at pH 7.4) (Grosjean et al. 2004). Modification yields were determined from phosphorimager scans (Imagequant).
ACKNOWLEDGMENTS
This work was supported by the DFG (HE 3397/4 and LY 14/3-3, Priority Programme Epigenetics). The authors thank Henri Grosjean and Jeff Patton for plasmids coding for recombinant Trm4 and Pus1, respectively, and Matthias Schäfer and Tamara Fries for providing Flag-Dnmt2 from transfected Drosophila cell lines.
Footnotes
Abbreviations: TLC, thin-layer chromatography; SAM, S-adenosylmethionine; Trm4, tRNA dependent methyltransferase 4; Dnmt2, DNA methyltransferase 2; Pus1, pseudouridine synthase 1; CMCT, N-cyclohexyl-N′-β-(4-methylmorpholinium)ethylcarbodiimide p-tosylate; MS, mass sprectroscopy.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.742708.
REFERENCES
- Bakin, A., Ofengand, J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: Analysis by the application of a new sequencing technique. Biochemistry. 1993;32:9754–9762. doi: 10.1021/bi00088a030. [DOI] [PubMed] [Google Scholar]
- Bakin, A.V., Ofengand, J. Mapping of pseudouridine residues in RNA to nucleotide resolution. Methods Mol. Biol. 1998;77:297–309. doi: 10.1385/0-89603-397-X:297. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant, I., Massenet, S., Immel, F., Patton, J.R., Motorin, Y., Branlant, C. A previously unidentified activity of yeast and mouse RNA:pseudouridine synthases 1 (Pus1p) on tRNAs. RNA. 2006;12:1583–1593. doi: 10.1261/rna.100806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brule, H., Grosjean, H., Giege, R., Florentz, C. A pseudoknotted tRNA variant is a substrate for tRNA (cytosine-5)-methyltransferase from Xenopus laevis . Biochimie. 1998;80:977–985. doi: 10.1016/s0300-9084(99)80003-0. [DOI] [PubMed] [Google Scholar]
- Buchhaupt, M., Peifer, C., Entian, K.D. Analysis of 2′-O-methylated nucleosides and pseudouridines in ribosomal RNAs using DNAzymes. Anal. Biochem. 2007;361:102–108. doi: 10.1016/j.ab.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Cairns, M.J., King, A., Sun, L.Q. Optimization of the 10–23 DNAzyme-substrate pairing interactions enhanced RNA cleavage activity at purine–cytosine target sites. Nucleic Acids Res. 2003;31:2883–2889. doi: 10.1093/nar/gkg378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Patton, J.R. Cloning and characterization of a mammalian pseudouridine synthase. RNA. 1999;5:409–419. doi: 10.1017/s1355838299981591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz, R.P., Withers, J.B., Li, Y. Dinucleotide junction cleavage versatility of 8-17 deoxyribozyme. Chem. Biol. 2004;11:57–67. doi: 10.1016/j.chembiol.2003.12.012. [DOI] [PubMed] [Google Scholar]
- Feldman, A.R., Sen, D. A new and efficient DNA enzyme for the sequence-specific cleavage of RNA. J. Mol. Biol. 2001;313:283–294. doi: 10.1006/jmbi.2001.5058. [DOI] [PubMed] [Google Scholar]
- Goll, M.G., Kirpekar, F., Maggert, K.A., Yoder, J.A., Hsieh, C.L., Zhang, X., Golic, K.G., Jacobsen, S.E., Bestor, T.H. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- Grosjean, H., Motorin, Y., Morin, A. RNA-modifying and RNA-editing enzymes: Methods for their identification. In: Grosjean H., Benne R., editors. Modification and editing of RNA. American Society of Microbiology Press; Washington, DC: 1998. pp. 21–46. [Google Scholar]
- Grosjean, H., Keith, G., Droogmans, L. Detection and quantification of modified nucleotides in RNA using thin-layer chromatography. Methods Mol. Biol. 2004;265:357–391. doi: 10.1385/1-59259-775-0:357. [DOI] [PubMed] [Google Scholar]
- Guymon, R., Pomerantz, S.C., Ison, J.N., Crain, P.F., McCloskey, J.A. Post-transcriptional modifications in the small subunit ribosomal RNA from Thermotoga maritima, including presence of a novel modified cytidine. RNA. 2007;13:396–403. doi: 10.1261/rna.361607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm, M., Attardi, G. Nuclear control of cloverleaf structure of human mitochondrial tRNALys . J. Mol. Biol. 2004;337:545–560. doi: 10.1016/j.jmb.2004.01.036. [DOI] [PubMed] [Google Scholar]
- Helm, M., Brule, H., Degoul, F., Cepanec, C., Leroux, J.P., Giege, R., Florentz, C. The presence of modified nucleotides is required for cloverleaf folding of a human mitochondrial tRNA. Nucleic Acids Res. 1998;26:1636–1643. doi: 10.1093/nar/26.7.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm, M., Florentz, C., Chomyn, A., Attardi, G. Search for differences in post-transcriptional modification patterns of mitochondrial DNA-encoded wild-type and mutant human tRNALys and tRNALeu(UUR) . Nucleic Acids Res. 1999;27:756–763. doi: 10.1093/nar/27.3.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm, M., Petermeier, M., Ge, B., Fiammengo, R., Jaschke, A. Allosterically activated Diels–Alder catalysis by a ribozyme. J. Am. Chem. Soc. 2005;127:10492–10493. doi: 10.1021/ja052886i. [DOI] [PubMed] [Google Scholar]
- Hou, Y.M., Li, Z., Gamper, H. Isolation of a site-specifically modified RNA from an unmodified transcript. Nucleic Acids Res. 2006;34:e21. doi: 10.1093/nar/gnj018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeltsch, A., Nellen, W., Lyko, F. Two substrates are better than one: Dual specificities for Dnmt2 methyltransferases. Trends Biochem. Sci. 2006;31:306–308. doi: 10.1016/j.tibs.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Keith, G. Mobilities of modified ribonucleotides on two-dimensional cellulose thin-layer chromatography. Biochimie. 1995;77:142–144. doi: 10.1016/0300-9084(96)88118-1. [DOI] [PubMed] [Google Scholar]
- Kuchino, Y., Hanyu, N., Nishimura, S. Analysis of modified nucleosides and nucleotide sequence of tRNA. Methods Enzymol. 1987;155:379–396. doi: 10.1016/0076-6879(87)55026-1. [DOI] [PubMed] [Google Scholar]
- Kurschat, W.C., Muller, J., Wombacher, R., Helm, M. Optimizing splinted ligation of highly structured small RNAs. RNA. 2005;11:1909–1914. doi: 10.1261/rna.2170705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama, S., Ueda, T., Crain, P.F., McCloskey, J.A., Watanabe, K. A novel wobble rule found in starfish mitochondria. Presence of 7-methylguanosine at the anticodon wobble position expands decoding capability of tRNA. J. Biol. Chem. 1998;273:3363–3368. doi: 10.1074/jbc.273.6.3363. [DOI] [PubMed] [Google Scholar]
- Motorin, Y., Grosjean, H. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: Identification of the gene and substrate specificity of the enzyme. RNA. 1999;5:1105–1118. doi: 10.1017/s1355838299982201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz, S.C., McCloskey, J.A. Detection of the common RNA nucleoside pseudouridine in mixtures of oligonucleotides by mass spectrometry. Anal. Chem. 2005;77:4687–4697. doi: 10.1021/ac058023p. [DOI] [PubMed] [Google Scholar]
- Saikia, M., Dai, Q., Decatur, W.A., Fournier, M.J., Piccirilli, J.A., Pan, T. A systematic, ligation-based approach to study RNA modifications. RNA. 2006;12:2025–2033. doi: 10.1261/rna.208906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro, S.W., Joyce, G.F. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberklang, M., Prochiantz, A., Haenni, A.L., Rajbhandary, U.L. Studies on the sequence of the 3′-terminal region of turnip-yellow-mosaic-virus RNA. Eur. J. Biochem. 1977;72:465–478. doi: 10.1111/j.1432-1033.1977.tb11270.x. [DOI] [PubMed] [Google Scholar]
- Silverman, S.K. In vitro selection, characterization, and application of deoxyribozymes that cleave RNA. Nucleic Acids Res. 2005;33:6151–6163. doi: 10.1093/nar/gki930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita, K., Ueda, T., Watanabe, K. The presence of pseudouridine in the anticodon alters the genetic code: A possible mechanism for assignment of the AAA lysine codon as asparagine in echinoderm mitochondria. Nucleic Acids Res. 1999;27:1683–1689. doi: 10.1093/nar/27.7.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y.T., Shu, M.D., Steitz, J.A. A new method for detecting sites of 2′-O-methylation in RNA molecules. RNA. 1997;3:324–331. [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., Yu, Y.T. Detection and quantitation of RNA base modifications. RNA. 2004;10:996–1002. doi: 10.1261/rna.7110804. [DOI] [PMC free article] [PubMed] [Google Scholar]