Abstract
A characteristic feature of tRNAs is the numerous modifications found throughout their sequences, which are highly conserved and often have important roles. Um44 is highly conserved among eukaryotic cytoplasmic tRNAs with a long variable loop and unique to tRNASer in yeast. We show here that the yeast ORF YPL030w (now named TRM44) encodes tRNASer Um44 2′-O-methyltransferase. Trm44 was identified by screening a yeast genomic library of affinity purified proteins for activity and verified by showing that a trm44-Δ strain lacks 2′-O-methyltransferase activity and has undetectable levels of Um44 in its tRNASer and by showing that Trm44 purified from Escherichia coli 2′-O-methylates U44 of tRNASer in vitro. Trm44 is conserved among metazoans and fungi, consistent with the conservation of Um44 in eukaryotic tRNAs, but surprisingly, Trm44 is not found in plants. Although trm44-Δ mutants have no detectable growth defect, TRM44 is required for survival at 33°C in a tan1-Δ mutant strain, which lacks ac4C12 in tRNASer and tRNALeu. At nonpermissive temperature, a trm44-Δ tan1-Δ mutant strain has reduced levels of tRNASer(CGA) and tRNASer(UGA), but not other tRNASer or tRNALeu species. The trm44-Δ tan1-Δ growth defect is suppressed by addition of multiple copies of tRNASer(CGA) and tRNASer(UGA), directly implicating these tRNASer species in this phenotype. The reduction of specific tRNASer species in a trm44-Δ tan1-Δ mutant underscores the importance of tRNA modifications in sustaining tRNA levels and further emphasizes that tRNAs undergo quality control.
Keywords: S. cerevisiae, tRNASer, TAN1, quality control
INTRODUCTION
tRNAs play a crucial role in the translation of mRNA into proteins, entering and leaving the ribosome about seven times per second in yeast, and faithfully decoding mRNA through interactions with the anticodon and commensurate addition of the amino acid to the growing peptide chain. This occurs on 300,000 ribosomes using a supply of 3,000,000 tRNAs to decode 60,000 mRNAs in one generation in yeast, and each tRNA is used slightly less than once every second (Waldron and Lacroute 1975; Ares et al. 1999).
A remarkable feature of tRNA is the numerous post-transcriptional base and sugar modifications found throughout the molecule. These modifications include a number of different methylations of distinct bases or of the ribose moiety as well as a number of more complex chemical modifications. In the yeast Saccharomyces cerevisiae, the 34 characterized cytoplasmic tRNAs each have an average of 12.7 modifications, and there are a total of 25 different modifications, including 12 distinct methylations of bases or ribose sugars (Sprinzl et al. 1998). Many tRNA modifications are highly conserved within one or more of the archaeal, eubacterial, and eukaryotic kingdoms and often occur at the same position in tRNAs from different organisms, suggesting important structural and functional roles of the modifications.
The roles of these modifications have been a subject of increasing study, as the genes catalyzing these modifications have been identified in the last several years. Thus, a number of modifications in the anticodon region have been shown to be important for translation (Laten et al. 1978; Dihanich et al. 1987; Lecointe et al. 1998; Gerber and Keller 1999; Bjork et al. 2001; Urbonavicius et al. 2001; Pintard et al. 2002; Kalhor and Clarke 2003). Several other modifications elsewhere in the tRNA have important roles, based on growth and translation defects of triple mutants such as Escherichia coli trmH trmA truB mutants (lacking Gm18, m5U54, and Ψ55) (Urbonavicius et al. 2002) and on synthetic lethality of double mutants such as pus1 pus4 mutants (lacking Ψ26–28, 34–36, 65,67 from Pus1 and Ψ55 from Pus4) (Grosshans et al. 2001).
There is also mounting evidence that certain modifications play important roles in tRNA folding or stability. Thus, trm6 ts (or trm61 ts) mutants, responsible for m1A58, have reduced levels of tRNAi Met at high temperature, due to nuclear degradation of pre-tRNAi Met by polyadenylation and exonucleolytic degradation, mediated by TRF4 and RRP6 (Anderson et al. 1998, 2000; Kadaba et al. 2004; LaCava et al. 2005; Vanacova et al. 2005; Kadaba et al. 2006). Similarly, trm8-Δ trm4-Δ mutants, which lack m7G46 and m5C30,40,48,49 in their tRNA, are temperature sensitive for growth, and their tRNAVal(AAC) is rapidly degraded and deacylated at high temperature by a pathway that is independent of TRF4/RRP6 (Alexandrov et al. 2006). Such tRNA quality control pathways are not confined to modification defects, since mutations in tRNAArg(CCG) that lead to misfolding of the anticodon stem–loop or disruption of the acceptor stem (Chakshusmathi et al. 2003) also have reduced tRNAArg(CCG) levels due to a pathway that is independent of TRF4 (Copela et al. 2006).
One common tRNA modification is 2′-O-methylation of the ribose sugar. This modification is found at 13 positions in tRNA species from different organisms and at 5 positions in yeast tRNAs (positions 4, 18, 32, 34, and 44). Each of the modifications assigned to a gene product is effected by a 2′-O-methyltransferase rather than by a guide RNA mechanism, as described for several archaeal tRNA modifications (Clouet d'Orval et al. 2001; Renalier et al. 2005) and numerous rRNA and snRNA modifications (Decatur and Fournier 2003; Yu et al. 2005; Piekna-Przybylska et al. 2007). Thus, Trm13 is responsible for formation of Nm4 in tRNAHis, tRNAGly, and tRNAPro (Wilkinson et al. 2007), Trm3 for Gm18 of numerous tRNAs (Cavaille et al. 1999), and Trm7 for Nm32 and Nm34 of tRNALeu, tRNAPhe, and tRNATrp (Pintard et al. 2002). The enzyme catalyzing 2′-O-methylation at position 44 remains to be identified and is the subject of this report.
There are several reasons why 2′-O-methylation at position 44 is of interest. First, this modification is uniquely found in class II tRNAs, which have a long variable loop and include species of tRNASer, tRNALeu, and tRNATyr, and is only observed in tRNA species that have a U44 residue (Fig. 1A). Second, the location of Um44 at the junction between the anticodon stem and the variable loop suggests an important role of this residue in tRNA structure and/or stability. Indeed, residue 44 is across from residue 26 at the junction of the D-stem and the anticodon stem, and Um44 is predicted to form tertiary interactions with the m2,2G26 (Sampson and Uhlenbeck 1988). Third, this modification is highly conserved among eukaryotes (Sprinzl et al. 1998). Thus, 21 out of 23 characterized eukaryotic cytoplasmic tRNASer species have Um44, and the two exceptions are chicken tRNASer, which lacks the long variable loop and has A44, and tRNASer(CAG) from Candida cylindracea, which is specific for CUG leucine codons in this organism because of its altered genetic code and itself has unusual features in its primary structure (Yokogawa et al. 1992). Um44 is also found in 10 of the 22 characterized eukaryotic cytoplasmic tRNALeu species (although all of these 22 tRNALeu species have U44 and a long variable loop), in all five characterized mitochondrial tRNALeu species from plants, and in mitochondrial tRNATyr from Tetrahymena pyriformis (Schnare et al. 1985), but not in the four other organellar tRNATyr species with a long variable arm and U44. In yeast, Um44 is only found in cytoplasmic tRNASer species (Zachau et al. 1966; Etcheverry et al. 1979; Olson et al. 1981).
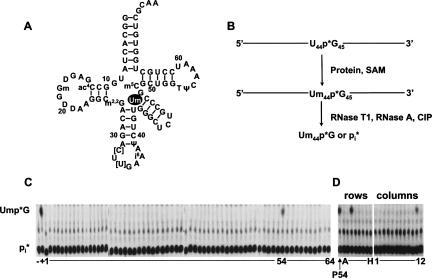
FIGURE 1.
Identification of yeast ORF associated with Um44 methyltransferase activity of tRNASer(UGA). (A) Two-dimensional structure of S. cerevisiae tRNASer(UGA). tRNA is numbered according to convention, in this case with no residue 17, an additional residue 20a, and an 11 residue variable arm between Um44 and G48. [C], [U], uncharacterized C and U modifications. Um44 is shown in white font within a black oval. (B) Assay scheme to detect 2′-O-methylation of tRNASer uniquely labeled at U44 (U44*Ser). Treatment with RNaseT1, RNaseA and calf intestinal phosphatase (CIP) yields Ump*G if U44 is methylated and inorganic phosphate (Pi) if the substrate is unmodified. (C) Assay of a genomic collection of GST-ORF pools for Um44 methyltransferase activity. GST-ORF fusion proteins were purified and assayed for tRNASer Um44 2′-O-methyltransferase using 2 μL of each GST-ORF fusion protein pool, 0.5 mM S-adenosyl methionine, and ∼1 nM U44*Ser tRNA in methyltransferase buffer at 30°C for 12 h, and then RNA was digested with RNase T1, RNase A, and CIP to produce Ump*G dinucleotide or inorganic phosphate, which were resolved by thin layer chromatography as described in Materials and Methods. (Lane −) no protein; (lane +), yeast crude extract. (D) Deconvolution of subpools of Plate 54 to identify the ORF associated with Um44 methyltransferase. (P54) GST-ORF proteins purified from pool 54.
In this study we identify the yeast tRNASer Um44 methyltransferase (Trm44), and show that trm44 mutants that also lack Tan1, required for ac4C12 in yeast tRNASer and tRNALeu (Johansson and Bystrom 2004), have a growth defect, and accumulate reduced levels of specific tRNASer species at elevated temperature.
RESULTS
Um44 methyltransferase activity copurifies with the product of ORF YPL030w
To identify the tRNASer Um44 2′-O-methyltransferase in S. cerevisiae, we screened the yeast proteome for this activity using yeast tRNASer(UGA) as the substrate (Fig. 1A), specifically labeled at the 3′ phosphate of U44 (U44*Ser). Formation of Um44 will generate a phosphodiester bond that is resistant to digestion with RNase T1, RNase A, and phosphatase, yielding the dinucleotide Um44p*G, whereas unreacted substrate will yield inorganic phosphate (Pi*) after this treatment, and these products are easily resolved by thin layer chromatography (Fig. 1B). We screened the yeast proteome for tRNASer Um44 2′-O-methyltransferase using pools of purified fusion proteins from a genomic collection of yeast strains expressing yeast ORF fusion proteins, as was done previously (Martzen et al. 1999; Wilkinson et al. 2007). We found 2′-O-methyltransferase activity in pool 54 (Fig. 1C), and deconvolution of this pool shows that activity derives from the strain in row C and column 11 of plate 54 (Fig. 1D), which expresses yeast ORF YPL030w. We show below that this ORF encodes the yeast tRNASer Um44 2′-O-methyltransferase, and hence assign the name TRM44 to this gene.
Trm44 is necessary and sufficient for 2′-O-methylation of U44 of tRNASer in vitro and in vivo
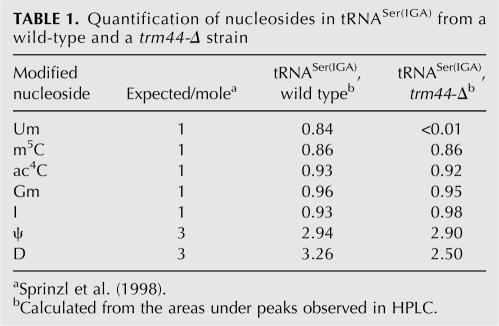
To determine if Trm44 is required for formation of Um44, we compared Um44 methyltransferase activity and tRNASer from a trm44-Δ deletion strain with that from an otherwise isogenic wild-type strain. TRM44 is required for tRNASer Um44 2′-O-methyltransferase activity in vitro, since crude extracts from a wild-type strain have readily detectable tRNA Um44 methyltransferase activity with as little as 0.54 μg extract (Fig. 2A, lanes c–h), whereas crude extracts from a trm44-Δ strain did not have detectable activity even with 14.6 μg extract (Fig. 2A, lanes i–m). Moreover, TRM44 is also required for the modification in vivo, since tRNASer(IGA) from a trm44-Δ strain specifically lacks Um44. As shown in Figure 2B (upper trace), tRNASer(IGA) from the wild-type control strain has all the expected modifications (Ψ, D, m5C, Um, I, Gm, and ac4C) as determined by retention times and UV absorbance spectra (Gehrke and Kuo 1989), whereas tRNASer(IGA) from a trm44-Δ deletion strain has all the same modifications except for Um (Fig. 2B, lower trace). Quantification shows that Um levels in tRNASer(IGA) are reduced from 0.84 mol/mole in the wild-type strain to less than 0.01 mol in the trm44-Δ strain (Table 1), whereas the levels of all other modifications are similar from both strains. The modest variation in dihydrouridine levels arises from incomplete resolution of the peaks in this system and the difficulty in quantifying dihydrouridine because of the lack of absorption peak (Xing et al. 2004). We also observed lack of Um in nucleosides from tRNASer(UGA) purified from the trm44-Δ strain (data not shown). Thus we conclude that TRM44 is required for formation of Um44 in tRNASer in vitro and in vivo.
FIGURE 2.
TRM44 is necessary and sufficient for the Um44 methyltransferase activity in vitro and in vivo. (A) trm44-Δ extracts lack tRNASer Um44 methyltransferase activity. Crude extracts from wild-type and trm44-Δ strains were assayed in 10 μL reactions containing 1 nM U44*Ser tRNA, by serial fivefold dilution of wild-type extracts (lanes c–g), beginning with 13.4 μg protein or trm44-Δ extracts (lanes i–m), beginning with 14.6 μg protein. (Lanes a,h) no protein; (lane b) yeast crude extract. (B) tRNASer(IGA) isolated from a trm44-Δ strain lacks Um. tRNASer(IGA) was purified from wild-type and trm44-Δ mutant strains, and nucleosides were prepared and resolved on HPLC as described in Materials and Methods. (C) Trm44 expressed in E. coli has Um44 methyltransferase activity. Trm44 was expressed in E. coli and purified as described in Materials and Methods and assayed for Um44 methyltransferase activity with U44*Ser tRNA substrate and 10-fold serial dilutions of the control extract (lanes c–e, beginning with 12 μg protein), Trm44p expressing extract (lanes f–k, beginning with 8.6 μg), or purified Trm44p (lanes l–q, beginning with 5.2 μg). (Lane a) no protein; (lane b) yeast crude extract (40 μg).
TABLE 1.
Quantification of nucleosides in tRNASer(IGA) from a wild-type and a trm44-Δ strain
Two lines of evidence suggest that Trm44 is sufficient for Um44 2′-O-methyltransferase activity. First, expression of TRM44 is the limiting factor for activity, since crude extracts made from the strain expressing GST-TRM44 have 10-fold more activity than the control extract (data not shown). Second, purified yeast Trm44 prepared after expression in E. coli has robust tRNASer Um44 2′-O-methyltransferase activity. Crude extracts from an E. coli strain expressing yeast Trm44 as a His6-Trm44 fusion protein have a 66-kDa polypeptide that is not observed in the control strain and that corresponds to the expected molecular weight of Trm44, and this polypeptide is substantially enriched after IMAC purification (data not shown). As shown in Figure 2C, extracts from the E. coli strain expressing His6-Trm44 have substantial activity (Fig. 2C, lanes f–k), easily detectable by assay of 0.86 ng protein (Fig. 2C, lane j), whereas the vector control extract (Fig. 2C, lanes c–e) has no detectable activity, even with 14,000-fold more protein (lane c). Moreover, purified His6-Trm44 has robust activity (Fig. 2C, lanes l–q). Thus, expression of Trm44 is sufficient for tRNASer Um44 2′-O-methyltransferase activity in E. coli. Furthermore, the 20% yield of activity after purification suggests that no dissociable cofactors are required for activity under these conditions, other than S-adenosylmethionine, which is included in the reaction mixture. S-adenosylmethionine (SAM) is required for the methyltransferase reaction, based on the complete absence of detectable activity (less than 0.3%) observed with 800 nM His6-Trm44 protein and 800 nM singly labeled tRNASer, compared with the conversion of 35% of the substrate to product when SAM is added under the same conditions.
The Trm44 protein family is highly conserved among metazoans and fungi but not plants
To assess the occurrence of Trm44 in other species we did a BLAST search of the NCBI translated nucleotide database and the nonredundant protein database. Trm44 protein is highly similar to predicted proteins from other fungi, with E values ranging from 0.0 for close relatives to e-37 for Ustilago maydis and somewhat similar to predicted proteins from a number of metazoans, including Anopheles gambiae (e−34), Drosophila melanogaster (e−33), Xenopus laevis (e−28), Homo sapiens (e−28), Strongylocentrotus purpuratatus (e−26), Caenorhabditis elegans (e−20), Danio rerio (e−19), and Schistosoma japonicum (e−8). None of these proteins has been characterized, although most are named as “putative methyltransferase.” The widespread occurrence of members of the Trm44 protein family among eukaryotes is consistent with the widespread occurrence of Um44 in eukaryotes. Surprisingly, however, plants do not appear to have a Trm44 homolog, although tRNASer and tRNALeu species in plants have Um44. Thus, plants may possess another mechanism for Um44 modification of tRNAs.
Most of the homology in the Trm44 protein family is restricted to an ∼200 amino acid region toward the C terminus of the protein, which is called DUF1613 by Pfam. An alignment from BLAST analysis is shown in Figure 3, which reveals the presence of a conserved methyltransferase domain, featuring the characteristic GXGXG amino acid sequence found in this superfamily (Kagan and Clarke 1994).
FIGURE 3.
Alignment of Trm44 homologs. Trm44 homologs were identified by BLAST search (Altschul et al. 1997), and several proteins representing yeast to humans were aligned using Multalin (Corpet 1988). Sequences with little or no homology are omitted from proteins for clarity, as indicated by numbers in parentheses. Residues shaded in dark shading have >70% consensus and light shading have >50% consensus.
Cells lacking both Trm44 and the tRNA modifying enzyme Tan1 have an aggravated growth defect
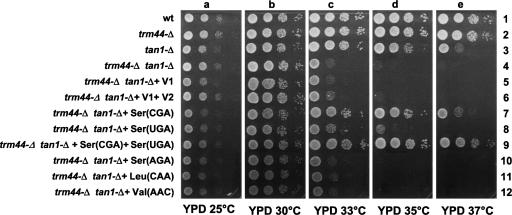
Although Um44 is highly conserved among eukaryote tRNASer species, trm44-Δ mutant strains do not have any observable growth defect at a range of temperatures from 18°C to 37°C in minimal or rich media containing either glucose or glycerol (data not shown). To further probe the role of Trm44, we created and tested the growth phenotypes of a set of double mutant strains, each bearing a trm44-Δ deletion as well as a deletion in a second tRNA modification gene that acts on tRNASer species, the only known substrate of Trm44 (Fig. 1A). We used a similar approach previously to assign a role for trm8 mutants (Alexandrov et al. 2006). We thus targeted TAN1, encoding tRNA cytidine N4 acetyltransferase (Johansson and Bystrom 2004), DUS1, DUS2, and DUS4, encoding dihydrouridine synthases for D16, D20, and D20a (Xing et al. 2002, 2004), TRM3, encoding Gm18 methyltransferase (Cavaille et al. 1999), TRM1, encoding m2,2G26 dimethyltransferase (Ellis et al. 1986), MOD5, encoding i6A isopentenyltransferase (Laten et al. 1978; Dihanich et al. 1987), TRM4, encoding m5C48 methyltransferase (Wu et al. 1998), and TRM2, encoding m5U54 methyltransferase (Hopper et al. 1982; Nordlund et al. 2000). These genes are all known to be nonessential, and all but MOD5 act outside the anticodon region. In addition, we mutated the yeast La homolog LHP1 (Lin-Marq and Clarkson 1995), which is known to affect 3′-end formation of tRNA and tRNA stability (Yoo and Wolin 1997; Chakshusmathi et al. 2003; Copela et al. 2006; Huang et al. 2006), and the 2′-O-methyltransferase TRM7 (Pintard et al. 2002) as a negative control.
As expected from previous published results, we found no obvious growth defects of single mutant strains lacking TRM2, TRM3, TRM4, MOD5, LHP1, DUS1, DUS2, and DUS4 (Fig. 4A, panels a,c,e,g), and we found that trm7-Δ mutant strains were quite sick under a variety of conditions (Fig. 4A, row 5; Pintard et al. 2002). In addition, we find that trm1-Δ mutants and tan1-Δ mutants each grow poorly on YP media containing glucose (YPD) at 37°C (Fig. 4A, rows 1,8), which has not been previously reported.
FIGURE 4.
trm44-Δ tan1-Δ double mutants have a synthetic growth phenotype. (A) Analysis of mutant strains for growth phenotypes. Homozygous diploid strains bearing single and double deletions (in conjunction with a trm44-Δ mutation) of genes encoding tRNA modifying enzymes were constructed as described in Materials and Methods, grown in YP+ Dextrose media overnight at 25°C, diluted to OD600 = 1, serially diluted (10-fold), spotted on plates containing YP+ Dextrose and YP+ Glycerol, and incubated at indicated temperatures for 2 d. (B) Complementation of the growth defect of trm44-Δ tan1-Δ mutant strains. trm44-Δ tan1-Δ mutant strains were transformed with CEN URA3 plasmids containing TAN1 or TRM44 or an empty vector, and transformants were grown in SD-uracil media at 25°C overnight, diluted to OD600 = 1, 10-fold serially diluted, and spotted on YPD plates and incubated for 2 d at indicated temperatures.
We find that TRM44 is essential for growth of tan1-Δ mutant strains in YPD media or in YP+ Glycerol media at 35°C (Fig. 4A, row 8) or 33°C (data not shown) and in synthetic media containing glucose (SD) or glycerol at 33°C or 35°C (data not shown). The synthetic growth phenotype of trm44-Δ tan1-Δ double mutant strains is complemented by introduction of either missing gene, expressed under control of its own promoter on a single-copy (CEN) plasmid (PTAN1-TAN1 or PTRM44-TRM44) but not by an empty vector, confirming that the phenotype is due to the TRM44 and TAN1 genes (Fig. 4B, panels b,c, rows 7–9). Notably, trm44-Δ mutants do not display a synthetic growth defect with any of the other tested deletions, including the controls LHP1 and TRM7.
Lack of TRM44 and TAN1 leads to reduced levels of tRNASer(CGA) and tRNASer(UGA) at restrictive temperature
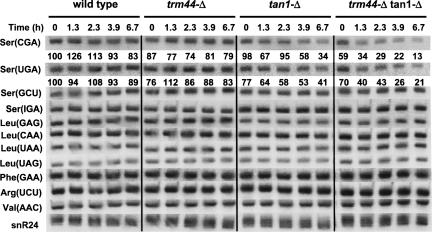
The conditional growth defect of trm44-Δ tan1-Δ double mutant strains is likely due to effects on tRNASer function, since in yeast only tRNASer is known to have Um44, and the ac4C12 product of Tan1 is also found in tRNASer as well as in tRNALeu. tRNASer function might be affected either by reduced activity of one or more tRNASer species or by reduced levels of these species at restrictive temperature. Our results indicate that levels of tRNASer(CGA) and tRNASer(UGA) are dramatically reduced in trm44-Δ tan1-Δ mutant strains grown in restrictive conditions (YP media containing glycerol at 36.5°C). tRNASer(CGA) levels are at 60% of wild-type levels before the temperature shift and are reduced to 34% 1.3 h after temperature shift, 22% after 3.9 h, and 13% after 6.7 h (Fig. 5). By contrast, tRNASer(CGA) levels are more modestly reduced in tan1-Δ single mutants and are nearly constant in wild-type cells and in trm44-Δ mutants. A similar reduction in tRNA levels is observed with tRNASer(UGA), although this is not quite as severe as that with tRNASer(CGA).
FIGURE 5.
Levels of tRNASer(CGA) and tRNASer(UGA) are reduced in a trm44-Δ tan1-Δ mutant strain at 36.5°C in YP+ Glycerol. Wild-type, trm44-Δ, tan1-Δ, and trm44-Δ tan1-Δ strains were grown in YP+ Glycerol media at 28°C and shifted to 36.5°C for indicated times, and RNA was analyzed by Northern blotting as described in Materials and Methods. Quantification of the levels of tRNASer(CGA) and tRNASer(UGA) is shown under each corresponding panel, as normalized first to snR24 (snRNA), and then to levels of the corresponding tRNA species in wild-type cells at permissive temperature.
Strikingly, there is no reduction in the other two tRNASer isoacceptors, tRNASer(IGA) and tRNASer(GCU) (Fig. 5), although tRNASer(IGA) is known to have the same modifications (Zachau et al. 1966), and tRNASer(GCU) is likely to have both modifications but is not characterized. Similarly, there is little observed decrease in any of the four different tRNALeu species (other than a modest reduction in tRNALeu(GAG) that is observed in tan1-Δ mutant strains), which are the only other tRNA species known to be modified by Tan1 (Sprinzl et al. 1998), and there is no observed decrease in each of several control tRNAs (Fig. 5).
The growth defect of trm44-Δ tan1-Δ double mutants is due to loss of tRNASer(CGA) and tRNASer(UGA)
Because Northern analysis of trm44-Δ tan1-Δ double mutants reveals specific loss of tRNASer(CGA) and tRNASer(UGA) species, but not other tRNAs, at restrictive temperature, it seemed likely that the loss of tRNASer(CGA) and/or tRNASer(UGA) was responsible for the growth defect of trm44-Δ tan1-Δ mutants. In agreement with this, we find that introduction of multicopy plasmids carrying tRNASer(CGA) and tRNASer(UGA) can completely suppress the growth defect of trm44-Δ tan1-Δ mutants (Fig. 6, row 9), whereas no suppression is observed after introduction of empty control plasmids or plasmids containing other tRNA species. Indeed, expression of tRNASer(CGA) alone is sufficient for nearly complete suppression of the growth defect of this strain at 37°C (Fig. 6, row 7), whereas expression of tRNASer(UGA) can only suppress the growth defect at 35°C (Fig. 6, row 8). This result provides strong evidence that the growth defect of trm44-Δ tan1-Δ double mutant strains at high temperatures is primarily due to loss of tRNASer(CGA) and secondarily to the loss of tRNASer(UGA).
FIGURE 6.
The synthetic growth phenotype of a trm44-Δ tan1-Δ mutant strain is suppressed by tRNASer(CGA) and tRNASer(UGA). A trm44-Δ tan1-Δ mutant strain was transformed with a multicopy plasmid(s), containing tRNASer(CGA) and tRNASer(UGA) or a vector control, and transformants were grown in synthetic media, diluted to OD600 = 1, spotted on YPD plates, and incubated at indicated temperatures for 2 d.
DISCUSSION
In this work we have identified the S. cerevisiae gene encoding tRNASer Um44 methyltransferase (Trm44) as the product of ORF YPL030w and demonstrated a role for this protein in helping to maintain levels of specific tRNASer species. We identified Trm44 as the Um44 2′-O-methyltransferase by assay of a genomic collection of purified yeast GST-ORF fusion proteins, using tRNASer(UGA) specifically labeled at U44 as the substrate and S-adenosylmethionine as the methyl donor. Trm44 is necessary for Um44 formation in vivo because Um44 is absent in tRNASer(IGA) and tRNASer(UGA) from a trm44-Δ strain, and Trm44 protein is sufficient for activity because yeast Trm44 protein purified from E. coli catalyzes 2′-O-methyltransferase activity. We have also shown that trm44-Δ tan1-Δ mutant strains (which lack both Um44 and ac4C12) are temperature sensitive and provided evidence that the loss of viability occurs because of loss of tRNASer(CGA) and tRNASer(UGA), since these species are specifically depleted at high temperature and since introduction of more copies of these tRNA genes rescues the temperature-sensitive defect of the strain. Moreover, since the growth defect at 37°C is due to the tan1-Δ mutation, this result suggests that the defect in tan1-Δ mutants is due to the same two tRNA species.
The two tRNASer species that are specifically reduced in trm44-Δ tan1-Δ mutants are remarkably similar to one another. Thus, tRNASer(CGA) and tRNASer(UGA) have 82 identical residues among their 85 bases, whereas tRNASer(CGA) has 13 differences with tRNASer(IGA) and 26 differences with tRNASer(GCU). Although this similarity explains why tRNA(Ser)CGA and tRNASer(UGA) behave similarly, it is not immediately obvious why tRNASer(CGA) is more sensitive than tRNASer(UGA) as measured by both the reduction in tRNA levels and by suppression of the temperature sensitivity of trm44-Δ tan1-Δ mutants with introduction of additional copies of tRNA genes. tRNASer(CGA) has a G28–C42 base pair in the anticodon stem instead of a C–G pair in tRNASer(UGA) and C34 rather than U34 at the anticodon, and these differences are predicted to preferentially stabilize the anticodon stem–loop from tRNASer(CGA) by only a very modest 0.2 kcal/mol (Mathews et al. 1999). The discrepancy in the loss rates of these tRNASer species may be related to differences in ongoing transcription or to subtle differences in cellular recognition of these undermodified tRNAs.
tRNASer(CGA) appears to be unusually sensitive to alterations in its modifications. Bystrom and coworkers (Johansson and Bystrom 2002, 2004) have shown that a strain with a mutation in the variable arm of tRNASer(CGA) has a synthetic growth defect when combined with deletion of any of several genes encoding modification enzymes acting on this tRNA, including TRM2, encoding m5U54 methyltransferase (Hopper et al. 1982; Nordlund et al. 2000), TRM1 (m2,2G26) (Ellis et al. 1986), TRM3 (Gm18) (Cavaille et al. 1999), PUS4 (Ψ55) (Becker et al. 1997), or TAN1 (ac4C12) (Johansson and Bystrom 2004), as well as LHP1, which affects 3′ processing of tRNAs and has a chaperone function (Yoo and Wolin 1997; Chakshusmathi et al. 2003). Moreover, each of these combinations of tRNASer(CGA) mutations and modification genes or LHP1 mutations affects levels of tRNASer(CGA) (Johansson and Bystrom 2002). This unusual sensitivity of tRNASer(CGA) may in part be due to the fact that it is a single copy essential tRNA gene (Etcheverry et al. 1982).
We note that tan1-Δ mutants also have reduced levels of both tRNASer(CGA) and tRNASer(UGA) at 36.5°C, albeit not as severely reduced as in trm44-Δ tan1-Δ double mutants. A likely explanation for this observation is that the instability of these tRNASer species in trm44-Δ tan1-Δ mutants is due more to the lack of Tan1 than to the lack of Trm44, consistent with the observed temperature sensitivity of tan1-Δ mutants at 37°C but not at the lower temperatures at which trm44-Δ tan1-Δ mutants are defective for growth (Fig. 4).
The mechanism by which 2′-O-methylation of U44 contributes to stability of tRNASer(CGA) is unclear. One possibility is that the effects are due to thermal stability conferred by Um44. It is known that a 2′-O-methyl group on uridine can stabilize the C3′-endo form of RNA by ∼0.8 kcal mol−1, attributed to decreased repulsion between the 2-carbonyl group of uridine, the 2′-O-methyl group, and the 3′-phosphate group relative to the C2′-endo form (Kawai et al. 1992). It is also known that the 2′-O-methyl group can confer increased stability, attributed to stacking, of poly-U polymers (Zmudzka and Shugar 1970) and of nucleotide dimers of form 5′NmpN3′, when the 5′ nucleoside is cytidine, uridine, or guanosine but not adenosine (Drake et al. 1974). It is interesting to note in this regard that the amount of 2′-O-methylated ribose moieties in tRNA increases threefold when cultures of Bacillus stearothermophillus are shifted from 50°C to 70°C (Agris et al. 1973). A second possibility is that the role of 2′-O-methylation at U44 is to stabilize a particular conformation for recognition purposes. Indeed, there is evidence that fixing the base at the wobble position of the tRNA anticodon (N34) at C3′-endo form or C2′-endo form can lead to proper recognition of codons (Yokoyama et al. 1985).
The finding that members of the Trm44 family are not found in plants is somewhat surprising. The Trm44 protein family is distributed widely among fungi and among metazoans, including the arthropods, chordates, echinoderms, nematodes, and platyhelminthes. This distribution is consistent with the occurrence of Um44 in tRNASer in all of the 14 characterized cytoplasmic tRNASer species from metazoan and fungal species. It is also likely consistent with the occurrence of Um44 in four tRNALeu species from metazoans and fungi, since most modifications at the same position in different tRNAs are formed by the same proteins (Hopper and Phizicky 2003). However, the lack of Trm44 family members in plants is not consistent with the presence of Um44 in each of seven cytoplasmic tRNASer species from three different orders within the class magnoliopsida (flowering plants), represented by Lupinus luteus, Nicotiana rustica, and Spinacia oleracea, or with the six tRNALeu species containing Um44 found in flowering plants. The complete absence of a recognizable Trm44 ortholog in plants might have two possible causes: First, the Trm44 protein family might be somewhat diverged in plants, consistent with the observation that although the Trm44 family is very highly conserved in fungi, it is much less conserved in metazoans. Second, 2′-O-methylation in plants might be catalyzed through appropriate Box C/D snoRNA guide molecules and the Nop1/Fibrillarin snoRNP complex (Galardi et al. 2002), much as 2′-O-methylation is catalyzed either enzymatically or by Box C/D snRNPs at residue N34 (Clouet d'Orval et al. 2001; Pintard et al. 2002) and at residue C56 (Renalier et al. 2005) in different groups of species.
The reduced levels of tRNASer(CGA) and tRNASer(UGA) observed in trm44-Δ tan1-Δ double mutants at restrictive temperature underscore the importance of modifications remote from the anticodon for tRNA stability in the cell and the importance of synergistic interactions among modifications. Lack of these tRNA modifications can lead to tRNA degradation by at least two pathways. First, cells lacking m1A58 have reduced tRNAi Met because pre-tRNAi Met is degraded by polyadenylation and exonucleolytic degradation, using the TRAMP complex containing Trf4, Air2, and Mtr4, as well as Rrp6 and the nuclear exosome (Kadaba et al. 2004, 2006; LaCava et al. 2005; Vanacova et al. 2005). Second, cells lacking m7G46 and m5C49 (trm8-Δ trm4-Δ mutants) have mature tRNAVal(AAC) that is rapidly degraded at high temperature by a pathway independent of the Trf4/Rrp6 degradation pathway (Alexandrov et al. 2006). The observed loss of tRNASer(CGA) and tRNASer(UGA) in trm44-Δ tan1-Δ mutants lacking Um44 and ac4C12 emphasizes the crucial importance of a number of modifications for tRNA stability, reinforcing the need to understand the mechanism and regulation of these tRNA quality control pathways.
MATERIALS AND METHODS
Strains and plasmids
Yeast strains deleted for specific genes and their isogenic parents (BY4741 and BY4742) were obtained from the yeast knockout collection (Open Biosystems). The kanMX cassette of MATa and MATα strains deleted for ORF YPL030w (trm44-Δ∷kanMX) were each replaced with the nourseothricin resistance cassette by transformation with linearized DNA derived from plasmid p4339 by EcoRI digestion (Tong et al. 2001) to generate yeast strains LY1050 (MATa trm44Δ∷natMX) and LY1023 (MATα trm44Δ∷natMX). The trm44-Δ∷kanMX, trm1-Δ∷kanMX, trm2-Δ∷kanMX, trm3-Δ∷kanMX, trm4-Δ∷kanMX, trm7-Δ∷kanMX, dus1-Δ∷kanMX, dus2-Δ∷kanMX, dus4-Δ∷kanMX, tan1-Δ∷kanMX, mod5-Δ∷kanMX, and lhp1-Δ∷kanMX alleles were PCR amplified from genomic DNA prepared from the corresponding knockout strains using upstream and downstream primers and transformed into either the trm44Δ∷natMX or wild-type haploid strains to generate the corresponding trm44Δ∷natMX geneX-Δ∷kanMX double mutant or geneX-Δ∷kanMX single mutant strains. All strains were confirmed by PCR with flanking primers and then mated to generate the corresponding homozygous diploid knockout strains and their isogenic parents (BY4741/BY4742). Strains are listed in Table 2.
TABLE 2.
Strains used in this study
For complementation, the TRM44 and TAN1 genes were PCR amplified together with ∼400 bp of upstream and downstream flanking sequence using primers TRM44 XbaI 5′ and TRM44 EcoR1 3′ or TAN1 HindIII Lac 5′ and TAN1 EcoR1 Lac 3′ and then digested accordingly and cloned into yCPlac33 URA3 (Gietz and Sugino 1988) to create plasmids pLK300 (CEN TRM44) and pLK313 (CEN TAN1), which were sequence verified. Multicopy plasmids containing tRNASer(UGA) (pLK328) and tRNASer(CGA) (pLK322), were constructed in a similar manner using primers Ser UGA HindIII 5′ and Ser UGA EcoRI 3′ to amplify the tS(UGA)E locus and ∼400 bp of flanking sequences and primer pairs tS(CGA)BamHI 5′ and tS(CGA)EcoRI 3′ to amplify tS(CGA)C locus followed by digestion and ligation into the 2 μ URA3 vector yEPlac195 (Gietz and Sugino 1988). Control multicopy plasmids were also created by similar cloning of tS(AGA)D3 and tL(CAA)A loci using primer pairs tS(AGA) HindIII 5′ and tS(AGA)EcoRI 3′ and tL(CAA) BamHI 5′ and tL(CAA) EcoRI 3′, respectively. The sequences of the primers are listed in Table 3. Construction of the multicopy plasmid containing tRNAVal(AAC) was previously described (Alexandrov et al. 2006).
TABLE 3.
DNA oligonucleotides used for cloning genes
S. cerevisiae TRM44 was cloned into the E. coli expression vector AVA421 (Alexandrov et al. 2004) by PCR amplification of TRM44 using primers YPL030W_Fwd (5′-GGGTCCTGGTTCGATGACTGGCGACGGTAGTGC-3′) and YPL030W_Rev (5′-CTTGTTCGTGCTGTTTATTAATGGTTTCTTGGGTTTCTTTTC-3′) followed by ligation-independent cloning to generate plasmid pLKD306 for expression of His6-Trm44 protein in E. coli.
Growth and preparation of extracts and purified proteins
The purification of GST-ORF fusion protein pools and subpools was previously described (Martzen et al. 1999; Phizicky et al. 2002).
Wild-type and trm44-Δ yeast strains were grown in YP media containing glucose to midlog phase, and 300 OD of cells were resuspended in 0.8 mL extraction buffer containing 50 mM Tris-HCl (pH 7.5), 1 mM EDTA, 4 mM MgCl2, 5 mM DTT, 10% glycerol, 1 M NaCl, and protease inhibitors (1 μg/mL pepstatin, 2 μg/mL leupeptin, and 1 mM pefabloc), lysed with glass beads (10 times for 20 sec in a bead beater), and centrifuged 10 min to remove debris and generate crude extract (20 mg/mL for wild type and 22 mg/mL for trm44-Δ mutants), as described (Jackman et al. 2003).
His6-Trm44 was purified from E. coli essentially as described (Steiger et al. 2001). BL21DE3pLysS cells transformed with pLKD306 were grown in 500 mL LB+Amp at 30°C to OD600 = 0.5 and induced with IPTG for 3 h, and harvested cells were resuspended in 10 mL buffer containing 20 mM HEPES (pH 7.5), 5% glycerol, 1 M NaCl, 2 mM β−mercaptoethanol, and protease inhibitors, and lysed by sonication, followed by centrifugation to produce crude extract (4.3 mg/mL). Then His6 tagged protein was purified by immobilized metal-ion chromatography (IMAC) using TALON resin (Clontech), as described (Steiger et al. 2001), followed by dialysis of eluted protein into buffer containing 20 mM HEPES (pH 7.5), 55 mM NaCl, 4 mM MgCl2, 1 mM DTT, and 50% glycerol. The purified protein (2.6 mg/mL) was estimated to be∼40% pure on SDS-PAGE by Coomassie staining (not shown).
Preparation of labeled tRNASer substrate and assay of U44 methyltransferase activity
To prepare tRNASer(UGA) substrate singly labeled at U44 (U44*Ser), tRNASer(UGA) was cloned in two fragments into a pUC13-derived vector, as a 5′ half molecule comprising bases 1–44 (plasmid LKD260) and a 3′ half molecule comprising bases 45–85 to generate (LKD261). The 5′ half molecule was assembled from synthetic oligonucleotides to contain, in order, 5′ PstI site, T7 promoter, tRNASer(UGA) 1–44, HpaI site, and BamH1, and the resulting plasmid was cut with HpaI for transcription. The 3′ half molecule was constructed in the same way except that the HpaI site was replaced by BstNI for BstNI-runoff transcripts. Transcripts made using T7 RNA polymerase were gel purified, and then the 3′ half molecule was treated with phosphatase and T4 PNK to label its 5′-phosphate, followed by ligation to the 5′ half molecule using T4 DNA ligase (USB) and a bridging DNA oligonucleotide (5′-GGCAGAGCCCAACAGATTTCAAGTC-3′), as previously described (Yu 1999; Jackman et al. 2003), The final ligated tRNASer(UGA) product was analyzed with RNase A and P1 nuclease to show that the junction sequence was UpG, as predicted.
The methyltransferase assay was performed with U44*Ser in 10-μL reaction mixtures containing 50 mM HEPES (pH 7.5), 50 mM ammonium acetate, 5 mM MgCl2, 1 mM DTT, 0.5 mM SAM, ∼1 nM U44*Ser, and a protein source (either crude extract or purified protein). Reactions were initiated by addition of protein and incubated for 2–12 h at 30°C. Then tRNA was supplemented with carrier RNA (20 μg), phenol extracted and ethanol precipitated, resuspended in 4 μL buffer containing 20 mM sodium acetate (pH 5.2), 1 mM EDTA, RNaseT1 (0.5 units), and RNaseA (1 μg), incubated for 30 min at 50°C, and treated with alkaline phosphatase (1 unit) for 30 min at 37°C in 1× buffer, and then the final product was applied to PEI cellulose TLC plates (EM Science) and resolved in solvent containing 0.5 M LiCl.
Preparation of low molecular weight RNA from yeast
Small molecular weight RNA was extracted from 300 OD of MATa haploid wild-type and trm44-Δ mutant trains, using hot phenol, followed by two ethanol preicpitations, and resuspension in 1 mL H2O, yielding 3–5 mg RNA. The concentration of RNA was calculated by assuming A260 of 40 μg/mL RNA = 1.
Purification of tRNASer(UGA) and tRNASer(IGA) from yeast
tRNASer(UGA) and tRNASer(IGA) were purified from small molecular weight RNA extracted from wild-type and trm44-Δ strains using 5′ biotinylated oligonucleotides (Integrated DNA Technologies) complementary to the residues between e23–73 for tRNASer(UGA) (5′Bio tRNASer1, 5′Bio-CGACACCAGCAGGATTTGAACCAGCGCGGG) and 1–32 for tRNASer(IGA) (5′Bio Ser3 5′ bio/ATCTTTCGCCTTAACCACTCGGCCAAGTTGCC) and streptavidin (SA) magnetic beads (Roche), as previously described (Jackman et al. 2003), using 2 mg of beads bound with oligomer and 1 or 3 mg of low-molecular-weight RNA to purify tRNASer(IGA) (10 μg) and tRNASer(UGA) (4 μg).
HPLC analysis of nucleosides
Purified tRNAs (1 μg) were analyzed for modifications by treatment of tRNA with Nuclease P1 (Calbiochem) followed by calf intestinal alkaline phosphatase, followed by resolution of nucleosides on a reverse phase C18 column (supelcosil LC-18-T, 25 cm × 4.6 μm; Supelco, Inc.) using an HPLC (Waters Alliance Model 2690, equipped with Waters 996 phosphodiode array detector) as described previously (Gehrke and Kuo 1989), and individual spectra of nucleosides were used to confirm the assignments and quantify nucleosides as described (Jackman et al. 2003).
Analysis of tRNA levels
Wild-type, trm44-Δ, tan1-Δ, and trm44-Δ tan1-Δ homozygous diploid yeast strains were grown in YP+ glycerol media at 25°C and switched to 36.5°C, cells were harvested at specific time points, and 2 μg of small molecular weight RNA from each sample was used for Northern analysis, as previously described (Alexandrov et al. 2006), with 5′ 32P-labeled oligonucleotide probes listed in Table 4.
TABLE 4.
Oligonucleotides used to analyze RNA expression in Northern blots
ACKNOWLEDGMENTS
We are grateful to J. Jackman, I. Chernyakov, M. Baker, and Y. Kon for valuable discussions and advice on this research. This research was supported by NIH grant GM52347 to E.M.P.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.811008.
REFERENCES
- Agris, P.F., Koh, H., Soll, D. The effect of growth temperatures on the in vivo ribose methylation of Bacillus stearothermophilus transfer RNA. Arch. Biochem. Biophys. 1973;154:277–282. doi: 10.1016/0003-9861(73)90058-1. [DOI] [PubMed] [Google Scholar]
- Alexandrov, A., Vignali, M., LaCount, D.J., Quartley, E., de Vries, C., De Rosa, D., Babulski, J., Mitchell, S.F., Schoenfeld, L.W., Fields, S., et al. A facile method for high-throughput coexpression of protein pairs. Mol. Cell. Proteomics. 2004;3:934–938. doi: 10.1074/mcp.T400008-MCP200. [DOI] [PubMed] [Google Scholar]
- Alexandrov, A., Chernyakov, I., Gu, W., Hiley, S.L., Hughes, T.R., Grayhack, E.J., Phizicky, E.M. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell. 2006;21:87–96. doi: 10.1016/j.molcel.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J., Phan, L., Cuesta, R., Carlson, B.A., Pak, M., Asano, K., Bjork, G.R., Tamame, M., Hinnebusch, A.G. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes & Dev. 1998;12:3650–3662. doi: 10.1101/gad.12.23.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J., Phan, L., Hinnebusch, A.G. The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae . Proc. Natl. Acad. Sci. 2000;97:5173–5178. doi: 10.1073/pnas.090102597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ares M., Jr, Grate, L., Pauling, M.H. A handful of intron-containing genes produces the lion's share of yeast mRNA. RNA. 1999;5:1138–1139. doi: 10.1017/s1355838299991379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, H.F., Motorin, Y., Planta, R.J., Grosjean, H. The yeast gene YNL292w encodes a pseudouridine synthase (Pus4) catalyzing the formation of psi55 in both mitochondrial and cytoplasmic tRNAs. Nucleic Acids Res. 1997;25:4493–4499. doi: 10.1093/nar/25.22.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork, G.R., Jacobsson, K., Nilsson, K., Johansson, M.J., Bystrom, A.S., Persson, O.P. A primordial tRNA modification required for the evolution of life? EMBO J. 2001;20:231–239. doi: 10.1093/emboj/20.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaille, J., Chetouani, F., Bachellerie, J.P. The yeast Saccharomyces cerevisiae YDL112w ORF encodes the putative 2′-O-ribose methyltransferase catalyzing the formation of Gm18 in tRNAs. RNA. 1999;5:66–81. doi: 10.1017/s1355838299981475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakshusmathi, G., Kim, S.D., Rubinson, D.A., Wolin, S.L. A La protein requirement for efficient pre-tRNA folding. EMBO J. 2003;22:6562–6572. doi: 10.1093/emboj/cdg625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouet d'Orval, B., Bortolin, M.L., Gaspin, C., Bachellerie, J.P. Box C/D RNA guides for the ribose methylation of archaeal tRNAs. The tRNATrp intron guides the formation of two ribose-methylated nucleosides in the mature tRNATrp. Nucleic Acids Res. 2001;29:4518–4529. doi: 10.1093/nar/29.22.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copela, L.A., Chakshusmathi, G., Sherrer, R.L., Wolin, S.L. The La protein functions redundantly with tRNA modification enzymes to ensure tRNA structural stability. RNA. 2006;12:644–654. doi: 10.1261/rna.2307206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur, W.A., Fournier, M.J. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 2003;278:695–698. doi: 10.1074/jbc.R200023200. [DOI] [PubMed] [Google Scholar]
- Dihanich, M.E., Najarian, D., Clark, R., Gillman, E.C., Martin, N.C., Hopper, A.K. Isolation and characterization of MOD5, a gene required for isopentenylation of cytoplasmic and mitochondrial tRNAs of Saccharomyces cerevisiae . Mol. Cell. Biol. 1987;7:177–184. doi: 10.1128/mcb.7.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake, A.F., Mason, S.F., Trim, A.R. Optical studies of the base-stacking properties of 2′-O-methylated dinucleoside monophosphates. J. Mol. Biol. 1974;86:727–739. doi: 10.1016/0022-2836(74)90349-0. [DOI] [PubMed] [Google Scholar]
- Ellis, S.R., Morales, M.J., Li, J.M., Hopper, A.K., Martin, N.C. Isolation and characterization of the TRM1 locus, a gene essential for the N2,N2-dimethylguanosine modification of both mitochondrial and cytoplasmic tRNA in Saccharomyces cerevisiae . J. Biol. Chem. 1986;261:9703–9709. [PubMed] [Google Scholar]
- Etcheverry, T., Colby, D., Guthrie, C. A precursor to a minor species of yeast tRNASer contains an intervening sequence. Cell. 1979;18:11–26. doi: 10.1016/0092-8674(79)90349-0. [DOI] [PubMed] [Google Scholar]
- Etcheverry, T., Salvato, M., Guthrie, C. Recessive lethality of yeast strains carrying the SUP61 suppressor results from loss of a transfer RNA with a unique decoding function. J. Mol. Biol. 1982;158:599–618. doi: 10.1016/0022-2836(82)90251-0. [DOI] [PubMed] [Google Scholar]
- Galardi, S., Fatica, A., Bachi, A., Scaloni, A., Presutti, C., Bozzoni, I. Purified box C/D snoRNPs are able to reproduce site-specific 2′-O-methylation of target RNA in vitro. Mol. Cell. Biol. 2002;22:6663–6668. doi: 10.1128/MCB.22.19.6663-6668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke, C.W., Kuo, K.C. Ribonucleoside analysis by reversed-phase high-performance liquid chromatography. J. Chromatogr. 1989;471:3–36. doi: 10.1016/s0021-9673(00)94152-9. [DOI] [PubMed] [Google Scholar]
- Gerber, A.P., Keller, W. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science. 1999;286:1146–1149. doi: 10.1126/science.286.5442.1146. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D., Sugino, A. New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base-pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Grosshans, H., Lecointe, F., Grosjean, H., Hurt, E., Simos, G. Pus1p-dependent tRNA pseudouridinylation becomes essential when tRNA biogenesis is compromised in yeast. J. Biol. Chem. 2001;276:46333–46339. doi: 10.1074/jbc.M107141200. [DOI] [PubMed] [Google Scholar]
- Hopper, A.K., Phizicky, E.M. tRNA transfers to the limelight. Genes & Dev. 2003;17:162–180. doi: 10.1101/gad.1049103. [DOI] [PubMed] [Google Scholar]
- Hopper, A.K., Furukawa, A.H., Pham, H.D., Martin, N.C. Defects in modification of cytoplasmic and mitochondrial transfer RNAs are caused by single nuclear mutations. Cell. 1982;28:543–550. doi: 10.1016/0092-8674(82)90209-4. [DOI] [PubMed] [Google Scholar]
- Huang, Y., Bayfield, M.A., Intine, R.V., Maraia, R.J. Separate RNA-binding surfaces on the multifunctional La protein mediate distinguishable activities in tRNA maturation. Nat. Struct. Mol. Biol. 2006;13:611–618. doi: 10.1038/nsmb1110. [DOI] [PubMed] [Google Scholar]
- Jackman, J.E., Montange, R.K., Malik, H.S., Phizicky, E.M. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA. 2003;9:574–585. doi: 10.1261/rna.5070303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson, M.J., Bystrom, A.S. Dual function of the tRNA(m5U54)methyltransferase in tRNA maturation. RNA. 2002;8:324–335. doi: 10.1017/s1355838202027851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson, M.J., Bystrom, A.S. The Saccharomyces cerevisiae TAN1 gene is required for N4-acetylcytidine formation in tRNA. RNA. 2004;10:712–719. doi: 10.1261/rna.5198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba, S., Krueger, A., Trice, T., Krecic, A.M., Hinnebusch, A.G., Anderson, J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae . Genes & Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba, S., Wang, X., Anderson, J.T. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA. 2006;12:508–521. doi: 10.1261/rna.2305406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan, R.M., Clarke, S. Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes. Arch. Biochem. Biophys. 1994;310:417–427. doi: 10.1006/abbi.1994.1187. [DOI] [PubMed] [Google Scholar]
- Kalhor, H.R., Clarke, S. Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol. Cell. Biol. 2003;23:9283–9292. doi: 10.1128/MCB.23.24.9283-9292.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai, G., Yamamoto, Y., Kamimura, T., Masegi, T., Sekine, M., Hata, T., Iimori, T., Watanabe, T., Miyazawa, T., Yokoyama, S. Conformational rigidity of specific pyrimidine residues in tRNA arises from posttranscriptional modifications that enhance steric interaction between the base and the 2′-hydroxyl group. Biochemistry. 1992;31:1040–1046. doi: 10.1021/bi00119a012. [DOI] [PubMed] [Google Scholar]
- LaCava, J., Houseley, J., Saveanu, C., Petfalski, E., Thompson, E., Jacquier, A., Tollervey, D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Laten, H., Gorman, J., Bock, R.M. Isopentenyladenosine deficient tRNA from an antisuppressor mutant of Saccharomyces cerevisiae . Nucleic Acids Res. 1978;5:4329–4342. doi: 10.1093/nar/5.11.4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecointe, F., Simos, G., Sauer, A., Hurt, E.C., Motorin, Y., Grosjean, H. Characterization of yeast protein Deg1 as pseudouridine synthase (Pus3) catalyzing the formation of psi 38 and psi 39 in tRNA anticodon loop. J. Biol. Chem. 1998;273:1316–1323. doi: 10.1074/jbc.273.3.1316. [DOI] [PubMed] [Google Scholar]
- Lin-Marq, N., Clarkson, S.G. A yeast RNA binding protein that resembles the human autoantigen La. J. Mol. Biol. 1995;245:81–85. doi: 10.1006/jmbi.1994.0008. [DOI] [PubMed] [Google Scholar]
- Martzen, M.R., McCraith, S.M., Spinelli, S.L., Torres, F.M., Fields, S., Grayhack, E.J., Phizicky, E.M. A biochemical genomics approach for identifying genes by the activity of their products. Science. 1999;286:1153–1155. doi: 10.1126/science.286.5442.1153. [DOI] [PubMed] [Google Scholar]
- Mathews, D.H., Sabina, J., Zuker, M., Turner, D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Nordlund, M.E., Johansson, J.O., von Pawel-Rammingen, U., Bystrom, A.S. Identification of the TRM2 gene encoding the tRNA(m5U54)methyltransferase of Saccharomyces cerevisiae . RNA. 2000;6:844–860. doi: 10.1017/s1355838200992422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, M.V., Page, G.S., Sentenac, A., Piper, P.W., Worthington, M., Weiss, R.B., Hall, B.D. Only one of two closely related yeast suppressor tRNA genes contains an intervening sequence. Nature. 1981;291:464–469. doi: 10.1038/291464a0. [DOI] [PubMed] [Google Scholar]
- Phizicky, E.M., Martzen, M.R., McCraith, S.M., Spinelli, S.L., Xing, F., Shull, N.P., Van Slyke, C., Montagne, R.K., Torres, F.M., Fields, S., et al. Biochemical genomics approach to map activities to genes. Methods Enzymol. 2002;350:546–559. doi: 10.1016/s0076-6879(02)50984-8. [DOI] [PubMed] [Google Scholar]
- Piekna-Przybylska, D., Decatur, W.A., Fournier, M.J. New bioinformatic tools for analysis of nucleotide modifications in eukaryotic rRNA. RNA. 2007;13:305–312. doi: 10.1261/rna.373107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintard, L., Lecointe, F., Bujnicki, J.M., Bonnerot, C., Grosjean, H., Lapeyre, B. Trm7p catalyses the formation of two 2′-O-methylriboses in yeast tRNA anticodon loop. EMBO J. 2002;21:1811–1820. doi: 10.1093/emboj/21.7.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renalier, M.H., Joseph, N., Gaspin, C., Thebault, P., Mougin, A. The Cm56 tRNA modification in archaea is catalyzed either by a specific 2′-O-methylase, or a C/D sRNP. RNA. 2005;11:1051–1063. doi: 10.1261/rna.2110805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson, J.R., Uhlenbeck, O.C. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl. Acad. Sci. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnare, M., Heinonen, T., Young, P., Gray, M. Phenylalanine and tyrosine transfer RNAs encoded by Tetrahymena pyriformis mitochondrial DNA: Primary sequence, post-transcriptional modifications, and gene localization. Curr. Genet. 1985;9:389–393. doi: 10.1007/BF00421610. [DOI] [PubMed] [Google Scholar]
- Sprinzl, M., Horn, C., Brown, M., Ioudovitch, A., Steinberg, S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger, M.A., Kierzek, R., Turner, D.H., Phizicky, E.M. Substrate recognition by a yeast 2′-phosphotransferase involved in tRNA splicing and by its Escherichia coli homolog. Biochemistry. 2001;40:14098–14105. doi: 10.1021/bi011388t. [DOI] [PubMed] [Google Scholar]
- Tong, A.H., Evangelista, M., Parsons, A.B., Xu, H., Bader, G.D., Page, N., Robinson, M., Raghibizadeh, S., Hogue, C.W., Bussey, H., et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Urbonavicius, J., Qian, Q., Durand, J.M., Hagervall, T.G., Bjork, G.R. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 2001;20:4863–4873. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbonavicius, J., Durand, J.M., Bjork, G.R. Three modifications in the D and T arms of tRNA influence translation in Escherichia coli and expression of virulence genes in Shigella flexneri . J. Bacteriol. 2002;184:5348–5357. doi: 10.1128/JB.184.19.5348-5357.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova, S., Wolf, J., Martin, G., Blank, D., Dettwiler, S., Friedlein, A., Langen, H., Keith, G., Keller, W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron, C., Lacroute, F. Effect of growth rate on the amounts of ribosomal and transfer ribonucleic acids in yeast. J. Bacteriol. 1975;122:855–865. doi: 10.1128/jb.122.3.855-865.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, M.L., Crary, S.M., Jackman, J.E., Grayhack, E.J., Phizicky, E.M. The 2′-O-methyltransferase responsible for modification of yeast tRNA at position 4. RNA. 2007;13:404–413. doi: 10.1261/rna.399607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, P., Brockenbrough, J.S., Paddy, M.R., Aris, J.P. NCL1, a novel gene for a nonessential nuclear protein in Saccharomyces cerevisiae . Gene. 1998;220:109–117. doi: 10.1016/s0378-1119(98)00330-8. [DOI] [PubMed] [Google Scholar]
- Xing, F., Martzen, M.R., Phizicky, E.M. A conserved family of Saccharomyces cerevisiae synthases effects dihydrouridine modification of tRNA. RNA. 2002;8:370–381. doi: 10.1017/s1355838202029825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, F., Hiley, S.L., Hughes, T.R., Phizicky, E.M. The specificities of four yeast dihydrouridine synthases for cytoplasmic tRNAs. J. Biol. Chem. 2004;279:17850–17860. doi: 10.1074/jbc.M401221200. [DOI] [PubMed] [Google Scholar]
- Yokogawa, T., Suzuki, T., Ueda, T., Mori, M., Ohama, T., Kuchino, Y., Yoshinari, S., Motoki, I., Nishikawa, K., Osawa, S., et al. Serine tRNA complementary to the nonuniversal serine codon CUG in Candida cylindracea: Evolutionary implications. Proc. Natl. Acad. Sci. 1992;89:7408–7411. doi: 10.1073/pnas.89.16.7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama, S., Watanabe, T., Murao, K., Ishikura, H., Yamaizumi, Z., Nishimura, S., Miyazawa, T. Molecular mechanism of codon recognition by tRNA species with modified uridine in the first position of the anticodon. Proc. Natl. Acad. Sci. 1985;82:4905–4909. doi: 10.1073/pnas.82.15.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, C.J., Wolin, S.L. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell. 1997;89:393–402. doi: 10.1016/s0092-8674(00)80220-2. [DOI] [PubMed] [Google Scholar]
- Yu, Y.T. Construction of 4-thiouridine site-specifically substituted RNAs for cross-linking studies. Methods. 1999;18:13–21. doi: 10.1006/meth.1999.0752. [DOI] [PubMed] [Google Scholar]
- Yu, Y.-T., Terns, R.M., Terns, M.P. Mechanisms and functions of RNA-guided RNA modifications. In: Grosjean H., editor. Fine-tuning of RNA functions by modification and editing. Vol. 12. Springer; New York: 2005. pp. 223–262. [Google Scholar]
- Zachau, H.G., Dutting, D., Feldmann, H. The structures of two serine transfer ribonucleic acids. Hoppe Seylers Z. Physiol. Chem. 1966;347:212–235. doi: 10.1515/bchm2.1966.347.1.212. [DOI] [PubMed] [Google Scholar]
- Zmudzka, B., Shugar, D. Role of the 2′-hydroxyl in polynucleotide conformation. Poly 2′-O-methyluridylic acid. FEBS Lett. 1970;8:52–54. doi: 10.1016/0014-5793(70)80223-x. [DOI] [PubMed] [Google Scholar]