Abstract
Errors in the mRNP biogenesis pathway can lead to retention of mRNA in discrete, transcription-site-proximal foci. This RNA remains tethered adjacent to the transcription site long after transcriptional shutoff. Here we identify Sus1, Thp1, and Sac3 as factors required for the persistent tethering of such foci (dots) to their cognate genes. We also show that the prolonged association of previously activated GAL genes with the nuclear periphery after transcriptional shutoff is similarly dependent on the Sac3-Thp1-Sus1-Cdc31 complex. We suggest that the complex associates with nuclear mRNP and that mRNP properties influence the association of dot-confined mRNA with its gene of origin as well as the post-transcriptional retention of the cognate gene at the nuclear periphery. These findings indicate a coupling between the mRNA-to-gene and gene-to-nuclear periphery tethering. Taken together with other recent findings, these observations also highlight the importance of nuclear mRNP to the mobilization of active genes to the nuclear rim.
Keywords: mRNA export, mRNP, Saccharomyces cerevisiae
INTRODUCTION
The export of eukaryotic mRNA to the cytoplasm is preceded by nuclear RNA processing. This includes covalent events (capping, splicing, and polyadenylation) as well as the noncovalent addition of hnRNP proteins, which facilitate binding and translocation of mRNA through nuclear pores. There are also elaborate quality-control mechanisms, which monitor the biogenesis of nuclear mRNP (Jensen et al. 2003; Fasken and Corbett 2005). Indeed, Saccharomyces cerevisiae strains bearing temperature-sensitive mutations in genes encoding nuclear processing factors manifest a pronounced nuclear mRNA retention. These genes include poly(A) polymerase PAP1 (Hilleren et al. 2001), the mRNA export receptor MEX67 (Jensen et al. 2001b), and 3′-end processing factors RNA14 and RNA15 (Libri et al. 2002), as well as the components of the nuclear THO/TREX complex SUB2, HPR1, and MFT1 (Jensen et al. 2001a; Libri et al. 2002). Upon a shift to nonpermissive conditions, these strains accumulate mRNA in discrete, transcription-site-proximal foci (hereafter referred to as “dots”) that can be visualized by fluorescent in situ hybridization (FISH) with gene-specific probes.
Nuclear mRNA dots can also be observed in wild-type cells under physiological conditions. For example, robust, intense dots arise from a GAL-driven GFP-encoding reporter construct, which bypasses normal 3′-end processing because it terminates in a hammerhead ribozyme (GAL-GFP-RZ). Dots also arise from a reporter with a wild-type GAL 3′-UTR (containing GAL1 3′-end formation signals; GAL-GFP-pA) and even from the endogenous GAL1 gene (Dower et al. 2004; Abruzzi et al. 2006). Therefore, dot formation likely reflects a regular feature of gene expression, which is quantitatively increased when nuclear mRNA processing is suboptimal or perturbed. Importantly, both GAL-GFP-RZ and GAL-GFP-pA reporters give rise to dots containing RNA that is largely post-transcriptional (i.e., non-nascent). This is because the dots are spatially distinct from their transcription sites and because they persist long after the transcriptional shutoff (Abruzzi et al. 2006). Moreover, dots remain adjacent to their transcription sites during the shutoff.
Intriguingly, dot formation correlates with the tendency of active genes to associate with the nuclear periphery (Abruzzi et al. 2006), which suggests mechanistic links between these two processes. Several mechanisms have been proposed to contribute to the recruitment, capture, and/or subperipheral retention of genes at the nuclear envelope. These include direct interactions between transcriptional activators and nucleoporins (Menon et al. 2005; Schmid et al. 2006), the act of transcription itself (Cabal et al. 2006; Taddei et al. 2006), transcription-associated chromatin remodeling (Brickner et al. 2007), as well as unspecified mRNA- and/or 3′-UTR-dependent interactions (Casolari et al. 2005; Taddei et al. 2006). Our own experiments have also highlighted the importance of 3′-end formation signals on gene-periphery associations. Moreover, these have a strong and parallel influence on dot formation (Abruzzi et al. 2006).
In this study, we conducted a FISH-based screen for dot tethering factors. Our strategy was based on the assumption that perturbation of the tether(s) between the dot and its gene would probably impact dot morphology. Indeed, we identified Sus1, Thp1, and Sac3 as factors affecting dot morphology as well as the persistent tethering of dots to their cognate genes after transcriptional shutoff. This strongly implicates the Sac3-Thp1-Sus1-Cdc31 complex in post-transcriptional dot-gene tethering. Remarkably, the association of the endogenous GAL1 locus to the nuclear periphery previously has been shown to be inhibited by the absence of this same complex (Cabal et al. 2006; Drubin et al. 2006). However, its dual association with the transcription coactivator SAGA as well as with NPCs has precluded discriminating between a transcriptional and a post-transcriptional role of the complex. The findings reported here favor the latter, because the retention of previously activated (but transcriptionally silent) GAL-promoter-driven reporter genes at the nuclear periphery similarly requires the activity of these three proteins. The parallel findings on these two tethering phenomena underscore the mechanistic coupling between the mRNA-to-gene and gene-nuclear periphery interactions and emphasize the contribution of post-transcriptional events to both processes.
RESULTS
Components of the Sac3-Thp1-Sus1-Cdc31 complex affect the morphology of nuclear mRNP dots arising because of the bypass of the 3′-end processing
To conduct a FISH-based screen for the dot tethering factors, we used a strain containing a single-copy, integrated reporter gene construct. It contains a TDH3-promoter-driven GFP ORF followed by a ribozyme (RZ) 3′-end formation sequence (TDH-GFP-RZ). The construct gives rise to a robust, constitutive nuclear dot in almost 100% of cells, which is visualized by FISH (note that this construct generates negligible amounts of GFP protein). We then examined dot morphology in a focused sublibrary of viable deletions (Tong et al. 2001), which was compiled via an extensive manual survey of genes implicated in mRNA biogenesis, processing, and export (Table 1).
TABLE 1.
A sublibrary of viable mRNA export-relevant deletion mutants used in the FISH-based screen
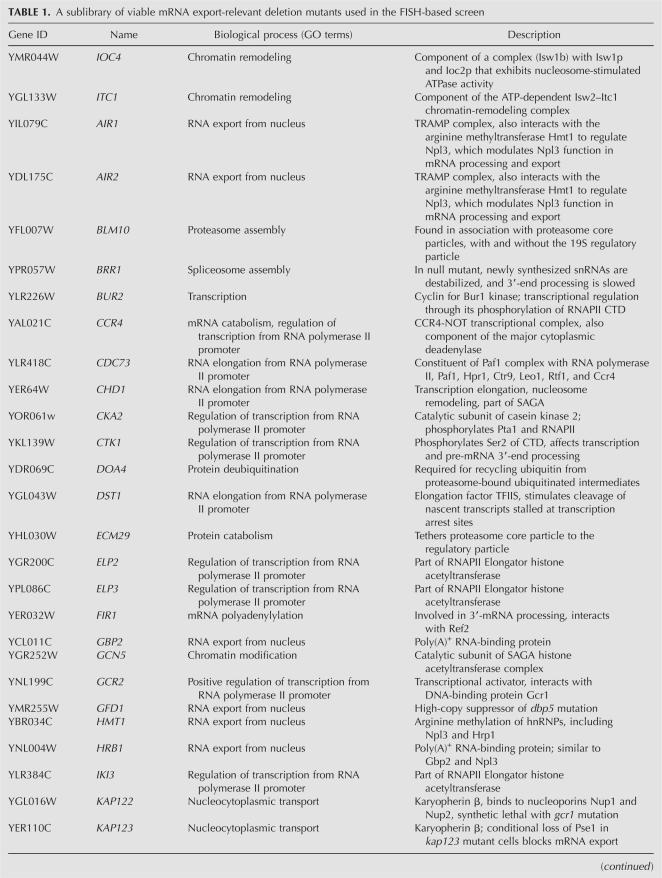
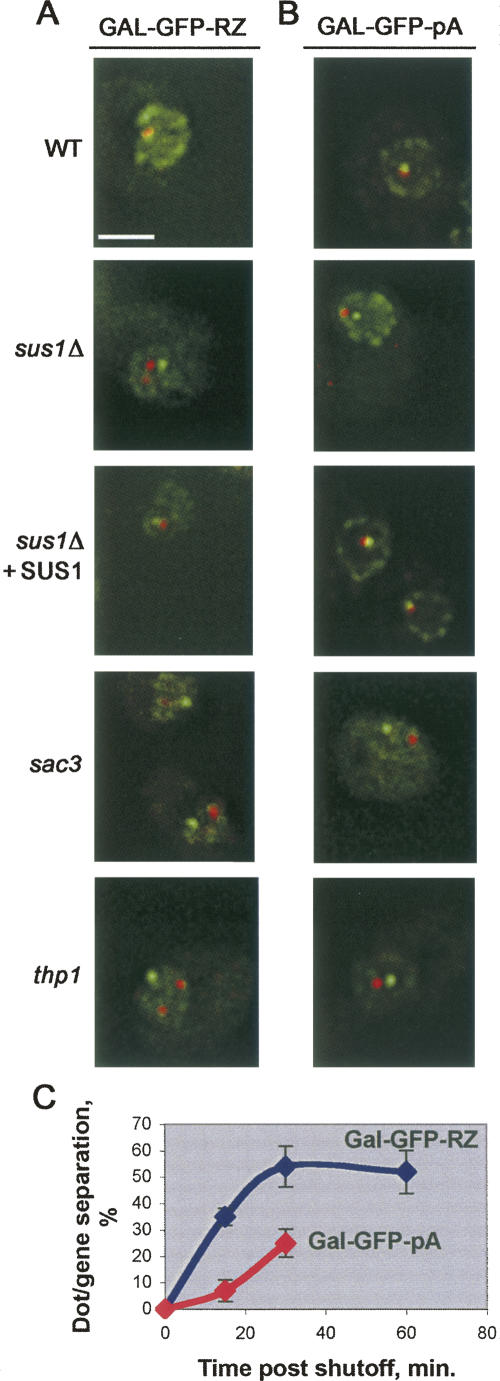
Deletion of SUS1 resulted in a striking enlargement and/or fragmentation of the dot (Fig. 1A,B), reproducibly observed in ≥80% of cells. Importantly, these effects were not dependent on the specific construct or on its integration site, because substituting the conditional GAL1 promoter for the TDH3 promoter as well as placing the reporter at a different genomic location led to an identical dot enlargement and fragmentation in an sus1Δ background (Fig. 1C). Moreover, simultaneous visualization of the GAL-GFP-RZ locus using TetR-GFP bound to a tandem array of 448 Tet operators integrated <5 kb from the reporter construct (TetR-GFP/TetO448 system) showed that one major dot was always adjacent to the reporter gene (Fig. 1C). (In these and subsequent experiments, the nuclear periphery was simultaneously visualized using Nup49-GFP, which by itself has no effect on the dot morphology.) This suggests a precursor–product relationship between the primary, locus-proximal mRNA dot and secondary dots that break off and diffuse away. Introducing a wild-type (WT) copy of the SUS1 gene into the sus1Δ strain fully rescued this phenotype (Fig. 1D) as well as the other associated phenotypes described below.
FIGURE 1.
Altered nuclear-retained (A,B,E,F) TDH-GFP-RZ and (C,D) GAL-GFP-RZ mRNA dot morphology in sus1Δ, sac3Δ, and thp1Δ cells. (A,B,E–H), fluorescent in situ hybridization (FISH) for the TDH-GFP-RZ mRNA in (A) wild-type (WT), (B) sus1Δ, (E) gcn5Δ, (F) spt20Δ, (G) sac3Δ, and (H) thp1Δ cells. (Left panels) FISH signal (red); (middle panels) DAPI staining for total DNA (blue); (right panels) overlay. Scale bar, 2 μm. (C,D) FISH for the GAL-GFP-RZ mRNA with simultaneous visualization of the TetO448 array integrated at the BMH1 locus on Chromosome V, <5kb away, using TetR-GFP in (C) sus1Δ cells and (D) sus1Δ cells expressing an ectopically integrated WT copy of SUS1 as well as a Nup49-GFP (the presence of Nup49-GFP has no effect on the dot morphology phenotype). (Left panels) FISH signal (red); (middle panels) FISH signal (red) overlaid on TetR-GFP signal (green); (right panels) triple overlay of FISH signal, TetR-GFP signal, and DAPI staining for the total DNA (blue). (I) In contrast to the GAL-GFP-RZ mRNA dot (C,D), no enlargement of the GAL-GFP-pA mRNA dot is observed in sus1Δ cells.
Sus1 is a component of the Sac3-Thp1-Sus1-Cdc31 complex, which is anchored to the nuclear pore via an Sac3–Nup1 interaction (Fischer et al. 2002). Sus1 also copurifies with the transcriptional coactivator SAGA and is believed to mediate transcription-coupled mRNA export (Rodriguez-Navarro et al. 2004). Indeed, loss of Sus1 compromises transcription of a subset of SAGA-regulated genes and also leads to increased accumulation of poly(A)+ RNA in the nucleus (Fischer et al. 2002, 2004; Rodriguez-Navarro et al. 2004). Because the TDH3 promoter used to drive the GFP-RZ reporter is SAGA-regulated (Basehoar et al. 2004), we determined whether the sus1Δ effect on dot morphology is principally due to the impaired function of SAGA or of the Sac3-Thp1-Sus1-Cdc31 complex. To this end, we tested by FISH four additional deletion strains missing components of these complexes.
We found that deletions of the SAGA genes GCN5 or SPT20 had no effect on TDH-GFP-RZ dot morphology (Fig. 1E,F; data not shown). The spt20Δ mutation causes severe disruption of SAGA complex integrity and function (Grant et al. 1997; Sterner et al. 1999) and prevents Sus1 from associating with the promoter of GAL1 (Kohler et al. 2006). Moreover, the effect of gcn5Δ on transcription is quantitatively identical to that of sus1Δ (Fig. 3C,D below; Dudley et al. 1999). We therefore conclude that the SAGA complex does not contribute substantially to mRNP dot morphology.
FIGURE 3.
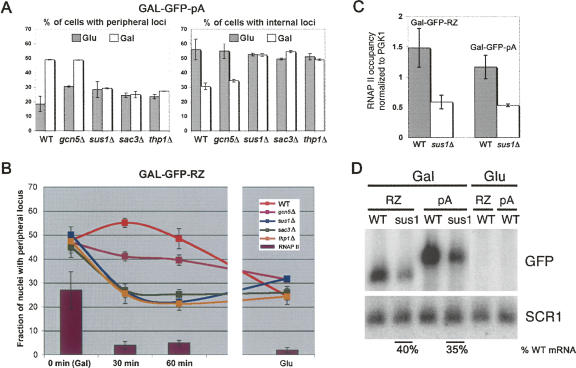
Deleting SUS1, SAC3, and THP1 has distinct transcription-independent effects on retention of the activated GAL-GFP-pA and GAL-GFP-RZ genes at the nuclear periphery. (A) Intranuclear positioning of the GAL-GFP-pA gene is indistinguishable in sus1Δ, sac3Δ, and thp1Δ cells under activating and repressing conditions. The proportion of cells with (left panel) peripherally and (right panel) internally positioned loci is plotted for galactose-grown (Gal) and glucose-grown (Glu) cells. The intranuclear position of the locus was revealed using the TetR-GFP/TetO448 system. The nuclear periphery was visualized using Nup49-GFP fusion coexpressed in the same strain. Subperipheral fraction remains unchanged at ∼20% (data not shown). Error bars represent standard deviation. (B) Dissociation of the GAL-GFP-RZ gene locus from the nuclear periphery in sus1Δ, sac3Δ, and thp1Δ cells is dramatically accelerated and parallels the kinetics of the RNAP II runoff from the gene. The proportion of cells with a peripherally positioned GAL-GFP-RZ gene locus after 0, 30, or 60 min of transcriptional shutoff or in steady-state glucose conditions is plotted. For comparison, RNAP II occupancy after transcriptional shutoff as well as in steady-state glucose conditions, measured by ChIP, is shown as vertical bars. Error bars represent standard deviation. (C) RNAP II occupancy of the GAL-GFP-pA and GAL-GFP-RZ reporter constructs in sus1Δ cells is affected to the same extent. The RNAP II levels are normalized to the RNAP II occupancy on the endogenous PGK1 gene, which is unaffected by the loss of SUS1. Error bars represent standard deviation. (D) Northern blot quantification of the steady-state GAL-GFP-pA and GAL-GFP-RZ mRNA levels using a GFP-specific probe in WT and sus1Δ cells grown on galactose and glucose. (SCR1) Loading control used for normalization.
In contrast, deletions of either THP1 or SAC3 genes resulted in effects indistinguishable from those observed in sus1Δ cells (Fig. 1G,H), that is, an exaggerated and/or fragmented dot. We conclude that compromising the function of the Sac3-Thp1-Sus1-Cdc31 complex perturbs the transcription-site-proximal GFP-RZ mRNP pool, which is visualized by FISH as an exaggerated and/or fragmented dot. Importantly, this effect is specific, since other mutants that affect poly(A)+ mRNA export to a comparable or greater extent (e.g., lrp1Δ) (Hieronymus et al. 2004; data not shown) had no effect on dot morphology.
The Sac3-Thp1-Sus1-Cdc31 complex impacts the dot-gene tether
To further explore the role of the Sac3-Thp1-Sus1-Cdc31 complex in the tethering of the mRNA dot to its cognate gene during transcription as well as after the transcriptional shutoff, we simultaneously monitored the locations of the dots with FISH and the reporter locus with the TetR-GFP/TetO448 system as described above. As previously reported, the dot as well as its tether to the reporter gene persist for at least 60 min after transcriptional shutoff in WT cells (Abruzzi et al. 2006). In the sus1Δ, sac3Δ, and thp1Δ mutant strains, however, we observed that the GAL-GFP-RZ dots progressively detach from their loci of origin after transcriptional shutoff (Fig. 2). To extend this observation, we also tested the GAL-GFP-pA reporter construct integrated at the same genomic location. GAL-GFP-pA construct possesses a normal GAL1 3′-UTR and polyadenylation signal (while its GFP chromophore has been inactivated to enable visualization of TetR-GFP/TetO448), and the GAL-GFP-pA dot appears normal during active transcription (Fig. 1I). However, it similarly detaches from its gene in the three deletion strains after transcriptional shutoff (Fig. 2). These data indicate that the Sac3-Thp1-Sus1-Cdc31 complex contributes to retention of a post-transcriptional dot near its gene of origin. Moreover, the lack of an effect before transcriptional shutoff suggests that there are additional, Sac3-, Thp1-, and Sus1-independent mechanisms contributing to the dot-gene tether during active transcription.
FIGURE 2.
Deletions of SUS1, SAC3, and THP1 lead to a loss of association of GAL-GFP-RZ and GAL-GFP-pA mRNA dots with their respective genes. (A) GAL-GFP-RZ mRNA or (B) GAL-GFP-pA mRNA is visualized by FISH (red); respective reporter loci are visualized using the TetR-GFP/TetO448 system (green); and nuclear periphery is visualized using Nup49-GFP (green). Images are taken at the 30-min time point after the transcriptional shutoff. Scale bar, 2 μm. (C) relative kinetics of the GAL-GFP-RZ and GAL-GFP-pA mRNA dots' separation from respective genes after transcriptional shutoff in sus1Δ cells. (Very few GAL-GFP-pA mRNA dots remain at 60 min in sus1Δ cells; dot/locus separation in WT never exceeded 5%.) Error bars represent standard deviation.
The Sac3-Thp1-Sus1-Cdc31 complex also impacts the gene–nuclear periphery tether
The difference in dot phenotype between GAL-GFP-pA and GAL-GFP-RZ in the mutant strains (Fig. 1) parallels a difference in dot-gene dissociation kinetics, as the GAL-GFP-RZ mRNA dot separates more quickly from the gene after transcriptional shutoff (Fig. 2C). Moreover, differences in dot phenotypes were previously observed in wild-type cells: the GAL-GFP-pA dots disappear more quickly than the GAL-GFP-RZ dots after transcriptional shutoff, which further correlates with differences in the dissociation of the two genes from the nuclear rim after transcriptional shutoff in WT cells: GAL-GFP-pA dot dissociates more quickly than the GAL-GFP-RZ dot (Abruzzi et al. 2006). These differences presumably reflect the different RNP compositions of the two transcripts (see Discussion).
Motivated by these observations, we examined the relationship of the reporter loci to the nuclear periphery during active transcription in the sus1Δ, sac3Δ, and thp1Δ strains as well as peripheral retention after transcriptional shutoff. In contrast to WT cells (Abruzzi et al. 2006), the GAL-GFP-pA gene locus completely failed to localize to the nuclear rim under activating conditions in the deletion strains (galactose) (Fig. 3A). This recapitulates perfectly the behavior of the endogenous GAL1 locus in these strains (Cabal et al. 2006; Drubin et al. 2006). In contrast, the GAL-GFP-RZ gene still localized to the nuclear periphery in the deletion strains. However, its association was rapidly lost upon transcriptional shutoff, with kinetics that paralleled that of RNAP II runoff (Fig. 3B). This contrasts with WT cells, in which the GAL-GFP-RZ locus dissociates from the nuclear rim only very slowly after glucose addition (>60 min) (Abruzzi et al. 2006). The results suggest the existence of additional transcription-dependent tether(s) that maintain an actively transcribing GAL-GFP-RZ locus at the nuclear periphery. They also suggest that the post-transcriptional retention of the GAL-GFP-RZ locus at the nuclear periphery requires the Sac3-Thp1-Sus1-Cdc31 complex.
Because Sus1 directly participates in transcription from the GAL1 promoter (Rodriguez-Navarro et al. 2004; Kohler et al. 2006), we addressed the possibility that the differences in intranuclear positioning between the active GAL-GFP-pA and GAL-GFP-RZ genes in the sus1Δ mutant is a consequence of differential transcriptional effects on these two genes. To this end, we compared the RNAP II occupancy on these two reporter constructs, using chromatin immunoprecipitation (ChIP). The magnitude of reduction in RNAP II occupancy of GAL-GFP-RZ and GAL-GFP-pA in sus1Δ relative to WT cells was identical (Fig. 3C), and it was also identical to that of the endogenous GAL1 gene in sus1Δ cells (data not shown). Moreover, the decrease was quantitatively mirrored by the steady-state mRNA level differences of both reporter genes between the two strains (Fig. 3D).
We also found that a loss of Gcn5, which associates with the GAL1 promoter (Dhasarathy and Kladde 2005) and impacts its transcriptional activity comparably to sus1Δ (approximately threefold) (Fig. 3C,D; Dudley et al. 1999), had no effect on peripheral retention of the GAL-GFP-pA gene (Fig. 3A). Moreover, it had no more than a modest effect on the dissociation kinetics of the GAL-GFP-RZ locus from the nuclear rim (Fig. 3B). We conclude that the differential impact of an Sus1 deletion on the morphology of GAL-GFP-RZ and GAL-GFP-pA mRNA dots as well as on the nuclear rim association of these genes is not due to its SAGA-related functions but to the Sac3-Thp1-Sus1-Cdc31 complex. We also speculate that post-transcriptional mRNP may more generally facilitate gene repositioning to the nuclear periphery.
DISCUSSION
Using a FISH-based genetic screen, we identified Sus1, Thp1, and Sac3 as factors that have an impact on mRNA dot morphology. They also affect the persistent tethering of dots to their cognate genes as well as to the nuclear rim after transcriptional shutoff. These findings reveal a novel, post-transcriptional function of the Sac3-Thp1-Sus1-Cdc31 complex in addition to its previously described roles in transcription itself, mRNA export, and the tethering of genes to the nuclear periphery during active transcription. Indeed, it is striking that the disruption of this complex affects the tethering of post-transcriptional mRNPs to their respective genes as well as the capture and retention of transcriptionally activated loci at the nuclear rim. Although we cannot rule out indirect effects, a parsimonious interpretation of this unexpected dual role is that the dot-to-gene and gene-to-nuclear periphery interactions are both mediated by post-transcriptional mRNP decorated with the Sac3-Thp1-Sus1-Cdc31 complex. Although the steady-state distribution of these proteins is predominantly at the nuclear rim, this does not preclude an association with mRNP. Indeed, Sus1 and its partners interact with mRNA and chromatin, in addition to their interactions with the nuclear pores (Fischer et al. 2002; Gallardo et al. 2003; Lei et al. 2003; Rodriguez-Navarro et al. 2004; Kohler et al. 2006).
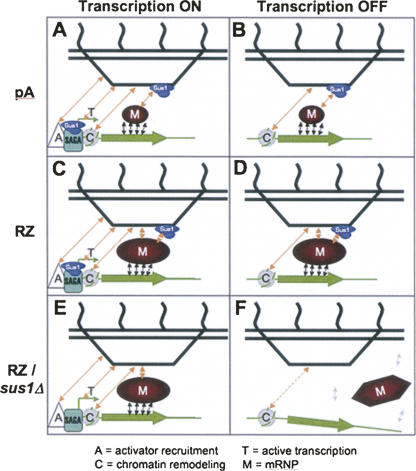
Characterization of the sus1Δ, thp1Δ, and sac3Δ strains indicates that fragmentation of the GAL-GFP-RZ mRNP dot is intimately related to weakening of the gene-dot tethering. This might indicate that dot integrity is dependent on contacts between dot mRNP, Sac3-Thp1-Sus1-Cdc31 complex, and chromatin. When these are diminished (e.g., in sus1Δ), dot fragmentation as well as separation of the dot and the gene occur (Figs. 1, 4). Notably, despite their fragmentation in the mutant backgrounds, the GAL-GFP-RZ dots do not completely disperse and disappear, indicating that there must be Sus1-, Sac3-, and Thp1-independent associations between the individual mRNP particles within the dot that contribute to its integrity.
FIGURE 4.
A model that integrates the multiple interactions between the activated GAL genes (green), non-nascent mRNP pools (red), and nuclear pores (black), emphasizing the dual role of the Sac3-Thp1-Sus1-Cdc31 complex (blue) at the site of transcription as well as at the nuclear periphery. (A) In WT cells, perinuclear repositioning of the GAL-GFP-pA locus occurs via a combined action of multiple mechanisms including transcriptional activator (A)-dependent recruitment as well as transcription (T)-, chromatin (C)-, and mRNP (M)-mediated capture and retention. (B) Upon shutoff of transcription, persistent chromatin- and mRNP-mediated retention prevents the immediate departure of the GAL-GFP-pA locus from the nuclear rim. (C) The abnormal and large GAL-GFP-RZ mRNP pool produced when mRNA 3′-end processing is bypassed by RZ makes additional contacts at the nuclear pore, which are not Sac3-Thp1-Sus1-Cdc31 complex-dependent, and hence is strongly retained at the nuclear rim. (D) Upon transcriptional shutoff, the synergy of these additional contacts with the NPC, acting together with the Sac3-Thp1-Sus1-Cdc31 complex-dependent and chromatin-dependent contacts, allows the GAL-GFP-RZ mRNP to persist at the nuclear periphery longer than GAL-GFP-pA. (E) Even in the absence of the Sac3-Thp1-Sus1-Cdc31 complex at the NPC, the synergy of the activator-mediated contacts, ongoing transcription, chromatin-mediated and the Sac3-Thp1-Sus1-Cdc31 complex-independent interactions of the exaggerated GAL-GFP-RZ mRNP with the NPC support a modest degree of peripheral tethering. (F) Upon transcriptional shutoff, this modest tethering fails immediately, while the aberrant mRNP remodeling (illustrated by the change in shape of the mRNP pool) and/or compromised export of the GAL-GFP-RZ mRNP in the absence of Sac3-Thp1-Sus1-Cdc31 complex causes its separation from the gene.
We find that deletions of Sus1, Sac3, and Thp1 affect the dot–gene interactions not only in the case of the 3′-end-impaired GFP-RZ construct, but also in the case of the GAL-GFP-pA mRNP, suggesting that Sus1, Sac3, and Thp1 are also components of the GAL-GFP-pA-containing dots. Moreover, sus1Δ, sac3Δ, and thp1Δ mutations strongly impact the association of the GAL-GFP-pA gene with the nuclear rim. Yet their effects on this reporter differ in two respects from their effects on GAL-GFP-RZ: the GAL-GFP-pA dots are neither enlarged nor fragmented in the mutants, and the association of the GAL-GFP-pA gene with the nuclear periphery is abolished completely, i.e., even during active transcription. As only the GAL-GFP-RZ gene remains associated with the nuclear rim during active transcription in the mutant strains, there must be additional transcription-dependent contacts that are stronger or more numerous between the GAL-GFP-RZ gene and the nuclear periphery.
Although we cannot rule out that such differential contacts may be mediated by chromatin per se, there are no known chromatin effects associated with replacing the GAL1 3′-UTR with the ribozyme. An alternative (or additional) possibility would be due to a difference between GAL-GFP-pA and GAL-GFP-RZ mRNP structure or composition, which can be qualitative, quantitative, or both. Indeed, we reported previously that the GAL-GFP-RZ gene shows an altered cotranscriptional recruitment profile of Yra1 (Abruzzi et al. 2006). Moreover, a quantitative difference is indicated by the consistently larger GAL-GFP-RZ dots than the GAL-GFP-pA dots (data not shown; D. Zenklusen and R. Singer, pers. comm.). Larger dots would provide more contact area between RZ mRNP and the nuclear periphery than between pA RNP and the periphery, which might contribute to RZ gene retention at the nuclear rim during active transcription even in a Sus1-deleted strain (Fig. 4).
This view implies that there are multiple contacts between a tethered gene and the NPC. Indeed, it has been shown that transcriptional activator binding (Schmid et al. 2006), transcription-associated chromatin remodeling (Brickner et al. 2007), and perhaps the act of transcription itself (Casolari et al. 2005; Cabal et al. 2006; Drubin et al. 2006) all contribute to the recruitment of the activated GAL genes to the nuclear periphery. Moreover, diminished perinuclear positioning during active transcription was observed for endogenous GAL genes in sus1Δ and sac3Δ cells (e.g., Cabal et al. 2006; Drubin et al. 2006). Our findings extend these conclusions by suggesting that this effect is mediated by the post-transcriptional mRNP and becomes relevant only after initial contact of the activated gene with the nuclear rim (modeled in Fig. 4). Whereas the initial encounter of the locus with the nuclear periphery is transcriptional activator-dependent, independent of Sus1 (Schmid et al. 2006), and precedes the onset of transcription (Brickner et al. 2007), stable retention becomes independent of active transcription and is facilitated by the Sac3-Thp1-Sus1-Cdc31 complex; it is more generally aided by intrinsic properties of post-transcriptional mRNP. We suggest that multiple tethering mechanisms serve to strengthen the association between the active gene and NPC and hence facilitate rapid mRNA export, as originally proposed by Blobel in the gene gating hypothesis (Blobel 1985).
MATERIALS AND METHODS
Strain design and growth conditions
Construction of strains bearing the TDH-GFP-RZ reporter loci marked with nourseothricin resistance and KanMX-marked deletions of selected nonessential genes was done using the approach originally developed for genome-wide scoring of synthetic lethal interactions (Tong et al. 2001). Reporter strains designed for mating with deletion array strains were in a Y7092 background (MATα can1Δ∷STE2pr-SpΔhis5 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 lyp1Δ trp1Δ∷GAL1-IpgB1-URA3) (Alto et al. 2006; gift from Charlie Boone).
Yeast strains other than the ones used in the FISH-based screen for altered dot morphology as well as their derivation are described in detail in Table 2. To delete the SUS1, SAC3, and THP1, the kanamycin (KanMX) cassette plus respective flanking regions were PCR-amplified from the respective KanMX-marked deletion strains in the BY4741 background (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), obtained from Open Biosystems. The fragment encompassing NUP49-GFP fusion and HIS3 (Huh et al. 2003) was amplified from the Invitrogen strain collection. Integrations were verified by PCR and/or Southern blotting.
TABLE 2.
Yeast strains other than the deletion sublibrary used in FISH-based screen
Yeast cell growth conditions for FISH, visualization of GFP fusion proteins, as well as for galactose-to-glucose shifts were as described (Abruzzi et al. 2006).
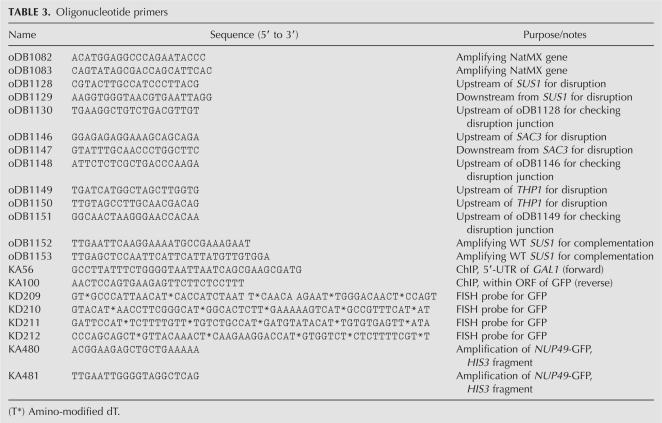
Plasmid constructs and oligonucleotide primers
The primers used in this study are listed in Table 3. Plasmid pDB700 was designed for integrating the TDH3-GFP-RZ reporter marked by nureseothricin resistance marker gene NatMX (Goldstein and McCusker 1999) into the genomic trp1 locus. To this end, NatMX was amplified with oDB1082/1083 and cloned into the AatII site of the pRS304/2μ bearing TDH3-GFP-RZ (Dower et al. 2004), thus replacing its 2μ origin of replication. Integration was conducted after linearization at the Bsu36I site within the TRP1 gene sequence.
TABLE 3.
Oligonucleotide primers
pDB716 and pDB719 (described in Abruzzi et al. 2006) were designed for inserting the GAL-GFP-RZ and GAL-GFP-pA, respectively, by the gamma-integration method (Sikorski and Hieter 1989) into the intergenic region between ECM32 and BMH1, after linearization with NotI and selecting for Trp+ transformants.
To generate pDB729, WT genomic SUS1 fragment was amplified by PCR with oDB1152 and oDB1153, cloned into the Not site of pDB700, followed by releasing the TDH-GFP-RZ reporter from the polylinker after digesting with SpeI + ApaI, repairing the ends with T4 DNA polymerase, and religating. Integration was conducted into TRP1 locus after linearization with Bsu36I, with selection for nurseothricin resistance.
Microscopy
GFP fusion proteins were observed in cells grown to OD ∼ 0.5 and fixed for 15 min in 4% paraformaldehyde (without acetic acid) using an Olympus IX70-based DeltaVision workstation (Applied Precision). Z-stacks were taken at 0.2 μm step size and subjected to constrained iterative deconvolution. Positions of the TetR-GFP marked locus were scored in the z-section that cuts through the middle of the nucleus as described (Brickner and Walter 2004) into intranuclear, peripheral, and subperipheral (i.e., locus touching the nuclear envelope but not coplanar with it). The subperipheral fraction varied little in all conditions and therefore is not reported in Figure 3. Each experiment was done in replicate, and between 100 and 150 cells were scored per sample per time point in each replicate in all experiments shown. FISH with Cy3-labeled oligonucleotide probes was carried out according to Dower et al. (2004). Red/green channel signal offset due to chromatic aberration alone, as estimated by imaging TetraSpec beads (100 nm diameter, Invitrogen/Molecular Probes) under identical conditions, was negligible compared to the separation of FISH (Cy3) and GFP signals.
Chromatin immunoprecipitation and RNA analyses
RNAP II ChIP was performed using monoclonal antibody 8WG16 (Covance). Target DNA levels in input and IP samples were determined by real-time PCR using RotorGene (Corbett Research), and results were normalized as described (Abruzzi et al. 2004). The Northern hybridization signals in Figure 3 obtained with a GFP-specific probe were normalized to respective signals for SCR1, a RNAP III transcript, using ImageQuant software. Signal ratios were identical to the ChIP signal ratios in the respective strains.
ACKNOWLEDGMENTS
We appreciate the technical assistance of Prabhat Mallik and Andrey Belostotsky. We are grateful to the Boone, Nasmyth, and Wente laboratories for strains, as well as to Kristine O'Brien, other members of the Melissa Moore laboratory, and Alexey Khodjakov for help with deconvolution microscopy. This work was supported in part by grants from NIH to D.A.B. and J.A.C. (#GM073872) and to M.R. (#GM23549) and from the NSF to D.A.B. (Grant MCB0424651).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.764108.
REFERENCES
- Abruzzi, K.C., Lacadie, S., Rosbash, M. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 2004;23:2620–2631. doi: 10.1038/sj.emboj.7600261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abruzzi, K.C., Belostotsky, D.A., Chekanova, J.A., Dower, K., Rosbash, M. 3′-End formation signals modulate the association of genes with the nuclear periphery as well as mRNP dot formation. EMBO J. 2006;25:4253–4262. doi: 10.1038/sj.emboj.7601305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alto, N.M., Shao, F., Lazar, C.S., Brost, R.L., Chua, G., Mattoo, S., McMahon, S.A., Ghosh, P., Hughes, T.R., Boone, C., et al. Identification of a bacterial type III effector family with G protein mimicry functions. Cell. 2006;124:133–145. doi: 10.1016/j.cell.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Basehoar, A.D., Zanton, S.J., Pugh, B.F. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- Blobel, G. Gene gating: A hypothesis. Proc. Natl. Acad. Sci. 1985;82:8527–8529. doi: 10.1073/pnas.82.24.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner, J.H., Walter, P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2:e342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner, D.G., Cajigas, I., Fondufe-Mittendorf, Y., Ahmed, S., Lee, P.C., Widom, J., Brickner, J.H. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabal, G.G., Genovesio, A., Rodriguez-Navarro, S., Zimmer, C., Gadal, O., Lesne, A., Buc, H., Feuerbach-Fournier, F., Olivo-Marin, J.C., Hurt, E.C., et al. SAGA interacting factors confine subdiffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- Casolari, J.M., Brown, C.R., Drubin, D.A., Rando, O.J., Silver, P.A. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes & Dev. 2005;19:1188–1198. doi: 10.1101/gad.1307205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhasarathy, A., Kladde, M.P. Promoter occupancy is a major determinant of chromatin remodeling enzyme requirements. Mol. Cell. Biol. 2005;25:2698–2707. doi: 10.1128/MCB.25.7.2698-2707.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower, K., Kuperwasser, N., Merrikh, H., Rosbash, M. A synthetic A tail rescues yeast nuclear accumulation of a ribozyme-terminated transcript. RNA. 2004;10:1888–1899. doi: 10.1261/rna.7166704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin, D.A., Garakani, A.M., Silver, P.A. Motion as a phenotype: The use of live-cell imaging and machine visual screening to characterize transcription-dependent chromosome dynamics. BMC Cell Biol. 2006;7:19. doi: 10.1186/1471-2121-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley, A.M., Rougeulle, C., Winston, F. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes & Dev. 1999;13:2940–2945. doi: 10.1101/gad.13.22.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasken, M.B., Corbett, A.H. Process or perish: Quality control in mRNA biogenesis. Nat. Struct. Mol. Biol. 2005;12:482–488. doi: 10.1038/nsmb945. [DOI] [PubMed] [Google Scholar]
- Fischer, T., Strasser, K., Racz, A., Rodriguez-Navarro, S., Oppizzi, M., Ihrig, P., Lechner, J., Hurt, E. The mRNA export machinery requires the novel Sac3p–Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 2002;21:5843–5852. doi: 10.1093/emboj/cdf590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, T., Rodriguez-Navarro, S., Pereira, G., Racz, A., Schiebel, E., Hurt, E. Yeast centrin Cdc31 is linked to the nuclear mRNA export machinery. Nat. Cell Biol. 2004;6:840–848. doi: 10.1038/ncb1163. [DOI] [PubMed] [Google Scholar]
- Gallardo, M., Luna, R., Erdjument-Bromage, H., Tempst, P., Aguilera, A. Nab2p and the Thp1p–Sac3p complex functionally interact at the interface between transcription and mRNA metabolism. J. Biol. Chem. 2003;278:24225–24232. doi: 10.1074/jbc.M302900200. [DOI] [PubMed] [Google Scholar]
- Goldstein, A.L., McCusker, J.H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae . Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Grant, P.A., Duggan, L., Cote, J., Roberts, S.M., Brownell, J.E., Candau, R., Ohba, R., Owen-Hughes, T., Allis, C.D., Winston, F., et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: Characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes & Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Hieronymus, H., Yu, M.C., Silver, P.A. Genome-wide mRNA surveillance is coupled to mRNA export. Genes & Dev. 2004;18:2652–2662. doi: 10.1101/gad.1241204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren, P., McCarthy, T., Rosbash, M., Parker, R., Jensen, T.H. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature. 2001;413:538–542. doi: 10.1038/35097110. [DOI] [PubMed] [Google Scholar]
- Huh, W.K., Falvo, J.V., Gerke, L.C., Carroll, A.S., Howson, R.W., Weissman, J.S., O'Shea, E.K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Jensen, T.H., Boulay, J., Rosbash, M., Libri, D. The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr. Biol. 2001a;11:1711–1715. doi: 10.1016/s0960-9822(01)00529-2. [DOI] [PubMed] [Google Scholar]
- Jensen, T.H., Patricio, K., McCarthy, T., Rosbash, M. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell. 2001b;7:887–898. doi: 10.1016/s1097-2765(01)00232-5. [DOI] [PubMed] [Google Scholar]
- Jensen, T.H., Dower, K., Libri, D., Rosbash, M. Early formation of mRNP: License for export or quality control? Mol. Cell. 2003;11:1129–1138. doi: 10.1016/s1097-2765(03)00191-6. [DOI] [PubMed] [Google Scholar]
- Kohler, A., Pascual-Garcia, P., Llopis, A., Zapater, M., Posas, F., Hurt, E., Rodriguez-Navarro, S. The mRNA export factor Sus1 is involved in Spt/Ada/Gcn5 acetyltransferase-mediated H2B deubiquitinylation through its interaction with Ubp8 and Sgf11. Mol. Biol. Cell. 2006;17:4228–4236. doi: 10.1091/mbc.E06-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, E.P., Stern, C.A., Fahrenkrog, B., Krebber, H., Moy, T.I., Aebi, U., Silver, P.A. Sac3 is an mRNA export factor that localizes to cytoplasmic fibrils of nuclear pore complex. Mol. Biol. Cell. 2003;14:836–847. doi: 10.1091/mbc.E02-08-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri, D., Dower, K., Boulay, J., Thomsen, R., Rosbash, M., Jensen, T.H. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell. Biol. 2002;22:8254–8266. doi: 10.1128/MCB.22.23.8254-8266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon, B.B., Sarma, N.J., Pasula, S., Deminoff, S.J., Willis, K.A., Barbara, K.E., Andrews, B., Santangelo, G.M. Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc. Natl. Acad. Sci. 2005;102:5749–5754. doi: 10.1073/pnas.0501768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro, S., Fischer, T., Luo, M.J., Antunez, O., Brettschneider, S., Lechner, J., Perez-Ortin, J.E., Reed, R., Hurt, E. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116:75–86. doi: 10.1016/s0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Arib, G., Laemmli, C., Nishikawa, J., Durussel, T., Laemmli, U.K. Nup-PI: The nucleopore–promoter interaction of genes in yeast. Mol. Cell. 2006;21:379–391. doi: 10.1016/j.molcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Sikorski, R.S., Hieter, P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner, D.E., Grant, P.A., Roberts, S.M., Duggan, L.J., Belotserkovskaya, R., Pacella, L.A., Winston, F., Workman, J.L., Berger, S.L. Functional organization of the yeast SAGA complex: Distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei, A., Van Houwe, G., Hediger, F., Kalck, V., Cubizolles, F., Schober, H., Gasser, S.M. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- Tong, A., Boone, C. High-throughput strain construction and systematic synthetic lethal screening in Saccharomyces cerevisiae . In: Stansfield I., Stark M.J.R., editors. Methods in microbiology: Yeast gene analysis. 2nd ed. Vol. 36. Elsevier; Amsterdam: 2007. pp. 369–386. 706–707. [Google Scholar]
- Tong, A.H., Evangelista, M., Parsons, A.B., Xu, H., Bader, G.D., Page, N., Robinson, M., Raghibizadeh, S., Hogue, C.W., Bussey, H., et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]