Abstract
Annotation of the complete genome of the extreme halophilic archaeon Haloarcula marismortui does not include a tRNA for translation of AUA, the rare codon for isoleucine. This is a situation typical for most archaeal genomes sequenced to date. Based on computational analysis, it has been proposed recently that a single intron-containing tRNA gene produces two very similar but functionally different tRNAs by means of alternative splicing; a UGG-decoding tRNATrp CCA and an AUA-decoding tRNAIle UAU. Through analysis of tRNAs from H. marismortui, we have confirmed the presence of tRNATrp CCA, but found no evidence for the presence of tRNAIle UAU. Instead, we have shown that a tRNA, currently annotated as elongator methionine tRNA and containing CAU as the anticodon, is aminoacylated with isoleucine in vivo and that this tRNA represents the missing isoleucine tRNA. Interestingly, this tRNA carries a base modification of C34 in the anticodon different from the well-known lysidine found in eubacteria, which switches the amino acid identity of the tRNA from methionine to isoleucine and its decoding specificity from AUG to AUA. The methods described in this work for the identification of individual tRNAs present in H. marismortui provide the tools necessary for experimentally confirming the presence of any tRNA in a cell and, thereby, to test computational predictions of tRNA genes.
Keywords: archaea, Haloarcula marismortui, minor isoleucine tRNA, base modification, lysidine
INTRODUCTION
Many archaeal tRNA genes contain introns that are located primarily between nucleotides 37 and 38 in the anticodon loop. In the pre-tRNA, the exon–intron boundaries are defined by a characteristic bulge–helix–bulge motif, or variants thereof, which are recognized by the archaeal splicing machinery (Tocchini-Valentini et al. 2005; Xue et al. 2006). Based on a study encompassing 800 archaeal tRNA genes (Marck and Grosjean 2003), ∼75% of all tRNA introns—termed canonical—are found at position 37/38. Twenty-five percent of tRNA introns are noncanonical and are located at various positions in the tRNA, including the anticodon loop at position 32/33, anticodon stem, variable loop, D- and T-arms, and acceptor stem. The discovery of split tRNA genes in Nanoarchaeum equitans, which generates functional tRNAs from two independently encoded tRNA halves, has added yet another dimension to the diversity of tRNA splicing pathways in archaea (Randau et al. 2005a,b).
tRNAs and the presence of canonical tRNA introns are predicted with high accuracy with algorithms such as tRNAscan-SE (Lowe and Eddy 1997) and ARAGORN (Laslett and Canback 2004). More recently, modified algorithms, which allow the prediction of tRNAs containing noncanonical introns, multiple introns, and split tRNA genes, for annotation of tRNAs at the genome level have been developed (Randau et al. 2005a; Ghosh et al. 2006; Sugahara et al. 2006, 2007). For example, the in silico search for missing tRNA genes in the genome of the extreme halophilic archaeon Haloarcula marismortui led to the identification of a putative tRNAIle UAU (Ghosh et al. 2006), enclosed completely within the gene coding for another tRNA, tRNATrp CCA. It was, therefore, proposed that the single intron-containing gene produces two different tRNAs by means of alternative splicing. More recently, additional examples of genes with such “overlapping” tRNA sequences were identified in silico in the methanogenic archaeon Methanopyrus kandleri (J. Chakrabarti, pers. comm.) and in a study of archaeal tRNA genes using the updated SPLITSX algorithm (Sugahara et al. 2007). SPLITSX has identified several tRNA genes with multiple noncanonial introns, including three different proline tRNAs, each with three introns.
In the work described here, we investigated whether the minor isoleucine tRNA responsible for translation of rare AUA codons in H. marismortui (3.6 out of 1000 codons) and tRNATrp CCA are generated by means of alternative splicing from a composite isoleucine–tryptophan tRNA gene transcript as proposed. Our results confirm the presence of the UGG-decoding tRNATrp CCA derived from this locus, but provide no evidence for the presence of tRNAIle UAU. In contrast, we show that the minor AUA-decoding isoleucine tRNA in H. marismortui and other archaeal species is most likely derived from a CAU anticodon-containing tRNA, currently annotated as methionine tRNA, in which C34 in the anticodon is post-transcriptionally modified as in the minor isoleucine tRNAs of eubacterial and organellar systems (Grosjean and Björk 2004, and references therein). In addition, we show that the post-transcriptional modification of the C at position 34 in the anticodon of this tRNA, responsible for the switch in amino acid and decoding specificity, is different from those present at position 34 of isoleucine tRNA species in eubacteria and in eukaryotes.
RESULTS
Analysis of H. marismortui tRNA for the presence of tRNATrp CCA and tRNAIle UAU
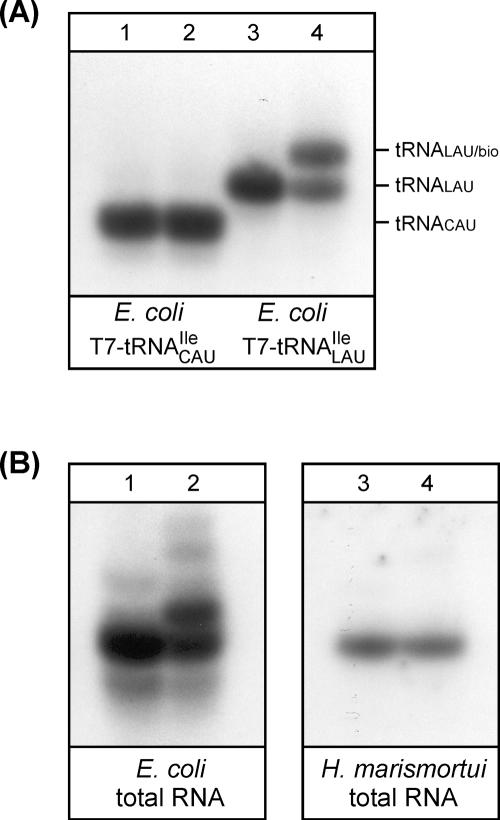
To investigate the question of whether alternative splicing in archaea produces both the tRNATrp CCA and tRNAIle UAU from a common transcript (Supplemental Fig. 1), total RNA from H. marismortui was analyzed for the presence of these tRNAs. RNA was fractionated by polyacrylamide gel electrophoresis (PAGE) and subjected to Northern hybridization using 32P-labeled DNA oligonucleotide probes against anticodon stem–loop regions of the putative tRNATrp CCA and tRNAIle UAU. We could confirm the presence of tRNATrp CCA, but could find no evidence for the presence of tRNAIle UAU (Supplemental Fig. 2A). tRNATrp CCA is annotated as tRNA_5 in the H. marismortui genome (The Genomic tRNA Database at http://lowelab.ucsc.edu/GtRNAdb) and can be identified by its anticodon sequence and recognition elements that are similar to those required for aminoacylation of eukaryotic tryptophan tRNAs by their cognate tryptophanyl-tRNA synthetases (TrpRS) (Xue et al. 1993; Guo et al. 2002). Supplemental Figure 2B shows that the deacylated tRNATrp CCA can indeed be re-aminoacylated with tryptophan using purified human TrpRS.
The H. marismortui genome includes another intron-containing tRNATrp CCA gene that is also annotated as tRNATrp CCA (tRNA_7 in The Genomic tRNA Database at http://lowelab.ucsc.edu/GtRNAdb; Supplemental Fig. 3A). Using Northern blot analysis with a probe specific for tRNA_7, we have detected such an RNA. However, tRNA_7 is not a tryptophan tRNA and may not even be a tRNA since it is not aminoacylated in vivo and cannot be aminoacylated with tryptophan using either purified human TrpRS (Supplemental Fig. 3B), Escherichia coli TrpRS, or TrpRS present in archaeal extracts (data not shown). Our results agree with the predictions of Sugahara et al. (2006, 2007), whose SPLITS and SPLITSX algorithms for identification of archaeal tRNA genes do not identify tRNA_7 as a tRNA gene. Nevertheless, the presence of such a tRNA-like RNA molecule in H. marismortui is interesting and raises the question of whether this RNA plays a role other than that of a typical tRNA.
Identification of a putative AUA codon-specific isoleucine tRNA derived from a gene encoding a CAU anticodon-containing tRNA
In the absence of a tRNAIle necessary to decode the rare AUA codons in H. marismortui, we explored the possibility of a tRNAIle derived from a tRNA with a CAU anticodon in which the C is subsequently modified, as it is in many eubacterial and organellar systems (for review, see Grosjean and Björk 2004, and references therein). In these systems, the C in the CAU anticodon is modified to lysidine (tRNAIle LAU; L, lysidine). Therefore, all tRNA genes that have CAU as the anticodon and that are currently annotated in the H. marismortui genome as methionine tRNAs were considered as potentially encoding the AUA-reading tRNAIle. Based on high sequence similarity with initiator tRNAs from other kingdoms and the presence of specific sequence features including three consecutive G-C pairs in the anticodon stem (Seong and RajBhandary 1987; RajBhandary 1994), one of the tRNAs was identified as the initiator methionine tRNA, tRNAi Met. The remaining two tRNAs (annotated as tRNA_12 and tRNA_34 in The Genomic tRNA Database at http://lowelab.ucsc.edu/GtRNAdb), which are different from each other, show the characteristics of typical elongator tRNAs (Fig. 1). Both tRNAs have the potential for aminoacylation by MetRS, and most notably, both tRNAs also have most of the identity elements necessary for recognition by a eubacterial-type IleRS (Nureki et al. 1994), which, besides the anticodon, include the discriminator base A73, nucleotides A37 and A38, and base-pairs C29–G41, U12–A23, and C4–G69. These identity elements are also present in the major tRNAIle GAU of H. marismortui, responsible for decoding AUC and AUU codons.
FIGURE 1.
Cloverleaf structures of CAU anticodon-containing elongator tRNAs in H. marismortui. In this study, tRNA_12 (left) has been confirmed as the elongator methionine tRNAMet CAU, while tRNA_34 (right) has been identified as the minor isoleucine tRNAIle XAU (X, unknown modified base) responsible for translation of AUA codons.
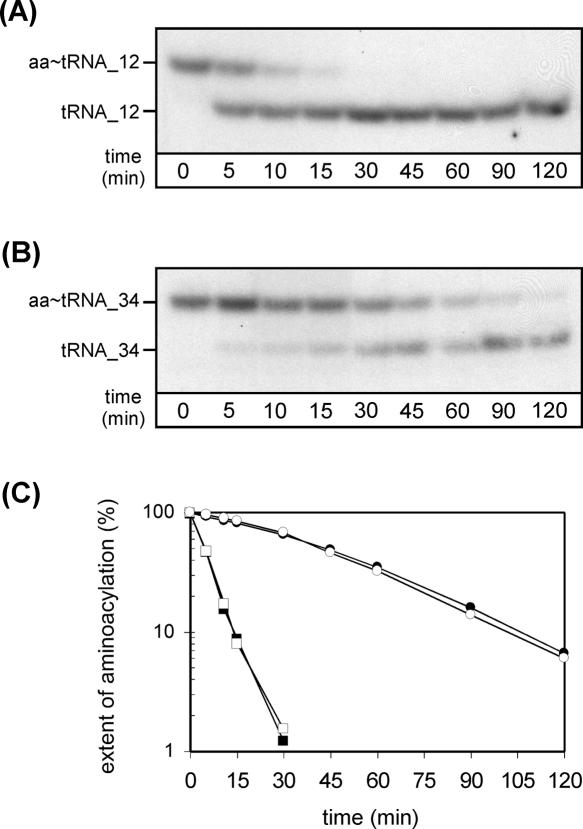
To identify which of the two remaining CAU anticodon-containing tRNAs of H. marismortui is possibly aminoacylated with isoleucine in vivo, we measured the kinetics of chemical deacylation of aminoacyl-tRNAs corresponding to tRNA_12 and tRNA_34 (Fig. 2). It is known that the stability of the ester link between tRNA and amino acid is determined by the nature of the amino acid attached to the tRNA (Matthaei et al. 1966; Drabkin and RajBhandary 1998; Ramesh and RajBhandary 2001), with isoleucine and valine being the most stable, and methionine being much less stable. Aminoacyl-tRNAs present in total RNA were isolated from H. marismortui under acidic conditions to retain the aminoacyl–ester linkage and were subjected to base-catalyzed deacylation (for details, see Materials and Methods). Samples were subsequently analyzed by acid urea PAGE, which separates tRNA from aminoacyl-tRNA, followed by Northern blotting using probes specific for tRNA_12 and tRNA_34, respectively. It can be seen that aminoacyl-tRNA_12 is almost completely deacylated within 15 min, whereas deacylation of aminoacyl-tRNA_34 is incomplete even after 120 min (Fig. 2A,B). The half-life of deacylation of aminoacyl-tRNA_12 is ∼5 min, which is the same as that of Met-tRNAi Met, and that of aminoacyl-tRNA_34 is ∼35 min, which is the same as that of Ile-tRNAIle GAU (Fig. 2C). These data show that the amino acids attached to tRNA_12 and tRNA_34 are different and that they are most likely methionine and isoleucine, respectively.
FIGURE 2.
Deacylation rates of tRNA_12 (tRNAMet CAU) and tRNA_34 (tRNAIle XAU) from H. marismortui. Total RNA was isolated under acidic conditions and subjected to deacylation by base treatment as described in Materials and Methods followed by acid urea PAGE/Northern analysis using probes specific for tRNA_12 (A) and tRNA_34 (B). (aa-tRNA) aminoacyl-tRNA. (C) Comparison of deacylation rates obtained for tRNA_12 (closed squares), tRNA_34 (closed circles), initiator tRNAi Met (open squares), and major isoleucine tRNAIle GAU (open circles).
FIGURE 5.
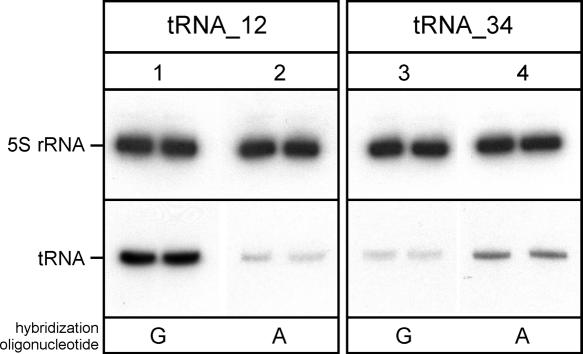
Hybridization preference of tRNA_12 (tRNAMet CAU) and tRNA_34 (tRNAIle XAU) from H. marismortui. Analysis of total RNA by denaturing PAGE and probes targeting the anticodon stem–loop of tRNA_12 and tRNA_34 (positions 30–49). (Lane 1) tRNA_12 probe with a perfectly matched G opposite tRNA position C34; (lane 2) tRNA_12 probe with a mismatched A opposite tRNA position C34; (lane 3) tRNA_34 probe with G opposite tRNA position X34; (lane 4) tRNA_34 probe with A opposite tRNA position X34. Samples were analyzed in duplicates. A probe against 5S rRNA was used as loading control.
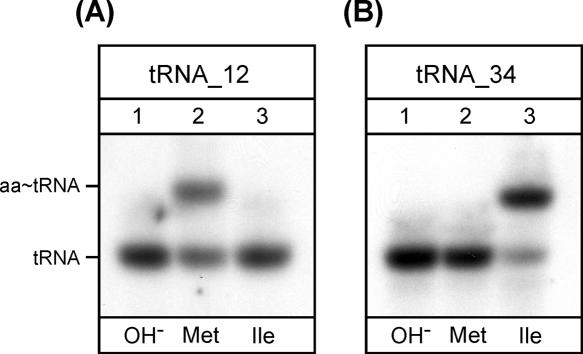
For final identification of tRNA_12 and tRNA_34 as tRNAMet and tRNAIle, respectively, the deacylated tRNAs were aminoacylated in vitro using either H. marismortui MetRS or IleRS, which have been overexpressed in E. coli and purified. Samples were then subjected to acid urea PAGE followed by Northern hybridization using probes specific for tRNA_12 and tRNA_34. The results show that tRNA_12 is aminoacylated by MetRS but not by IleRS (Fig. 3A), whereas tRNA_34 is aminoacylated by IleRS but not by MetRS (Fig. 3B). Similar results were obtained with purified E. coli MetRS and IleRS (data not shown). In contrast, a T7 transcript corresponding to tRNA_34 and lacking all base modifications could be aminoacylated efficiently with methionine but not with isoleucine (Fig. 4), indicating that a base modification is most likely responsible for the amino acid identity switch of tRNA_34 from tRNAMet to tRNAIle. These data show that tRNA_12 is the elongator methionine tRNAMet CAU and that tRNA_34 is most likely the tRNAIle responsible for translation of AUA codons. tRNA_34 will, henceforth, be called tRNAIle XAU, X representing an unknown modified base.
FIGURE 3.
Aminoacylation specificity of tRNA_12 (tRNAMet CAU) and tRNA_34 (tRNAIle XAU) from H. marismortui using purified H. marismortui MetRS and IleRS. Analysis in vitro of aminoacylation of total RNA by acid urea PAGE/Northern hybridization using probes specific for tRNA_12 (A) and tRNA_34 (B). (Lane 1) tRNA after deacylation by base treatment (OH−); (lanes 2,3) in vitro aminoacylation of deacylated total RNA with methionine using H. marismortui MetRS (Met) or isoleucine using H. marismortui IleRS (Ile), respectively. (aa-tRNA) Aminoacyl-tRNA.
FIGURE 4.
Aminoacylation specificity of unmodified T7 transcripts corresponding to H. marismortui tRNA_12 (tRNAMet CAU) and tRNA_34 (tRNAIle XAU). In vitro aminoacylation of T7 transcripts with methionine or isoleucine using an S30 cell extract prepared from H. marismortui. Incorporation of [3H]-isoleucine (black bars) or [35S]-methionine (gray bars) into 0.1 A260 of T7 transcript (tRNA_12, tRNA_34) or 1 A260 of total RNA isolated from H. marismortui. The 60 min time point is shown.
A modification of C at position 34 in tRNAIle XAU of H. marismortui is responsible for a switch in base-pairing properties
The quantitative assessment of hybridization signals provides valuable information about the stability and specificity of a hybrid between a DNA oligonucleotide and its RNA target. Total RNA from H. marismortui was subjected to Northern blot analysis using probes directed against the anticodon stem–loop region nucleotides 30–49 of tRNAMet CAU and tRNAIle XAU. These probes carried either a G or an A opposite to position 34 in the tRNA target (Fig. 5). tRNAMet CAU shows a strong signal with a DNA oligonucleotide carrying a G that matches the tRNA perfectly, allowing a standard G-C base pair between the hybridization probe and its target (Fig. 5, lane 1). The signal is significantly weaker with a probe that introduces an AxC mismatch at C34 (Fig. 5, cf. lanes 1 and 2). In contrast, tRNAIle XAU shows a stronger signal with an oligonucleotide that contains an A opposite position 34 in the tRNA (Fig. 5, cf. lanes 3 and 4). Thus, consistent with the idea that a post-transcriptional modification at position 34 of tRNAIle XAU is responsible for the switch in both the amino acid specificity from methionine to isoleucine and codon specificity from AUG (methionine) to AUA (isoleucine), a significant difference in the base-pairing preferences was observed for the two tRNAs.
Primer extension analysis confirms the presence of a post-transcriptional modification at position 34 in tRNAIle XAU of H. marismortui and other archaea
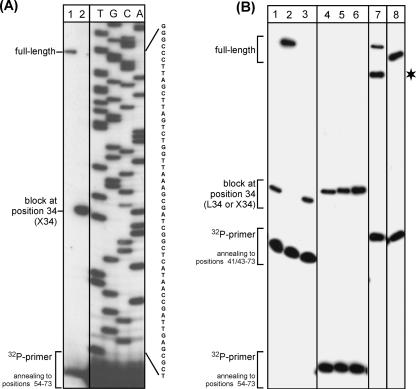
To confirm the presence of a post-transcriptional modification at position 34 of tRNAIle XAU of H. marismortui and other archaeal species, primer extension analysis was performed (Fig. 6). With total RNA isolated from H. marismortui, a clear block was observed at position 34 hindering the reverse transcriptase from further extension (Fig. 6A, lane 2). In contrast, a T7 transcript derived from the H. marismortui tRNAIle XAU sequence allows extension to the very first position of the tRNA (Fig. 6A, lane 1). Parallel experiments were performed with isoleucine tRNAs from E. coli, Haloferax volcanii, Halobacterium salinarum, and Saccharomyces cerevisiae containing specific post-transcriptional modifications. Lysidine, which is present in most eubacterial minor isoleucine tRNAs, causes a strong block at position 34 (Fig. 6B, lanes 1,3), similar to the one observed in other haloarchaeal tRNAs (Fig. 6B, lanes 4–6). In contrast, inosine and pseudouridine present in the yeast major and minor isoleucine tRNAs (tRNAIle IAU and tRNAIle ΨAΨ), respectively, do not interfere with the extension of the reverse transcriptase (Fig. 6B, lanes 7,8).
FIGURE 6.
Primer extension analysis of post-transcriptional modifications present at position 34 in various isoleucine tRNAs from pro- and eukaryotic origin. (A) Analysis of tRNA_34 (tRNAIle XAU) from H. marismortui. (Lane 1) Unmodified T7 transcript; (lane 2) total RNA from H. marismortui; (lanes TGCA) sequencing ladder. (B) Comparison of isoleucine tRNAs from E. coli, archaea and yeast. (Lanes 1–3) Minor isoleucine tRNAIle LAU from E. coli (lane 1, T7 transcript after in vitro modification with lysidine using TilS; lane 2, unmodified T7-transcript; lane 3, total RNA). (Lanes 4–6) Minor isoleucine tRNAIle XAU in total RNA from various archaea (lane 4, H. marismortui; lane 5, H. salinarum; lane 6, H. volcanii). (Lanes 7,8) Isoleucine tRNAs in total tRNA from S. cerevisiae (lane 7; minor isoleucine tRNAIle Ψ AΨ; lane 8, major isoleucine tRNAIle IAU); asterisk indicates a partial premature stop of reverse transcriptase in tRNAIle ΨAΨ.
The base modification present in the anticodon of tRNAIle XAU of H. marismortui and other archaea is not lysidine
The modified base lysidine that determines both the codon and amino acid specificities of the eubacterial tRNAIle LAU and the enzyme TilS (tRNAIle–lysidine synthetase) responsible for the modification have been identified and studied in detail (Harada and Nishimura 1974; Muramatsu et al. 1988a,b; Soma et al. 2003; Ikeuchi et al. 2005; Nakanishi et al. 2005). To investigate whether the modified nucleoside at position 34 in tRNAIle XAU of H. marismortui is also lysidine as in eubacteria, we reacted total RNA from H. marismortui with sulfo-NHS-LC-biotin (N-hydroxysuccinimide [NHS] ester of biotin). In parallel, total RNA from E. coli and a T7 transcript derived from the E. coli tRNAIle LAU sequence were used as controls. Lysidine contains a free α-NH2 group, which reacts with the NHS-activated biotin. Both in vitro lysidinylation of the control T7 transcript and subsequent biotinylation of tRNAs can be monitored by acid urea PAGE followed by Northern blot analysis using tRNA specific probes (Fig. 7). A clear mobility shift is caused by the biotinylation of lysidine; a T7 transcript lacking lysidine does not show a shift (Fig. 7A, cf. lanes 1 and 2), whereas the same T7 transcript does, following in vitro lysidinylation with TilS (Fig. 7A, cf. lanes 3 and 4). Introduction of lysidine into the T7 transcript also causes a mobility shift by itself (Fig. 7A, cf. lanes 1 and 3). This is expected since lysidine contains an α-NH2 group, which carries a positive charge under the acidic conditions of the gel electrophoresis.
FIGURE 7.
The post-transcriptional modification present at position 34 in archaeal minor isoleucine tRNAs is not lysidine. In vitro biotinylation of free α-NH2 groups present in modified nucleosides in minor isoleucine tRNAs from E. coli and H. marismortui followed by acid urea PAGE/Northern analysis. (A) T7 transcript corresponding to tRNAIle LAU from E. coli. (Lanes 1,2) Unmodified T7 transcript; (lanes 3,4) T7 transcript after in vitro modification with lysidine using TilS. (B) Analysis of minor tRNAIle in total RNA from E. coli and H. marismortui. (Lanes 1,2) tRNAIle LAU from E. coli; (lanes 3,4) tRNA_34 (tRNAIle XAU) from H. marismortui. Comparison of tRNAs before (lanes 1,3) and after (lanes 2,4) in vitro biotinylation.
Based on the sequence similarity between tRNAIle XAU from H. marismortui and H. volcanii, one of the few organisms for which a full set of tRNA sequences has been determined experimentally (Gupta 1984), we can assume that similar base modifications are present in both the H. marismortui and the H. volcanii tRNAs. None of these modifications contains a free α-NH2 group. Therefore halobacterial tRNAIle XAU should not be biotinylated unless a modification at position 34 introduces a free α-NH2 group. The results of Northern hybridization show that biotinylation of the lysidine-containing T7 transcript (Fig. 7A, cf. lanes 3 and 4) and the mature tRNAIle LAU of E. coli (Fig. 7B, cf. lanes 1 and 2) can be followed by an electrophoretic mobility shift in acid urea polyacrylamide gels. The mature E. coli tRNAIle LAU showed a major and several minor shifts upon biotinylation (Fig. 7B, lane 2). These probably correspond to reaction primarily with the free α-NH2 group present in lysidine and partial reaction with other base modifications such as 3-(3-amino-3-carboxypropyl) uridine. Interestingly, while tRNAIle LAU from E. coli exhibits a typical shift upon biotinylation, tRNAIle XAU from H. marismortui does not (Fig. 7B, cf. lanes 3 and 4). Thus, the base modification at C34 in the archaeal tRNA is not lysidine. Similar experiments using total RNA isolated from H. volcanii, H. salinarum, and Methanocaldococcus jannaschii gave the same results (data not shown). These data are also consistent with control experiments demonstrating that a total cell extract isolated from H. marismortui does not contain TilS activity and, therefore, could not introduce lysidine into T7 transcripts corresponding to the minor isoleucine tRNAs from H. marismortui or E. coli (data not shown).
DISCUSSION
In a computational search for a tRNAIle necessary for reading the AUA codon in H. marismortui, Ghosh et al. (2006) identified a putative tRNAIle UAU embedded within the gene coding for tRNATrp CCA and proposed that a pre-tRNA derived from this locus was alternatively spliced to produce both the tRNATrp CCA and tRNAIle UAU. Work described in this article has confirmed the presence of the pre-tRNA and tRNATrp CCA encoded by this tRNA gene (Supplemental Figs. 1, 2). However, we obtained no indication for the presence of tRNAIle UAU, suggesting that, at least in tRNAs isolated from H. marismortui cells grown under standard conditions, alternative splicing is not used to produce two tRNAs from the same transcript.
In the absence of an annotated tRNAIle capable of decoding the AUA codon in H. marismortui, we focused our attention on tRNA genes with the CAU anticodon. In eubacteria, a CAU anticodon leads to aminoacylation with methionine, but modification of C34 to lysidine (LAU) leads to aminoacylation of the tRNA with isoleucine. This modified tRNA now reads the AUA codon for isoleucine instead of AUG for methionine. In H. marismortui, we have also identified a tRNAIle with a CAU anticodon in which the C at position 34 is subsequently modified (tRNAIle XAU; X, unknown modified base). This tRNA is aminoacylated with isoleucine in H. marismortui in vivo (Fig. 2) and can be aminoacylated in vitro with isoleucine but not with methionine (Fig. 3). We showed that C34 in the anticodon has a base modification that results in a strong stop at this site in primer extension experiments (Fig. 6A). Furthermore, using an NHS-ester specific for primary amino groups, we showed that the modified C in H. marismortui tRNAIle XAU is different from lysidine found in eubacterial tRNAIle LAU (Fig. 7). These results are consistent with: (1) absence, thus far, of a TilS ortholog in archaeal genomes; (2) absence of TilS activity in crude extracts from H. marismortui (data not shown); and (3) results from sequence analysis of tRNAs from the archaeon H. volcanii (Gupta 1984) showing an unknown modified base at position 34 in the putative minor isoleucine tRNA.
Different organisms have developed different strategies for generating isoleucine tRNAs that read the AUA codon for isoleucine. In most eubacteria, this is achieved by modification of CAU to LAU. In eukaryotes, the tRNAIle that reads exclusively the AUA codon has a UAU anticodon which, at least in S. cerevisiae, is modified to ψAψ (tRNAIle ΨAΨ; ψ, pseudouridine) (Szweykowska-Kulinska et al. 1994). Another tRNAIle with the anticodon IAU (tRNAIle IAC; I, inosine) can also read the AUA codon in addition to the major isoleucine codons AUU and AUC (Crick 1966; Grosjean et al. 1996; Senger et al. 1997). Our finding that yeast tRNAIle ΨAΨ and tRNAIle IAU do not cause a block in primer extension with reverse transcriptase, whereas the H. marismortui tRNAIle XAU does, shows that X is not pseudouridine or inosine. Based on our results for yeast tRNAIle ΨAΨ, we can rule out the possibility of deamination of C34 in the anticodon of H. marismortui tRNAIle XAU to U34 followed by modification of U34 to ψ34. Our data suggest that the H. marismortui tRNAIle XAU has a modified base at position 34 that is different from those found in the major and minor tRNAIle of eubacteria and eukaryotes. Results from primer extension experiments with tRNAIle from H. volcanii and H. salinarum (Fig. 6B) and lack of reaction of H. volcanii, H. salinarum, and M. jannaschii tRNAs with an NHS-ester specific for primary amino groups (data not shown) suggest that these tRNAs may also contain the same modified C34. Thus it is possible that most minor isoleucine tRNAs in archaea, with the exception of N. equitans, which has a gene for tRNAIle UAU (Randau et al. 2005b), have a common base modification of C34 in the anticodon. It is clearly of much interest to identify the nature of this base modification, and attempts are currently underway toward this aim.
Analysis of almost 30 archaeal genomes available to date shows clearly that most archaea contain one or two genes coding for isoleucine tRNAs with a GAU anticodon, which is used for translating the major AUC and AUU isoleucine codons, but do not contain an isoleucine tRNA for translation of the rare AUA codon. However, these archaea contain several genes encoding CAU anticodon-containing tRNAs, one of which could play the role of the minor isoleucine tRNA as shown in this study. The implications are quite interesting from an evolutionary point of view; the overall pathway for supplying the cell with a minor isoleucine tRNA appears to be similar in eubacteria and most archaea and is based on post-transcriptional modification of a tRNA carrying a CAU anticodon to achieve amino acid and codon specificity. Nevertheless, the base modification and the enzyme(s) responsible for this switch appear to be different between eubacteria and archaea.
Identification of tRNAs present in H. marismortui, including the AUA-reading H. marismortui tRNAIle XAU, was based on analysis of total RNA and not of individual purified tRNA species. We used (1) PAGE under denaturing conditions and acid urea PAGE followed by Northern blot analysis to identify specific tRNAs and their aminoacylation status; (2) measurement of chemical deacylation rates of aminoacylated tRNA to identify the nature of amino acids attached to the tRNA; and (3) in vitro aminoacylation of tRNAs with purified aminoacyl-tRNA synthetases followed by identification of the aminoacylated tRNA species by acid urea PAGE and Northern blot hybridization. To identify individual tRNAs including tRNATrp CCA and tRNAIle XAU, we also attempted the use of reverse transcription followed by amplification of DNA by PCR, but with no success, due to stops in cDNA synthesis caused by base modifications present in these mature tRNAs.
Finally, as noted above, identification of tRNA genes in archaea is made difficult by the fact that introns in archaeal pre-tRNAs can be located at one of many positions and there can be more than one intron in some pre-tRNAs, particularly in crenarchaeota (Marck and Grosjean 2002, 2003; Sugahara et al. 2007). Consequently, many tRNA genes are missing or misidentified in automated annotations of archaeal genomes. Several algorithms have been developed recently to identify complete sets of tRNA genes necessary to read all 61 codons of the genetic code (Randau et al. 2005a,b; Ghosh et al. 2006; Sugahara et al. 2006, 2007). In contrast to earlier algorithms, such as tRNAscan-SE (Lowe and Eddy 1997), which assume the presence of introns exclusively at position 37/38, the more recently developed algorithms SPLITS and SPLITSX include a search for bulge–helix–bulge splicing motifs at the genome level to identify the location of possible introns and allow the presence of one or more introns throughout the tRNA gene (Sugahara et al. 2006, 2007). After removal of predicted introns, the resulting sequences are reanalyzed by the tRNAscan-SE algorithm. With the increased reliance on computational tools to identify tRNAs present in archaeal organisms, it is essential to verify the presence of such tRNAs in cells experimentally. For example, two different algorithms led to prediction of two different sequences for tRNAHis of Pyrobaculum aerophilum: One assumed the location of an intron between nucleotides 37 and 38 (Lowe and Eddy 1997; Marck and Grosjean 2003) and the other between nucleotides 44 and 45 (Sugahara et al. 2007). The two predicted tRNAHis have similar COVE scores of 66.7 and 70.9, based on a standard covariance model calculated by tRNAscan-SE (Lowe and Eddy 1997), respectively, but which of these two tRNAHis variants is actually present in P. aerophilum is not known. The methods described in this work for the identification of tRNAs present in H. marismortui including tRNATrp CCA, the AUA-reading tRNAIle XAU, and other tRNAs provide the tools necessary for experimentally confirming the presence of any tRNA in a cell and, thereby, test tRNA gene predictions based on various algorithms.
MATERIALS AND METHODS
General
General manipulations of E. coli and H. marismortui ATCC 43,049, H. salinarum, and H. volcanii were performed according to standard procedures (Sambrook et al. 1989; DasSarma and Fleischmann 1995). H. marismortui ATCC 43,049 was kindly provided by Dr. Peter Moore (Yale University). E. coli strain XL1-Blue (recA1 endA1 gyr96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)]) from Stratagene was used for plasmid propagation and isolation. Oligonucleotides were obtained from IDT and Operon.
Cloning and purification of E. coli TilS, E. coli IleRS, and human TrpRS
The genes for E. coli TilS and E. coli IleRS were amplified by PCR from genomic DNA from E. coli JM109 and inserted into pET15b (Novagen). The sequences of E. coli TilS and IleRS genes containing an NH2-terminal His6-tag were confirmed by DNA sequencing. E. coli TilS, E. coli IleRS, and human TrpRS (plasmid was kindly provided by the laboratory of Dr. Paul Schimmel, The Scripps Research Institute) were expressed in E. coli BL21(DE3) by induction with 1 mM IPTG. After induction, cultures were shifted to 30°C and cultivated for 3 h. Proteins were purified by Talon chromatography (Clontech) following the manufacturer's instructions for batch/gravity flow-purification. Fractions containing pure protein were pooled, dialyzed against 20 mM sodium phosphate (pH 7.0), 150 mM NaCl, 5 mM β-mercaptoethanol, 50% glycerol and stored at −20°C. The protein concentration was determined by BCA protein assay (Pierce) using BSA as standard. The purity of proteins was assessed by SDS-PAGE/Coomassie staining and Western blot analysis using a His4-antibody (Qiagen) following the instructions of the manufacturer.
Cloning and purification of H. marismortui IleRS and MetRS
The genes for H. marismortui IleRS and MetRS were amplified by PCR from genomic DNA from H. marismortui and inserted into pET28a (Novagen). The sequences of H. marismortui IleRS and MetRS genes containing an NH2-terminal His6-tag were confirmed by DNA sequencing. H. marismortui IleRS and MetRS were expressed in E. coli BL21(DE3) by induction with 0.2 mM IPTG. After 3 h of cultivation at 37°C, cells were harvested, resuspended in buffer A (40 mM Tris-Cl at pH 7.5, 20 mM MgCl2, 0.5 mM EDTA, 10% glycerol, 10 mM β-mercaptoethanol, 2 mM PMSF) and lysed by sonication. Inclusion bodies containing H. marismortui IleRS and MetRS were recovered by centrifugation, resuspended in buffer B (50 mM sodium phosphate at pH 7.5, 500 mM NaCl, 5 M urea, 10 mM β-mercaptoethanol) and kept on ice for 2 h with repeated mixing. After centrifugation at 20,000g for 15 min, the supernatant was diluted with 1 volume of buffer B supplemented with 10 mM imidazole and applied onto Ni-NTA resin (Qiagen) equilibrated with buffer C (50 mM sodium phosphate at pH 7.5, 500 mM NaCl, 2.5 M urea, 5 mM imidazole). Proteins were purified following the manufacturer's instructions for batch/gravity flow purification. After elution with 200–500 mM imidazole, fractions containing pure protein were pooled, first dialyzed against buffer D (50 mM sodium phosphate at pH 7.5, 500 mM NaCl) containing 1 M urea and then against buffer D. Final dialysis was carried out in buffer E (10 mM Tris-HCl at pH 7.5, 100 mM magnesium acetate, 3.4 M KCl). Protein samples were stored in small aliquots at −80°C.
Preparation of S30 cell extract from H. marismortui
A crude extract containing aminoacyl-tRNA synthetases from H. marismortui was prepared as described by Gupta (1995) with slight modifications. Briefly, ∼10 g of late-log-phase cells of H. marismortui were pelleted and washed once in solubilization buffer (10 mM Tris-HCl at pH 7.5, 100 mM magnesium acetate, and 3.4 M KCl). The cells were then resuspended in 20 mL of ice-cold solubilization buffer containing 5 mM β-mercaptoethanol and lysed in a French Press by two passages at 10,000 psi. The lysate was treated with 500 units of RNase-free DNase I (Roche Applied Science) for 30 min in ice for digestion of genomic DNA. The mixture was centrifuged at 4500g for 15 min at 4°C, and the resulting supernatant was centrifuged further for 2 h at 30,000g. The S30 cell extract was stored in small aliquots at −80°C.
Isolation of total tRNA from H. marismortui
Total RNA was extracted under acidic conditions using Trizol (Invitrogen). A pellet of H. marismortui cells obtained from ∼25 mL of culture was resuspended in 200 μL of ice-cold 0.1 M sodium acetate (pH 5.0); 1 mL of Trizol and 200 μL of chloroform were added immediately. Samples were mixed gently to avoid shearing of chromosomal DNA and placed on ice for 5 min with repeated mixing. Samples were centrifuged for 15 min, the aqueous layer was removed, and the phenol phase was reextracted with 100 μL of 0.1 M sodium acetate (pH 5.0). The aqueous layers of both centrifugation steps were combined and precipitated with ethanol. The tRNA was resuspended in 10 mM sodium acetate (pH 5.0) and stored at −80°C. A portion of the material was subjected to deacylation by addition of Tris-HCl (pH 9.5) to a final concentration of 0.2 M. The deacylation reaction was performed at 37°C for 90–120 min; deacylated tRNAs were reprecipitated with ethanol and stored in 10 mM HEPES-NaOH (pH 7.4) at −80°C. Alternatively, total tRNA was isolated as follows: A pellet of H. marismortui late-log phase cells from a 500 mL culture was lysed in 40 mL of cold sterile water. The lysate was mixed with an equal volume of phenol saturated with 0.1 M sodium acetate (pH 5.0) (acidic phenol). Samples were mixed and placed on ice for 15 min with repeated mixing. Samples were centrifuged for 15 min, the aqueous layer was removed, and the phenol phase was reextracted with 20 mL of acidic phenol. The aqueous layers of both centrifugation steps were combined and precipitated with an equal volume of isopropanol at 4°C overnight. The precipitate was recovered by centrifugation at 10,000g, washed twice in 70% ethanol, and then dissolved in 5 mL of buffer A (20 mM Tris-HCl at pH 7.4, 10 mM MgCl2, 0.2 M NaCl). tRNAs were further purified by chromatography on DE52-cellulose (Whatman) (RajBhandary and Ghosh 1969). The fractions containing the enriched tRNAs were pooled and precipitated with 2.5 volumes of ethanol overnight at 4°C. The precipitate was recovered by centrifugation, washed with ethanol, and then dissolved in 1 mL of TE buffer (10 mM Tris-HCl at pH 8.0, 1 mM EDTA). Samples were treated with 20 units of RNase-free DNase I (Roche Applied Science, 5 U/μL) for 3 h at 37°C. DNase I was removed by extraction with phenol-chloroform, and tRNAs were recovered by precipitation with ethanol, dissolved in 0.5 mL of TE buffer, and dialyzed extensively against buffer B (10 mM Tris-HCl at pH 8.0, 5 mM MgCl2, and 10 mM NaCl). The total tRNA was then quick-frozen and stored at −20°C in small aliquots.
Acid urea PAGE and Northern blot analysis of tRNA
tRNAs were analyzed by acid urea PAGE followed by Northern blotting (Varshney et al. 1991). tRNAs were visualized by hybridization using 32P-labeled DNA oligonucleotides according to standard procedures (Sambrook et al. 1989). Oligonucleotides were 5′-end labeled with γ-[32P]-ATP (3000 Ci/mmol; PerkinElmer) using T4-polynucleotide kinase (New England Biolabs). For quantitative assessment of hybridization signals, nonincorporated γ-[32P]-ATP was removed by G-25 Sephadex chromatography (MicroSpin G-25 columns; GE Healthcare). Northern blots were analyzed by autoradiography followed by PhosphorImaging using Imagequant software. Oligonucleotides targeting the D-stem (positions 1–20), anticodon stem–loop (positions 30–49), and T-stem (positions 54–73) of the respective tRNAs were used in this study.
Measurement of deacylation rates of aminoacyl-tRNAs
Total tRNA was isolated under acidic conditions as described above and resuspended in ice-cold sterile water. Deacylation was performed in 0.2 M Tris-HCl (pH 9.2) at 37°C. Aliquots corresponding to 0.1 A260 were removed at 0, 5, 10, 15, 30, 45, 60, 90, and 120 min, mixed with an equal volume of acid urea loading dye containing 0.1 M sodium acetate (pH 5.0), 8 M urea, bromophenol blue, and xylene cyanol FF at 0.05% each and quick-frozen on dry ice. Reaction products were subsequently analyzed by acid urea PAGE/Northern blot as described.
In vitro aminoacylation of tRNA
tRNA (0.1–1.0 A260) was aminoacylated in vitro with L-tryptophan, L-isoleucine, or L-methionine as described below using purified human TrpRS, E. coli IleRS, or E. coli MetRS at a final concentration of 0.5 μM, 0.04 μg/μL of purified H. marismortui MetRS or IleRS, or 0.1–0.5 μg/μL of a S30 extract prepared from H. marismortui. Reaction mixtures were as follows: (1) 50 mM imidazole (pH 7.6), 150 mM KCl, 15 mM MgCl2, 10 mM ATP, 0.1 μg/μl BSA, 2.5 mM DTT, and 100 μM L-tryptophan; (2) 50 mM imidazole (pH 7.6), 15 mM MgCl2, 10 mM ATP, 0.1 μg/μL BSA, and 100 μM L-isoleucine; or (3) 50 mM imidazole (pH 7.6), 150 mM NH4Cl, 15 mM MgCl2, 10 mM ATP, 0.1 μg/μl BSA, and 100 μM L-methionine. Aminoacylation reactions using purified halobacterial enzymes or halobacterial S30 extract contained 3 M KCl. Samples were incubated at 37°C for 30–60 min. Reaction products were examined by acid urea PAGE/Northern blot analysis as described. Alternatively, in vitro aminoacylations were carried out in the presence of radiolabeled amino acids using an S30 extract from H. marismortui in a buffer containing 5 mM Tris-HCl (pH 7.5), 2.3 M KCl, 15 mM MgCl2, 30 mM Mg(OAc)2, 5 mM ATP, and 100 μM L-tryptophan/L-[3H]-tryptophan, L-isoleucine/L-[3H]-isoleucine, or L-methionine/L-[35S]-methionine (Perkin Elmer, GE Healthcare). At various time points, aliquots were removed and analyzed by precipitation with TCA followed by liquid scintillation counting of TCA-precipitable counts.
Preparation of T7 transcripts
The templates carrying the tRNA gene under the control of the T7 promoter for in vitro transcription were amplified by PCR from synthetic DNA (Sampson and Uhlenbeck 1988). In vitro transcription was performed using a T7 AmpliScribe T7 High Yield Transcription Kit (Epicentre) or a MEGAshortscript Kit (Ambion) in a total volume of 40 μL for 120 min at 37°C. The DNA template was removed by treatment with RNAse-free DNase I (2 U, 20 min at 37°C; Epicentre). Transcripts were purified by denaturing electrophoresis on a 10% polyacrylamide gel containing 7 M urea, followed by dialysis and ethanol precipitation. T7 transcripts were stored in 10 mM HEPES-NaOH (pH 7.4) at −80°C. Prior to in vitro aminoacylation or modification (lysidinylation or biotinylation), T7 transcripts were renatured as follows: After denaturation at 65°C for 2 min, the transcript was added directly to the appropriate prewarmed reaction mixture containing 15 mM MgCl2 and allowed to renature for 15 min at 37°C; T7 transcripts were then kept at room temperature for another 15 min.
In vitro modification of C34 to lysidine in tRNA
In vitro modification of T7 transcripts with lysidine using TilS was carried out following the protocol of Ikeuchi et al. (2005) with minor modifications. Reactions containing 0.1–2 A260 of tRNA, 100 mM Tris-HCl (pH 7.4), 10 mM KCl, 15 mM MgCl2, 10 mM ATP, 5 mM L-lysine, and 1–5 μM of E. coli TilS were incubated for 60 min at 37°C. The modified T7 transcript was subjected to phenol extraction, dialyzed, and subsequently concentrated by precipitation with ethanol.
In vitro biotinylation of tRNA
Biotinylation of tRNA was carried out using the NH2-reactive sulfo-NHS-LC-biotin (Pierce) following the protocol of the manufacturer with modifications. A standard 10 μL reaction contained 10 mM sodium phosphate (pH 7.2) or 50 mM HEPES-NaOH (pH 8.0), 15 mM NaCl, 5 mM MgCl2, 4 mM sulfo-NHS-LC-biotin, and 0.01 A260 of T7 transcript or 0.5 A260 of total RNA. Biotinylation was carried out for 2–4 h at room temperature. Biotinylated tRNAs were precipitated with ethanol and analyzed by acid urea PAGE followed by Northern hybridization.
Primer extension to detect post-transcriptional modifications in tRNA
Primer extension was carried out using Superscript III (Invitrogen). Total RNA (0.25 A260) or 0.025 A260 of T7 transcript were mixed with 7.5 pmol of 5′-32P labeled primer in the reaction buffer supplied by the manufacturer, heated for 5 min at 95°C, and then chilled on ice. For primer extension, a 10 μL reaction contained the above tRNA/primer mix, 1.25 mM of each of the four dNTPs, 5 mM DTT, and 100 U of Superscript III reverse transcriptase. Incubation was at 42°C for 15 min. The reverse transcriptase was inactivated by addition of stop solution (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol FF) and denaturation at 95°C for 5 min. The tRNA gene sequences were determined in the presence of dideoxynucleoside triphosphates using standard protocols. Reaction products were resolved on a 12.5% denaturing polyacrylamide gel containing 7 M urea and analyzed by autoradiography. Oligonucleotides used for primer extension of various tRNAs were complementary to positions 54–73 (H. marismortui, H. salinarum, H. volcanii tRNAIle XAU), 43–73 (E. coli tRNAIle LAU, yeast tRNAIle IAU), and 41–73 (yeast tRNAIle ΨAΨ), respectively.
SUPPLEMENTAL DATA
Supplemental Materials are available online at http://web.mit.edu/biology/www/facultyareas/facresearch/pdfs/Kohrer_etal_SupplementaryFigures_RNA2007.pdf.
ACKNOWLEDGMENTS
H. marismortui ATCC 43049 was kindly provided by Dr. Peter Moore (Yale University). Total tRNA from H. volcanii was obtained from Eric Sullivan (this laboratory); total tRNA from M. jannaschii was a generous gift from Dr. Ya-Ming Hou (Jefferson University). A clone for expression and purification of human TrpRS was obtained from Dr. Xianglei Yang (Laboratory of Dr. Paul Schimmel, The Scripps Research Institute). We would like to thank the members of this laboratory for valuable discussions and comments, in particular Dr. Jae-Ho Yoo. The work of G.S. was supported by a National Research Scholarship Award from the National Institute of General Medical Sciences (N.I.H.). Funding for this work was provided by grant GM17151 of the National Institutes of Health to U.L.R.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.795508.
REFERENCES
- Crick, F.H. Codon–anticodon pairing: The wobble hypothesis. J. Mol. Biol. 1966;19:548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- DasSarma, S., Fleischmann, E.M. Archaea—A laboratory manual. Halophiles. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1995. [Google Scholar]
- Drabkin, H.J., RajBhandary, U.L. Initiation of protein synthesis in mammalian cells with codons other than AUG and amino acids other than methionine. Mol. Cell. Biol. 1998;18:5140–5147. doi: 10.1128/mcb.18.9.5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, Z., Chakrabarti, J., Mallick, B., Das, S., Sahoo, S., Sethi, H.S. tRNA–isoleucine–tryptophan composite gene. Biochem. Biophys. Res. Commun. 2006;339:37–40. doi: 10.1016/j.bbrc.2005.10.183. [DOI] [PubMed] [Google Scholar]
- Grosjean, H., Björk, G.R. Enzymatic conversion of cytidine to lysidine in anticodon of bacterial tRNAIle—An alternative way of RNA editing. Trends Biochem. Sci. 2004;29:165–168. doi: 10.1016/j.tibs.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Grosjean, H., Auxilien, S., Constantinesco, F., Simon, C., Corda, Y., Becker, H.F., Foiret, D., Morin, A., Jin, Y.X., Fournier, M., et al. Enzymatic conversion of adenosine to inosine and to N1-methylinosine in transfer RNAs: A review. Biochimie. 1996;78:488–501. doi: 10.1016/0300-9084(96)84755-9. [DOI] [PubMed] [Google Scholar]
- Guo, Q., Gong, Q., Tong, K.L., Vestergaard, B., Costa, A., Desgres, J., Wong, M., Grosjean, H., Zhu, G., Wong, J.T., et al. Recognition by tryptophanyl-tRNA synthetases of discriminator base on tRNATrp from three biological domains. J. Biol. Chem. 2002;277:14343–14349. doi: 10.1074/jbc.M111745200. [DOI] [PubMed] [Google Scholar]
- Gupta, R. Halobacterium volcanii tRNAs. J. Biol. Chem. 1984;259:9461–9471. [PubMed] [Google Scholar]
- Gupta, R. Preparation of transfer RNA, aminoacyl-tRNA synthetases, and tRNAs specific for an amino acid from extreme halophiles. In: Robb F.T., et al., editors. Archaea: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1995. pp. 119–131. [Google Scholar]
- Harada, F., Nishimura, S. Purification and characterization of AUA specific isoleucine transfer ribonucleic acid from Escherichia coli B . Biochemistry. 1974;13:300–307. doi: 10.1021/bi00699a011. [DOI] [PubMed] [Google Scholar]
- Ikeuchi, Y., Soma, A., Ote, T., Kato, J., Sekine, Y., Suzuki, T. Molecular mechanism of lysidine synthesis that determines tRNA identity and codon recognition. Mol. Cell. 2005;19:235–246. doi: 10.1016/j.molcel.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Laslett, D., Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, T.M., Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marck, C., Grosjean, H. tRNomics: Analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA. 2002;8:1189–1232. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marck, C., Grosjean, H. Identification of BHB splicing motifs in intron-containing tRNAs from 18 archaea: Evolutionary implications. RNA. 2003;9:1516–1531. doi: 10.1261/rna.5132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthaei, J.H., Voigt, H.P., Heller, G., Neth, R., Schöch, G., Kübler, H., Amelunxen, F., Sander, G., Parmeggiani, A. Specific interactions of ribosomes in decoding. Cold Spring Harb. Symp. Quant. Biol. 1966;31:25–38. doi: 10.1101/sqb.1966.031.01.009. [DOI] [PubMed] [Google Scholar]
- Muramatsu, T., Nishikawa, K., Nemoto, F., Kuchino, Y., Nishimura, S., Miyazawa, T., Yokoyama, S. Codon and amino acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature. 1988a;336:179–181. doi: 10.1038/336179a0. [DOI] [PubMed] [Google Scholar]
- Muramatsu, T., Yokoyama, S., Horie, N., Matsuda, A., Ueda, T., Yamaizumi, Z., Kuchino, Y., Nishimura, S., Miyazawa, T. A novel lysine-substituted nucleoside in the first position of the anticodon of minor isoleucine tRNA from Escherichia coli . J. Biol. Chem. 1988b;263:9261–9267. doi: 10.1351/pac198961030573. [DOI] [PubMed] [Google Scholar]
- Nakanishi, K., Fukai, S., Ikeuchi, Y., Soma, A., Sekine, Y., Suzuki, T., Nureki, O. Structural basis for lysidine formation by ATP pyrophosphatase accompanied by a lysine-specific loop and a tRNA-recognition domain. Proc. Natl. Acad. Sci. 2005;102:7487–7492. doi: 10.1073/pnas.0501003102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nureki, O., Niimi, T., Muramatsu, T., Kanno, H., Kohno, T., Florentz, C., Giegé, R., Yokoyama, S. Molecular recognition of the identity-determinant set of isoleucine transfer RNA from Escherichia coli . J. Mol. Biol. 1994;236:710–724. doi: 10.1006/jmbi.1994.1184. [DOI] [PubMed] [Google Scholar]
- RajBhandary, U.L. Initiator transfer RNAs. J. Bacteriol. 1994;176:547–552. doi: 10.1128/jb.176.3.547-552.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RajBhandary, U.L., Ghosh, H.P. Studies on polynucleotides. XCI. Yeast methionine transfer ribonucleic acid: Purification, properties, and terminal nucleotide sequences. J. Biol. Chem. 1969;244:1104–1113. [PubMed] [Google Scholar]
- Ramesh, V., RajBhandary, U.L. Importance of the anticodon sequence in the aminoacylation of tRNAs by methionyl-tRNA synthetase and by valyl-tRNA synthetase in an Archaebacterium. J. Biol. Chem. 2001;276:3660–3665. doi: 10.1074/jbc.M008206200. [DOI] [PubMed] [Google Scholar]
- Randau, L., Münch, R., Hohn, M.J., Jahn, D., Söll, D. Nanoarchaeum equitans creates functional tRNAs from separate genes for their 5′- and 3′-halves. Nature. 2005a;433:537–541. doi: 10.1038/nature03233. [DOI] [PubMed] [Google Scholar]
- Randau, L., Pearson, M., Söll, D. The complete set of tRNA species in Nanoarchaeum equitans . FEBS Lett. 2005b;579:2945–2947. doi: 10.1016/j.febslet.2005.04.051. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., Maniatis, T. Molecular cloning—A laboratory manual. 2nd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Sampson, J.R., Uhlenbeck, O.C. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl. Acad. Sci. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger, B., Auxilien, S., Englisch, U., Cramer, F., Fasiolo, F. The modified wobble base inosine in yeast tRNAIle is a positive determinant for aminoacylation by isoleucyl-tRNA synthetase. Biochemistry. 1997;36:8269–8275. doi: 10.1021/bi970206l. [DOI] [PubMed] [Google Scholar]
- Seong, B.L., RajBhandary, U.L. Escherichia coli formylmethionine tRNA: Mutations in GGG:CCC sequence conserved in anticodon stem of initiator tRNAs affect initiation of protein synthesis and conformation of anticodon loop. Proc. Natl. Acad. Sci. 1987;84:334–338. doi: 10.1073/pnas.84.2.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soma, A., Ikeuchi, Y., Kanemasa, S., Kobayashi, K., Ogasawara, N., Ote, T., Kato, J., Watanabe, K., Sekine, Y., Suzuki, T. An RNA-modifying enzyme that governs both the codon and amino acid specificities of isoleucine tRNA. Mol. Cell. 2003;12:689–698. doi: 10.1016/s1097-2765(03)00346-0. [DOI] [PubMed] [Google Scholar]
- Sugahara, J., Yachie, N., Sekine, Y., Soma, A., Matsui, M., Tomita, M., Kanai, A. SPLITS: A new program for predicting split and intron-containing tRNA genes at the genome level. In Silico Biol. 2006;6:411–418. [PubMed] [Google Scholar]
- Sugahara, J., Yachie, N., Arakawa, K., Tomita, M. In silico screening of archaeal tRNA-encoding genes having multiple introns with bulge–helix–bulge splicing motifs. RNA. 2007;13:671–681. doi: 10.1261/rna.309507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szweykowska-Kulinska, Z., Senger, B., Keith, G., Fasiolo, F., Grosjean, H. Intron-dependent formation of pseudouridines in the anticodon of Saccharomyces cerevisiae minor tRNAIle . EMBO J. 1994;13:4636–4644. doi: 10.1002/j.1460-2075.1994.tb06786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocchini-Valentini, G.D., Fruscoloni, P., Tocchini-Valentini, G.P. Coevolution of tRNA intron motifs and tRNA endonuclease architecture in Archaea. Proc. Natl. Acad. Sci. 2005;102:15418–15422. doi: 10.1073/pnas.0506750102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney, U., Lee, C.P., RajBhandary, U.L. Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J. Biol. Chem. 1991;266:24712–24718. [PubMed] [Google Scholar]
- Xue, H., Shen, W., Giegé, R., Wong, J.T. Identity elements of tRNATrp: Identification and evolutionary conservation. J. Biol. Chem. 1993;268:9316–9322. [PubMed] [Google Scholar]
- Xue, S., Calvin, K., Li, H. RNA recognition and cleavage by a splicing endonuclease. Science. 2006;312:906–910. doi: 10.1126/science.1126629. [DOI] [PubMed] [Google Scholar]