Abstract
The 3′ cap-independent translation element (BTE) of Barley yellow dwarf virus RNA confers efficient translation initiation at the 5′ end via long-distance base pairing with the 5′-untranslated region (UTR). Here we provide evidence that the BTE functions by recruiting translation initiation factor eIF4F. We show that the BTE interacts specifically with the cap-binding initiation factor complexes eIF4F and eIFiso4F in a wheat germ extract (wge). In wge depleted of cap-interacting factors, addition of eIF4F (and to a lesser extent, eIFiso4F) allowed efficient translation of an uncapped reporter construct (BLucB) containing the BTE in its 3′ UTR. Translation of BLucB required much lower levels of eIF4F or eIFiso4F than did a capped, nonviral mRNA. Both full-length eIF4G and the carboxy-terminal half of eIF4G lacking the eIF4E binding site stimulated translation to 70% of the level obtained with eIF4F, indicating a minor role for the cap-binding protein, eIF4E. In wge inhibited by either BTE in trans or cap analog, eIF4G alone restored translation nearly as much as eIF4F, while addition of eIF4E alone had no effect. The BTE bound eIF4G (Kd = 177 nm) and eIF4F (Kd = 37 nm) with high affinity, but very weakly to eIF4E. These interactions correlate with the ability of the factors to facilitate BTE-mediated translation. These results and previous observations are consistent with a model in which eIF4F is delivered to the 5′ UTR by the BTE, and they show that eIF4G, but not eIF4E, plays a major role in this novel mechanism of cap-independent translation.
Keywords: 3′ untranslated region, long-distance RNA interactions, plant virus gene expression, RNA binding protein, translation initiation
INTRODUCTION
Initiation is the most regulated step in translation of eukaryotic messages. The least abundant, and thus rate-limiting, initiation factor is the cap-binding complex eIF4F (Gingras et al. 1999; Hershey and Merrick 2000; Gallie 2007; Hinnebusch et al. 2007; Mathews et al. 2007; Pestova et al. 2007). In plants, eIF4F is a heterodimer composed of the cap-binding protein, eIF4E, and the multifunctional scaffolding protein, eIF4G (Lax et al. 1986; Gingras et al. 1999; Prevot et al. 2003a; Gallie 2007). In animals, the helicase eIF4A is also considered part of eIF4F (Gingras et al. 1999), but eIF4A is more loosely associated in plants.
Eukaryotic mRNAs normally contain a cap structure (Pestova et al. 2007) [m7G(5′)ppp(5′)N] at the 5′ end and a poly(A)-tail at the 3′ end. The cap is recognized by the eIF4E subunit of eIF4F (Marcotrigiano et al. 1997; Matsuo et al. 1997; von der Haar et al. 2004; Monzingo et al. 2007). eIF4G interacts with the mRNA (Prevot et al. 2003b), eIF4E (von Der Haar et al. 2000; Marcotrigiano et al. 2001), eIF4A, and poly(A)-binding protein (PABP), which is bound to the poly(A) tail (Tarun and Sachs 1996; Le et al. 1997; Imataka et al. 1998). Simultaneous binding of eIF4E to the 5′ cap and eIF4G, and of PABP to the poly(A) tail and eIF4G, leads presumably to circularization of the mRNA (Wells et al. 1998) during initiation. In this state, eIF4G apparently attracts the multisubunit complex eIF3, which docks the 43S ribosome complex (40S ribosomal subunit, eIF1, eIF1A, eIF2, tRNAi Met, eIF3) to the mRNA and associated initiation factors (Kapp and Lorsch 2004; Marintchev and Wagner 2004; Hinnebusch 2006; Gallie 2007; Hinnebusch et al. 2007; Pestova et al. 2007). Because even weak structures in the mRNA prevent the 43S complex from binding the 5′ UTR and scanning, eIF4A, with its RNA-helicase activity in conjunction with stimulatory factors eIF4G and eIF4B, is needed to melt any secondary structure close to the 5′ cap (Pause et al. 1994; Pestova and Kolupaeva 2002). Next, the 43S complex, perhaps while still bound to eIF3 and eIF4F (Marintchev and Wagner 2004; Poyry et al. 2004; Pestova et al. 2007) scans the mRNA in the 3′ direction until the correct AUG initiation codon is reached. At this point, most of the initiation factors are released and the 60S ribosomal subunit joins to begin protein synthesis (Kozak 1980; Kapp and Lorsch 2004; Marintchev and Wagner 2004; Merrick 2004; Jackson 2005; Hinnebusch et al. 2007; Pestova et al. 2007).
eIF4G plays a crucial role in ribosome recruitment and scanning. This scaffolding protein contains binding sites for PABP, eIF4E, eIF4A, eIF3, regulatory proteins, and RNA (Prevot et al. 2003a). The C-terminal two-thirds of mammalian eIF4G (p100), has the ability to recruit the 43S complex independently of eIF4E to both capped and uncapped mRNAs (Ali et al. 2001) or to picornaviral IRESes (Pestova et al. 1996; Pestova and Kolupaeva 2002). The N-terminal part of mammalian eIF4G is mostly unstructured, but the regions accepting PABP and eIF4E fold upon binding to their corresponding partners. These conformational changes are transferred to more distant regions of eIF4G, which affects its properties. Binding of eIF4E reduces the ability of eIF4G to promote translation of uncapped mRNAs (Tarun et al. 1997; De Gregorio et al. 1998).
Plants express at least two distinct isoforms of eIF4F (Browning et al. 1992; Gallie 2007). In wheat, eIF4E (26 kDa) pairs with eIF4G (165 kDa) to form eIF4F, and eIFiso4E (28 kDa) pairs with eIFiso4G (86 kDa) to form eIFiso4F (Browning 1996, 2004; Gallie 2007). eIF4G and eIFiso4G share only ∼30% similarity to each other in a central conserved domain. eIF4G contains a long N-terminal portion that is absent in eIFiso4G. Plant eIF4G is more similar to mammalian eIF4G in size than eIFiso4G. The cap-binding subunits, eIF4E and eIFiso4E, share ∼50% similarity to each other. Wheat eIFiso4F exhibits preference for capped, unstructured mRNAs (Gallie and Browning, 2001). In contrast, eIF4F efficiently supports translation of both capped mRNAs with structured leaders and uncapped messages and promotes internal initiation on a plant viral RNA (Gallie and Browning 2001). These functional differences result at least partly from the stronger RNA-dependent ATPase activity of eIF4G compared with that of eIFiso4G (Gallie and Browning 2001; Gallie 2007).
Viral mRNAs have evolved numerous unconventional means of recruiting translational machinery. These allow them to compete with host mRNAs and to avoid defense mechanisms that act at the level of translation. For example, uncapped RNAs of dicistroviruses (Sasaki and Nakashima 1999; Wilson et al. 2000; Jan 2006), picornaviruses (Jang et al. 1988; Pelletier and Sonenberg 1988), potyviruses (Levis and Astier-Manifacier 1993; Basso et al. 1994; Niepel and Gallie 1999), and capped mRNAs of retroviruses (Herbreteau et al. 2005; Nicholson et al. 2006), recruit ribosomes at an internal ribosome entry site (IRES). The IRES structure engages the 40S ribosomal subunit, independent of the 5′ end of the mRNA and brings it directly to close proximity of the initiation codon. Different IRESes function by very different mechanisms, with factor requirements ranging from all canonical initiation factors to almost none (Jackson 2005; Doudna and Sarnow 2007).
Cap-independent translation occurs by different mechanisms in plant viral RNAs that lack both a cap and a poly(A) tail. Translation of these mRNAs initiates at the 5′ end, but is mediated by a cap-independent translation element (CITE) residing in the 3′ UTR (Miller and White 2006). The CITEs fall into at least six structural classes, which share no apparent sequence or structural similarity with each other (Miller et al. 2007). These include the Barley yellow dwarf virus (BYDV)-like cap-independent translation elements (BTEs) in the Luteovirus, Necrovirus, Umbravirus, and Dianthovirus genera. BTEs contain a 17-nucleotide (nt) sequence that fits the consensus, GGAUCCUGGGAAACAGG, and a stem–loop capable of base pairing to the 5′ end of the mRNA (Kneller et al. 2006). Other structurally unrelated CITEs reside in the 3′ UTRs of Satellite tobacco necrosis virus (STNV) (Danthinne et al. 1993; Timmer et al. 1993; Meulewaeter et al. 1998; van Lipzig et al. 2002), Tomato bushy stunt virus (TBSV) (Fabian and White 2004, 2006), Maize necrotic streak virus (Scheets and Redinbaugh 2006), Panicum mosaic virus (Batten et al. 2006), Turnip crinkle virus (Qu and Morris 2000), Hibiscus chlorotic ringspot virus (Koh et al. 2002, 2003), and Blackcurrant reversion virus (BRV) (Karetnikov et al. 2006; Karetnikov and Lehto 2007). Communication with the 5′ UTR is known (BYDV, TBSV, BRV) or predicted to be mediated in most cases by long-distance base pairing, as most CITEs have the potential to base pair with structures within 5′ UTRs (Miller and White 2006). The cap-independent translation enhancer domain (TED) in STNV interacts specifically with the cap-binding subunits of eIF4F and eIFiso4F (Gazo et al. 2004).
The 5.7-kb genome of BYDV has a complex 869 nt 3′UTR, which contains cap-independent and poly(A) tail-independent translation elements and other regulators of gene expression and viral replication (Miller et al. 1988; Miller and Rasochova 1997; Miller and White 2006). The BTE sequence that is necessary and sufficient for cap-independent translation of BYDV RNA in wheat germ extract (wge) resides within a 105-nt tract (BTE105) between bases 4814 and 4918 at the 5′ end of the 3′UTR (Fig. 1A; Wang and Miller 1995; Wang et al. 1997, 1999; Guo et al. 2000). The BTE105 sequence alone represses translation of viral and nonviral mRNAs when added in trans (Wang et al. 1999; Shen et al. 2006). Addition of exogenous eIF4F restores translation to the trans-inhibited extract, suggesting that BTE105 binds factors such as eIF4F, required for both cap-dependent and BTE-dependent translation (Wang et al. 1997, 1999). Free cap analog also inhibits BTE-mediated translation, but much higher levels are required for inhibition of BTE-mediated translation than for cap-dependent translation (Wang and Miller 1995). The components of the translational machinery with which the BTE interacts to recruit the ribosome and allow efficient translation initiation are unknown. Here, we provide biochemical evidence that the BTE binds canonical translation initiation factor eIF4F, and that this binding correlates with ability of the BTE to facilitate cap-independent translation. The BTE interacts preferentially with eIF4G and shows distinct preference for eIF4F over eIFiso4F. This differs from the STNV TED, which shows only a twofold preference for eIF4F over eIFiso4F and binds via eIF4E and eIFiso4E (Gazo et al. 2004).
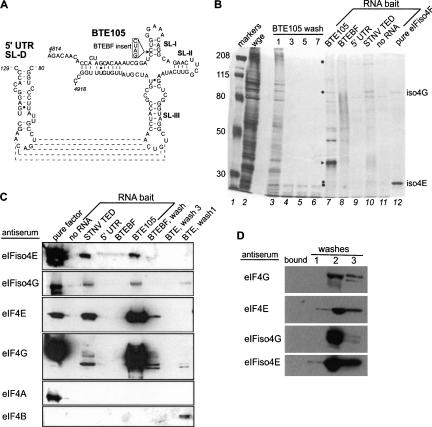
FIGURE 1.
Identification of BTE-interacting proteins from wheat germ extract. (A) Secondary structure of BTE105 RNA (right) and the stem–loop in the 5′UTR (SL-D) to which it base pairs (indicated by dashed lines) (Guo et al. 2001). In mutant BTEBF (inset), the BamHI4837 site (nucleotide 4837) is disrupted by insertion of a GAUC duplication that abolishes BTE function. (B) BTE-interacting proteins (BTEIPs) from wheat germ extract (wge) identified using biotin-labeled RNAs as bait and magnetic streptavidin beads to pull down the BTEIPs. Proteins were separated by 5% PAGE and silver stained. (Lane 1) Molecular weight markers of the indicated kDa at left; (lane 2) total wge protein; (lanes 3–6) unbound proteins obtained after indicated number of low-salt washes of bead-bound BTE RNA. RNA bait (lanes 7–11) show proteins that remained bound to the indicated RNA after washes. STNV TED consists of nucleotides 621–741 of STNV RNA. (Lane 12) Recombinant eIFiso4E and eIFiso4G. Dots to the left of lane 7 indicate BTE105-interacting proteins that comigrate with eIFiso4E and eIFiso4G. Expected Mobilities of eIF4G (> 200 kDa) and eIF4E (26 kDa) are indicated by squares. Arrowhead indicates BTE RNA. (C) Western blots using antibodies to known initiation factors on proteins pulled down by the indicated RNAs. Proteins were separated by SDS PAGE prior to blotting on PVDF membrane. Each panel represents a different gel and blot. Washes indicate proteins not bound to the indicated RNA. No pure eIF4B was used as positive control (first lane), but efficacy of antisera was evident by detection of eIF4B in the low-salt wash. Cleavage products of the labile eIF4G are visible. (D) Western blots against wheat germ extract proteins eluted from biotinylated, nonviral (166 nt vector-derived) RNA complexes. (bound) Proteins interacting with vector sequence (none detected in these Western blots). Unbound proteins obtained in washes (lanes 1–3) as indicated.
RESULTS
eIFiso4E, eIFiso4G, eIF4E, and eIF4G, and unidentified proteins interact specifically with the BTE
To identify proteins that may play a role in cap-independent translation, we used the 105-nt BTE (BTE105) as bait to pull down BTE-interacting proteins (BTEIPs) from the wheat germ translation extract (wge). As a negative control, we used a nonfunctional mutant, BTEBF, which differs from BTE105 only by the presence of a four-base GAUC insertion in the BamHI4837 site. BTEBF is completely inactive in facilitating cap-independent translation in cis, and in inhibiting translation in trans (Wang et al. 1997; Guo et al. 2000). We also tested as bait the BYDV 5′UTR and, as a positive control, the STNV 3′TED RNA. The STNV TED functions similarly to the BTE, even though its structure is unrelated. It is known to bind eIF4E and eIFiso4E (Gazo et al. 2004).
Several proteins were pulled down by BTE105 RNA that were not apparent among proteins pulled down by the negative control RNAs, as seen in the silver-stained gel in Figure 1B. Among the bands that appeared only in the BTE and STNV TED pull-down lanes were two that comigrated with eIFiso4E (28 kDa) and eIFiso4G (83 kDa) markers (Fig. 1B, dots and right lane). In the BTE105 pull-down lane, faint bands were also visible that migrated as expected for eIF4G (migrates >200 kDa), and eIF4E (26 kDa) (Fig. 1B, squares). The most prominent band in the BTE105 lane (Fig. 1B, triangle) proved to be BTE RNA itself, which coeluted with the bound proteins. The BTEBF, 5′ UTR, and STNV TED RNAs ran as smears in their respective lanes, and may have been degraded. Note that this SDS–polyacrylamide gel system is not denaturing for RNA, so RNAs forming multimers and heterogenous structures would not migrate as discrete bands.
To test for the presence of known initiation factors among the BTEIPs, we used factor-specific antibodies as probes in immunoblots of the BTEIPs. The immunoblots revealed the components of eIFiso4F (eIFiso4E and eIFiso4G), and of eIF4F (eIF4E and eIF4G), but not eIF4A or eIF4B, among the BTEIPs (Fig. 1C). In contrast, none of the subunits of eIF4F or eIFiso4F interacted with the nonfunctional mutant BTEBF RNA. As an additional negative control, we also tested proteins pulled down by a nonviral, vector-derived RNA flanking the firefly luciferase reporter, pGEMluc (Promega). This RNA did not pull down eIFiso4F or eIF4F subunits (bound lane, Fig. 1D), which were instead found in the unbound wash fractions (Fig. 1D). Thus, the BTE interacts directly or indirectly, but specifically, with eIFiso4F and eIF4F. The same set of factors bound to STNV TED (Fig. 1C) as reported previously (Gazo et al. 2004).
To determine whether eIF4F and/or eIFiso4F associate closely and thus directly with the BTE, a UV-cross-linking experiment was performed. Radiolabeled BTE105 or BTEBF RNAs were mixed with wheat germ extracts and subjected to UV cross-linking to stabilize protein–RNA interactions. Antisera to eIF4F or eIFiso4F were used to immunoprecipitate those proteins and associated RNA. BTE cross-linked proteins of the expected size for the components of eIF4F and eIFiso4F were indeed detected (Fig. 2). An additional BTE cross-linked protein(s), detected by both antisera, migrates at 70 kDa. This may be a degradation product of eIF4G and eIFiso4G. No cross-linking of BTEBF RNA to eIF4F or eIFiso4F was detected (Fig. 2). Taken together, the data support a specific and direct interaction between initiation factors eIF4F, eIFiso4F, and the BTE in the wheat germ extract.
FIGURE 2.
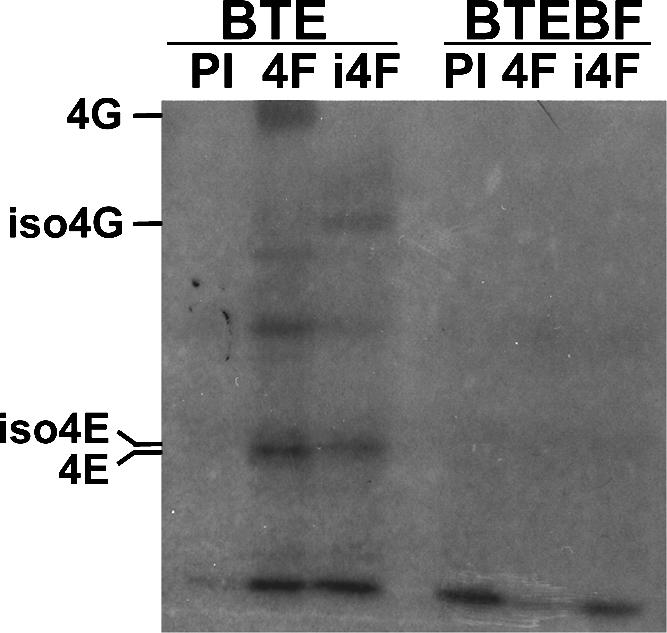
UV cross-linking between BTE RNAs and cap-binding proteins in wheat germ extracts. [32P]-labeled BTE105 or BTEBF RNAs were UV cross-linked to wheat germ extract proteins, RNase-treated, and immunoprecipitated with either preimmune antisera (PI) or antiserum to eIF4F (4F) or eIFiso4F (i4F). RNA–protein mixtures were then run on a 12.5% SDS–polyacrylamide gel and visualized by autoradiography. Mobilities of pure factors on the same gel are indicated at left.
BTE-mediated translation requires lower concentrations of cap-binding factors than cap-mediated translation and less eIF4F than eIFiso4F
Having established interaction of the BTE with eIF4F and eIFiso4F, we next determined the roles of these factors and their subunits in BTE-mediated translation. We made the wheat germ extract dependent on cap-binding factors (eIF4F/iso4F and any associated proteins) by passage through a m7GTP-Sepharose column. This process was shown previously to deplete the extract of nearly all eIFiso4E, eIFiso4G, eIF4E, and eIF4G, along with some eIF4A, eIF4B, and PABP (Gallie 2001; Gallie and Browning 2001). We observed the effect of adding purified factors back to the depleted extract programmed with a BTE-dependent reporter mRNA. This RNA, called BlucB, consists of the firefly luciferase ORF flanked by the 5′ and 3′ UTRs of BYDV RNA, including the complete 3′ BTE. BLucB (LUC869 in a previous report) (Wang et al. 1999) was shown previously to have all of the viral sequence necessary for cap- and poly(A) tail-independent translation in vitro and in vivo (Wang et al. 1997). The ability of added recombinant eIF4F and eIFiso4F to restore BTE-mediated translation was determined by measuring luciferase activity after BLucB-programmed translation in the depleted wheat germ extract.
We determined the dependence upon eIF4F or eIFiso4F of uncapped BLucB relative to a capped polyadenylated reporter lacking any viral sequence (capVLucV(A)68). In the absence of added factors, translation of BLucB in the depleted wge extract was very low, confirming functional depletion of these essential translation factors (Fig. 3A,B, y intercept). At the lowest concentration of added eIF4F (6.75 nM), translation of BLucB was stimulated 20-fold above the residual level obtained in the absence of factors, while translation of capVLucV(A)68 mRNA rose only about 2.5-fold (Fig. 3A). The difference in stimulation of translation for these two mRNAs decreased with increasing eIF4F concentration. At the highest concentration (108 nM), the increases in luciferase activity for BLucB and capVLucV(A)68 were 63-fold and 56-fold, respectively, above levels obtained in the absence of added factors. However, BLucB translation was about fivefold greater than that of capVLucV(A)68, even at this high level of eIF4F. Similarly, low concentrations of eIFiso4F stimulated BLucB translation much more than for cap-dependent translation (Fig. 3B). At the highest concentration of eIFiso4F, the increases in luciferase expression for the two mRNAs over background were similar, but unlike with eIF4F, BLucB translation was only two-thirds greater than that of capVLucV(A)68 (Fig. 3B). When compared in the same experiment, 54 nM eIF4F stimulated translation of BLucB 50-fold, whereas the same concentration of eIFiso4F increased translation by only 10-fold above background levels (Fig. 3C), revealing that eIF4F was fivefold more stimulatory than eIFiso4F. In summary, BLucB required much smaller concentrations of eIF4F or eIFiso4F for significant translation, and eIF4F stimulated BLucB translation to a greater extent than did eIFiso4F.
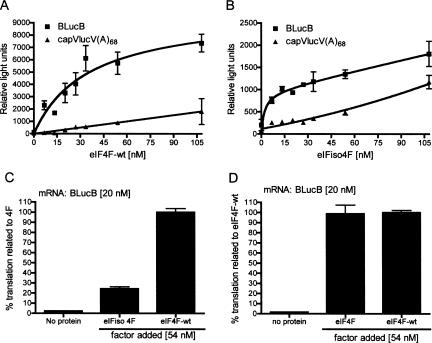
FIGURE 3.
Effect of added factors on translation in cap-binding factor-depleted wheat germ extract. Wheat germ extract (wge) was passed over a m7GTP-Sepaharose column to remove cap-binding protein complexes prior to in vitro translation. Indicated amounts of recombinant eIF4F (A) or eIFiso4F (B) were added to extracts prior to programming with 20 nM uncapped BLucB or capped vector with a 68-nt poly(A) tail (capVLucV(A)68) mRNA. The reaction mixtures were incubated for 1 h at 25°C and firefly luciferase activity was measured in relative light units. (C) Direct comparison of effects of eIFiso4F and eIF4F on BLucB translation. The data shown are averages of at least two experiments. Percent translation indicates relative light units normalized to the readings obtained with 54 nM eIF4F (4F). (D) Comparison of stimulation of translation by recombinant eIF4F containing his-tagged eIF4E used throughout this work (4F) with eIF4F containing native eIF4E (4F-wt).
The eIF4F preparations used in most experiments were reconstituted from N-terminally histidine-tagged eIF4E and wild-type eIF4G; both were expressed in, and purified from, Escherichia coli. To determine whether the additional histidine residues affected eIF4F function, we compared stimulatory activity of his-tagged eIF4F with eIF4F composed of both wild-type eIF4E and eIF4G (eIF4F-wt). His-tagged eIF4F and wild-type eIF4F both stimulated translation of BLucB to similar levels (Fig. 3D). Thus, the his-tag at the N terminus of eIF4E has no influence on the ability of eIF4F to facilitate BTE-mediated translation.
Taken together, these results show that (1) the BTE requires eIF4F or eIFiso4F to facilitate cap-independent translation; (2) BTE-containing mRNA and capped mRNA both use eIF4F more efficiently than eIFiso4F; and (3) BTE-mediated translation requires smaller amounts of these factors than does cap-dependent translation.
BTE-mediated translation is primarily eIF4G dependent
The key role of eIF4F in BTE-dependent translation prompted us to test the functional importance of its individual subunits. By adding increasing quantities of eIF4E, eIF4G, or eIF4F to factor-depleted extract programmed with BLucB mRNA, we found that eIF4E alone does not stimulate translation, whereas eIF4G alone stimulated translation significantly, and the two factors together (eIF4F) conferred significantly more translation than eIF4G alone (Fig. 4A).
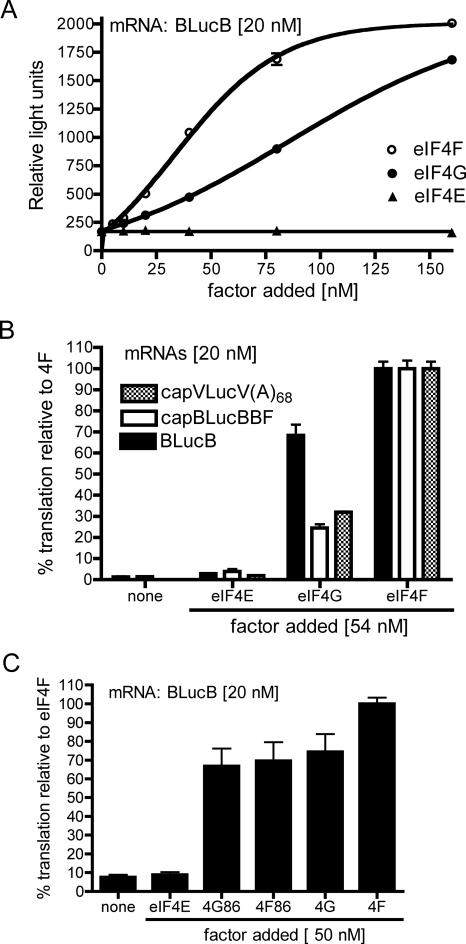
FIGURE 4.
Effect of added eIF4F and its individual subunits on translation in factor-depleted extracts. (A) Wge depleted of cap-binding complex (50 μL) (as in Fig. 3) was supplemented with indicated amounts of eIF4F or its subunits (eIF4E, eIF4G) and programmed with BLucB mRNA (20 nM). (B) Depleted wge (50 μL) was supplemented with indicated factors and programmed with BLucB, capped BLucBBF mRNA (cap-BLucBBF), and capped reporter RNA containing vector-derived UTRs and a 68-nt poly(A) tail [capVLucV(A)68] (final concentration 20 nM). The level of translation was expressed as a percentage of relative light units (RLU) obtained from luciferase translated in each extract supplemented with eIF4F. In the presence of eIF4F (defined as 100% for each mRNA), luciferase expression for capVLucV(A)68 was 3000 RLU, while capBLucBBF, and uncapped BLucB RNAs yielded between 5000 and 6000 RLU. (C) Depleted wge (50 μL) was supplemented with indicated factors and programmed with BLucB (20 nM). The 86-kDa truncation mutant of eIF4G, lacking the eIF4E recognition site is indicated as 4G86. The 1:1 mixture of eIF4E and 4G86 is indicated as 4F86. The level of translation is expressed as the percentage of luciferase produced in the extract supplemented with eIF4F. The data are averages of triplicates from at least two experiments, and error bars represent standard error.
To compare the effects of these factors on BLucB with their effects on cap-dependent translation, uncapped BLucB, capped BLucBBF, and capped VLucV(A)68 RNAs were translated in factor-depleted extracts in the presence of 54 nM eIF4E, eIF4G, or eIF4F. BLucBBF mRNA differs from BLucB only by the presence of the nonfunctional BTE (BTEBF) in its 3′UTR. BLucBBF (previously known as Luc869BF) is translated efficiently only when capped (Wang et al. 1997, 1999). VLucV(A)68 RNA was transcribed from linearized pGEMLuc vector and contains 48- and 200-nt vector-derived 5′- and 3′-UTRs, respectively, with a 68-nt poly(A) tail. In these depleted extracts, translation of uncapped BLucB and capped BLucBBF RNAs gave similar levels of luciferase activity in the presence of eIF4F, but translation of capped VLucV was about 40% lower. To compare effects of the eIF4F subunits on translation of the different mRNAs, for each mRNA, luciferase levels obtained in the presence of eIF4E or eIF4G were plotted as a percentage of that obtained in the presence of eIF4F for that mRNA. eIF4E alone had little or no stimulatory effect on translation of any RNA, while added eIF4G alone facilitated translation of BLucB to 75% of the level obtained in the presence of eIF4F (Fig. 4B). Addition of eIF4E to eIF4G (to form eIF4F) stimulated translation by a further 30%. In contrast, translation of cap-dependent RNAs, such as capBLucBBF and capVLucV(A)68, was also supported by eIF4G, but only to 25%–30% of the level obtained with eIF4F (Fig. 4B), showing strong dependence of capped mRNAs on eIF4E in addition to eIF4G, as expected. These results demonstrate that uncapped BLucB RNA translates efficiently in the absence of eIF4E and that BTE-mediated translation is eIF4G dependent.
To determine whether eIF4E must interact with eIF4G to stimulate BLucB translation, we tested the effect of deleting the eIF4E-binding site from eIF4G. We expressed and purified a truncated, 86-kDa fragment of eIF4G lacking the N-terminal 765 amino acids, including the eIF4E-binding motif that spans amino acids 710–720 (RKKYSRDFLLT) (Gallie and Browning 2001). This C-terminal half of eIF4G (4G86) facilitated cap-independent translation of BLucB mRNA in depleted extracts as efficiently as did the wild-type eIF4G (Fig. 4C). However, addition of eIF4E to 4G86 did not stimulate translation further, as it did in the presence of wild-type eIF4G (4F86) (Fig. 4C). Thus, in the absence of the eIF4E-binding site in eIF4G, eIF4E is unable to stimulate BTE-mediated translation. Importantly, the C-terminal half of eIF4G has full ability to stimulate translation to the same level as wild-type eIF4G alone.
Trans-inhibition of BTE-mediated translation is reversed by eIF4G
Previously, it was found that high concentrations of the cap analog, m7GTP, inhibited BTE-mediated translation (Wang and Miller 1995). Also, the BTE inhibited both cap-independent and cap-dependent translation when added in trans to wheat germ extracts, and this inhibition is reversed by addition of eIF4F (Wang et al. 1997). We predict that the trans-inhibition is due to competition for factor binding by the free BTE. To further analyze the factor dependence of BTE-mediated translation, we determined which factors can restore translation after trans-inhibition by added BTE or m7GTP. This approach allows us to assess translation in whole, rather than depleted, wge. In extracts inhibited with a 40-fold molar excess of BTE, addition of bovine serum albumin (BSA) or eIF4E did not restore translation of BLucB, whereas addition of eIF4G or eIF4F boosted translation to 65% and 90%, respectively, of the level in uninhibited extracts (Fig. 5A). Similarly, in extracts inhibited with 80 μM m7GTP, neither BSA nor eIF4E affected translation, while eIF4G restored 45% of translation and eIF4F boosted translation to 70% of the level in the absence of m7GTP (Fig. 5B). In general, the pattern of translation recovery by eIF4G and eIF4F in inhibited extracts is similar to that observed in factor-depleted wge (Fig. 4).
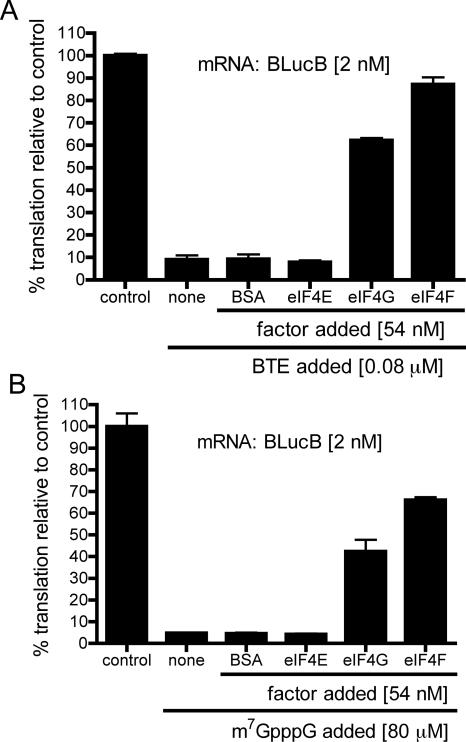
FIGURE 5.
Restoration of BTE-dependent translation by eIF4F in wheat germ extract inhibited by addition of BTE or cap analog. BTE105 RNA (A) or m7GpppG (B) and indicated factors were added to wheat germ extract, which was next programmed with BLucB RNA. The level of translation is expressed as a percentage of luciferase produced in the uninhibited extract (control). The data shown are averages of triplicates from at least two experiments and error bars represent standard error.
BTE interacts with eIF4F via direct eIF4G binding
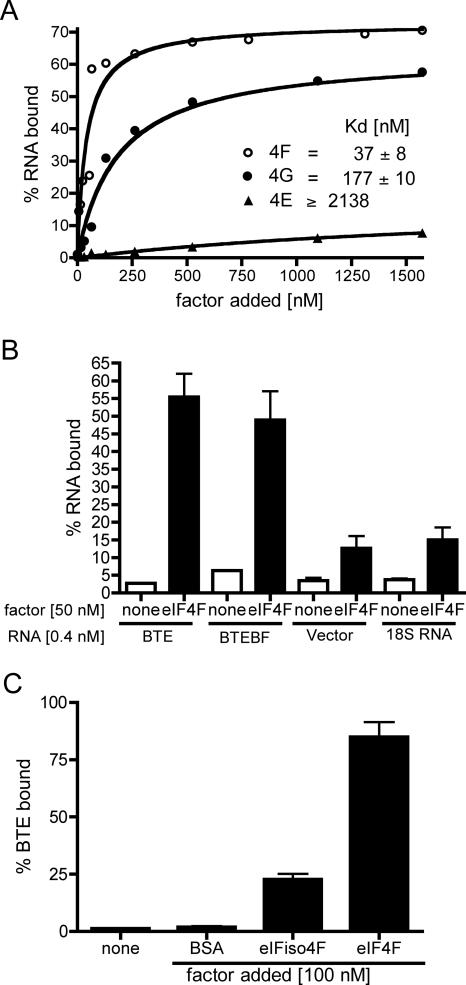
Given the key role of eIF4F and its eIF4G subunit, we quantitatively determined their affinity for the BTE using a double-membrane filter-binding assay. The BTE-binding curves yielded apparent dissociation constants of approximately > 2000, 177, and 37 nM for eIF4E, eIF4G, and eIF4F, respectively (Fig. 6A). These binding curves are remarkably similar to the translation stimulation curves in Fig. 4A. Thus, there is a strong correlation between binding affinity of the factors to the BTE and their ability to stimulate BTE-mediated translation.
FIGURE 6.
Binding of the BTE to initiation factors. (A) Binding curves to calculate apparent equilibrium dissociation constants (Kd) of eIF4F and its subunits for BTE RNA. [α-32P]-labeled BTE (0.4 nM) was mixed with indicated amounts of factors. Both protein-bound and unbound RNAs were measured in a double membrane filter-binding assay as described in Materials and Methods. Each point represents an average from at least three independent experiments. Data were fitted using GraphPad software and Kd values were calculated from equations of the best-fitted curves. (B) Filter binding of eIF4F (50 nM) to [α-32P]-labeled BTE, BTEBF, a 200-nt vector-derived RNA, and a 100-nt fragment of 18S rRNA. Binding of α-32P-labeled RNAs to nitrocellulose in the absence of eIF4F is shown in the bar indicated as none. Final concentration of all RNAs was 0.4 nM. The data are averages from at least two independent experiments performed in triplicate. Error bars indicate standard error. (C) Binding of bovine serum albumin (BSA), eIFiso4F, and eIF4F to [α-32P]-labeled BTE (0.4 nM) was performed as described in B.
To determine the specificity of binding, we measured the binding affinity of eIF4F to other RNAs. Unexpectedly, eIF4F bound the nonfunctional BTEBF with nearly identical affinity as the BTE (Fig. 6B; data not shown). In contrast, eIF4F had a much lower affinity for a vector-derived RNA and a fragment of 18S ribosomal RNA (Fig. 6B). Thus, binding is specific in these assays, but the GAUC duplication in BTEBF that knocks out cap-independent translation and ability to interact with factors in wheat germ extract (Figs. 2, 3) does not weaken binding of pure eIF4F to the RNA.
We also compared BTE binding with eIF4F with binding to eIFiso4F. At 100 nM factor concentrations, eIFiso4F bound 28% as much BTE as did eIF4F (Fig. 6C). This correlates remarkably well with the relative efficiencies of the two factors in facilitating translation in factor-depleted wge, where eIFiso4F was one-fourth as efficient as eIF4F (Fig. 3C).
DISCUSSION
The evidence presented here supports a mechanism in which the 3′ BTE facilitates translation by recruiting initiation factors and delivering them to the 5′ UTR, where translation begins. We showed previously that the long-distance interaction occurs by base pairing of the BTE to the 5′ UTR (Guo et al. 2001; Rakotondrafara et al. 2006). Here, we show that eIF4F and eIFiso4F are among the components of the translation machinery recruited to the BTE. This is consistent with previous results showing that exogenous eIF4F restores translation in extracts inhibited by BTE added in trans (Wang et al. 1997). Evidence for a role for eIF4F includes: (1) the BTE specifically pulled down eIF4F and eIFiso4F subunits from wge; (2) the BTE specifically cross-links to eIF4F and eIFiso4F; (3) low levels of eIF4F (and to a lesser extent, eIFiso4F) facilitate BTE-dependent translation in extracts depleted of cap-associated factors; (4) eIF4F restores translation of extracts inhibited by addition of excess BTE RNA or cap analog; and (5) purified eIF4F binds to the BTE with a high affinity (Kd ≅ 37 nm).
Roles of eIF4F subunits: eIF4E and eIF4G
Unlike in cap-dependent translation, eIF4G is necessary and sufficient for BTE-mediated translation, while eIF4E appears to play a lesser role in enhancing the activity of eIF4G. This is revealed by the mere 30%–50% stimulation of translation of BLucB by adding eIF4E together with eIF4G (i.e., eIF4F). In contrast, translation of cap-dependent messages was three- to fourfold more efficient in the presence of eIF4F relative to eIF4G alone (Fig. 4B). The high level of BTE-mediated translation is likely due to the high binding affinity of the factors for the BTE. There is a striking correlation between BTE-binding affinity of eIF4E (virtually none), eIF4G (Kd = 177 nm) and eIF4F (Kd = 37 nm) for the BTE (Fig. 6A), and their ability to stimulate translation (Fig. 4A). Thus, binding affinity to translation factors is at least one key component of the mechanism by which the BTE mediates translation.
It appears that eIF4E must bind eIF4G to stimulate translation, because deletion of the N-terminal half of eIF4G, including the eIF4E-binding site, eliminates stimulation by eIF4E (Fig. 4C). It is noteworthy that the C-terminal half of eIF4G facilitates BTE-mediated translation as efficiently as full-length eIF4G. Thus, we deduce that the C-terminal half of eIF4G contains the BTE binding site(s). This is consistent with the positions of the functional domains identified in wheat eIF4G (Kim et al. 1999; Gallie and Browning 2001), and the well-characterized mammalian eIF4G (Prevot et al. 2003a; Hinton et al. 2007; Pestova et al. 2007). Most of the key domains required for eIF4G function, including domains for binding RNA, eIF4A, and eIF3 are located in the C-terminal half. The missing N-terminal half includes the eIF4E-binding domain and the predicted PABP-binding site (amino acids 44–62, K. Treder, unpubl.), which is unlikely to be necessary because poly(A) tails do not affect mRNA translation efficiency in wge.
The BTE may have a particularly high affinity for either the known (Kim et al. 1999) or a second predicted (Allen et al. 1992; Gallie and Browning 2001) RNA-binding domain (RBD) in the C-terminal half of wheat eIF4G. In the C-terminal half of human eIF4GI, a 40 amino acid RBD is essential for general binding to the 5′ end of capped and uncapped mRNAs (Prevot et al. 2003b), and its central domain binds picornaviral IRESes specifically (Pestova et al. 1996; Lomakin et al. 2000).
In view of the minor role played by eIF4E, it is intriguing that addition of cap analog (m7GpppG) to wge inhibits BTE-mediated translation (Wang and Miller 1995), because this implies a need for an empty cap-binding pocket in eIF4E. Similarly, Hepatitis A virus (HAV) IRES-mediated translation is stimulated by eIF4E and sensitive to cap analog (Jackson 2005). So how does eIF4E influence eIF4G in BTE-dependent translation? It is likely that eIF4E binding to eIF4G induces conformational changes that enhance the affinity of eIF4G for the BTE. It has been reported that apo-eIF4E and the eIF4E-binding domain of eIF4G are mostly unstructured until they bind each other, inducing large folding transitions in both proteins (Gross et al. 2003; von der Haar et al. 2006). Accommodation of the cap analog to the cap-binding pocket of eIF4E bound to eIF4G induces minor additional changes throughout the entire complex (Gross et al. 2003; von der Haar et al. 2006). Thus, it is possible that eIF4E may place eIF4G in a conformation that is optimal both for BTE binding and for recruitment of the ribosome (e.g., eIF3 binding).
Functional evidence for eIF4G and eIFiso4G interacting with the BTE was shown previously in a study of pokeweed antiviral protein (PAP). PAP is an N-glycosidase that depurinates ribosomal RNA and certain viral RNAs (Wang and Hudak 2006). Wang and Hudak (2006) showed that PAP is recruited to the target mRNA via binding eIF4G or eIFiso4G. In wge, this interaction facilitates degradation of capped viral RNAs as well as an uncapped mRNA containing the BTE. Uncapped RNA containing the nonfunctional BTEBF mutation was not degraded by PAP (Wang and Hudak 2006), suggesting that eIF4G could not bind, which is consistent with our observation that BTEBF did not interact with eIF4F or eIFiso4F in wge (Figs. 1, 2).
The eIF4F-binding domain on the BTE is unknown. One candidate is stem–loop I (SL-I) in the conserved 17-nt sequence. The loop of SL-I, GGAAA, fits the consensus of the pentaloop in bacteriophage λ boxB RNA: GNRNA. Like the BTE, boxB RNA recruits a complex of two proteins, in this case the λ N protein, followed by E. coli NusA (Legault et al. 1998). We speculate that the GGAAA loop of the BTE may function similarly to recruit eIF4G and eIF4F.
RNA-binding properties of eIF4G and eIF4F
It is interesting that purified eIF4F (Fig. 6B) (as well as pure eIF4G, data not shown) bound the nonfunctional BTEBF RNA in the filter-binding assay with the same affinity with which it bound the functional BTE. In contrast, in wheat germ extract these factors did not bind BTEBF, nor did the BTEBF facilitate translation. There are at least two possibilities to explain this apparent discrepancy. First, there may be proteins or conditions in the wge that prevent binding of eIF4F to BTEBF RNA, or which specifically enhance the affinity of BTE, but not BTEBF RNA, for eIF4F. Binding assays are performed with purified factors in buffer, whereas translation assays take place in crude cell lysates that contain many proteins that may affect factor-BTE binding. Indeed, many proteins other than eIF4F and eIFiso4F were pulled down by the BTE (Fig. 1B). Their identity and role in BYDV translation, if any, remains unknown. Furthermore, depletion of the wge of cap-associated factors by m7GTP-Sepharose chromatography removes translation activity that is not restored fully even by adding high levels of eIF4F. Thus, while eIF4F is crucial, other components participate in BTE-mediated translation. The BTEBF RNA differs from the BTE only by presence of a four-base duplication (Fig. 1A) and it forms a very similar secondary structure in buffer similar to that used for filter binding (Guo et al. 2000). Thus, it is not entirely unexpected that pure eIF4G or eIF4F cannot discriminate between BTE and BTEBF outside of the cell or wheat germ extract.
A second possibility for the apparent lack of discrimination between BTE and BTEBF by pure eIF4G or eIF4F is that the kinetics may control the specificity of the eIF4F–BTE interaction. Our filter binding assays detect only the equilibrium dissociation constant, not changes in on–off rates. An example of the importance of binding kinetics was seen in mutagenesis of the neuronal protein HuD. Removal of one of its three RRM domains did not change the overall equilibrium of the interaction with its target RNA. However, this deletion increased the association–dissociation rate and rendered HuD nonfunctional (Katsamba et al. 2002). Future experiments will be necessary to determine whether this is the case with interactions of eIF4F/eIF4G with the BTE and BTEBF RNAs. It should be noted that eIF4G has a nonspecific RNA-binding domain (Kim et al. 1999), which explains why we also see some binding to 18S rRNA and vector RNA (Fig. 6B).
Roles of translation factors in other 3′ cap-independent translation elements
Like the BTE, the IRES in the 5′UTR of Tobacco etch virus (TEV) binds preferentially to eIF4G (Kd = 100 nm) with much higher affinity than eIFiso4G (Kd = 2.25 μm) (Ray et al. 2006). Moreover, eIF4G was determined as the primary factor for restoring translation of TEV IRES-containing constructs in eIF4F/iso4F-depleted wge (Gallie 2001). Interestingly, we see no structural similarities between the BTE and the TEV IRES.
The TED element in the 3′ UTR of STNV RNA is functionally very similar to the BTE, despite the lack of apparent sequence or structural similarity (van Lipzig et al. 2002; Gazo et al. 2004; Kneller et al. 2006). TED differs from the BTE by binding eIF4E and eIFiso4E in the absence of eIF4G or eIFiso4G (Gazo et al. 2004). This does not involve the cap-binding pocket, because STNV is insensitive to cap-analog inhibition (Smith and Clark 1979; Fletcher et al. 1990). However, binding to eIF4E/iso4E is more than 10 times tighter in the presence of eIF4G or iso4G. TED binds eIF4F and eIFiso4F with Kd's of 17–30 and 33–50 nM, respectively, depending on the assay (Gazo et al. 2004). These are similar to the BTE–eIF4F interaction (37 nM) (Fig. 6A), but TED differs from the BTE by its higher affinity for eIFiso4F. Corresponding to the difference in binding affinities, in vitro translation of STNV-1 RNA requires only twofold more eIFiso4F than eIF4F (Browning et al. 1992). These differences in the interactions with eIF4F and eIFiso4F, and in dependence on these factors, suggest that BYDV and STNV have evolved CITEs that achieve the same goal, usurpation of host factors for viral cap-independent translation by different mechanisms.
The different CITEs may act as natural initiation factor-binding aptamers. Indeed, some human eIF4GI-binding aptamers, obtained by SELEX, inhibit translation (Miyakawa et al. 2006), possibly by the same mechanism as the BTE in trans. SELEX has also yielded 3′ UTR sequences that modestly stimulate cap-independent translation in cis (Nagao and Obokata 2006). By analogy, the BTE and the TEV IRES may have evolved as aptamers for eIF4G, while STNV TED is an eIF4E/eIFiso4E-binding aptamer.
Genetic evidence consistent with a role for eIF4F in cap-independent translation of plant viral RNAs has accumulated recently. A growing number of plant viruses with uncapped RNAs has been found to be unable to infect hosts containing mutations or deletions in components of eIF4F or eIFiso4F (Diaz-Pendon et al. 2004; Robaglia and Caranta 2006). Natural mutations in eIF4G and eIF4E confer recessive resistance to Turnip crinkle virus and Melon necrotic spot virus, respectively (Yoshii et al. 2004; Nieto et al. 2006). These viruses are in the Tombusviridae family, to which BYDV is closely related (Miller et al. 2002), and they rely on a 3′ CITE for translation (Qu and Morris 2000; Diaz et al. 2004). While the mechanism of resistance is unknown, it is possible that these mutations prevent the factor from interacting with the viral CITE and facilitating cap-independent translation.
A model for BTE-mediated cap-independent translation
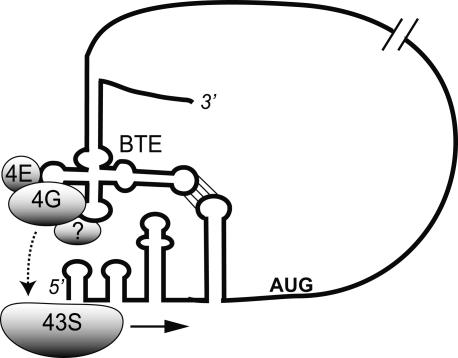
In summary, it is clear that the BTE interacts with and requires eIF4G for cap-independent translation and probably does so in the complex with eIF4E. We propose that the BTE recruits ribosomes by the mechanism shown in Figure 7. This model is based on data presented here and our previous work, which demonstrated a requirement for base pairing between the 3′ BTE (or adjacent 3′ UTR sequence) and the 5′ UTR to facilitate cap-independent translation initiation at the 5′-proximal AUG (Guo et al. 2001; Rakotondrafara et al. 2006). We also showed that 3′ BTE-mediated translation requires ribosome scanning through the 5′ UTR (Guo et al. 2001; Rakotondrafara et al. 2006). According to our model, eIF4F, and possibly other host proteins, interact directly with the BTE and are delivered to the 5′ UTR by the long-distance base pairing. This places eIF4F in close proximity to the 5′ end. This, in turn, facilitates entry of the 43S ribosomal complex, which scans by the normal eIF4G-facilitated process (Pestova et al. 2007) to the first AUG codon, where protein synthesis ensues. We predict that the 43S complex is not recruited directly to the BTE because, in that case, the BTE would be expected to function as an IRES, which is not supported by previous evidence (Allen et al. 1999). However, the exact ribosome-binding site remains to be determined. The proposed recognition of the 5′ end of uncapped mRNA by the 43S complex is supported by observations that it can bind and scan on unstructured 5′ UTRs in the absence of eIF4 factors (Pestova and Kolupaeva 2002). The BYDV genomic 5′ UTR is highly structured (Fig. 7) (Guo et al. 2001). Therefore, BLucB is eIF4F, and thus, BTE dependent. Finally, observations that some plant and mammalian virus IRESes can stimulate translation of upstream AUGs (Jaag et al. 2003; Herbreteau et al. 2005; Nicholson et al. 2006; Junemann et al. 2007) may be explained by mechanisms similar to that shown in Figure 7.
FIGURE 7.
Proposed model for 3′ BTE-mediated recruitment of translational machinery to viral mRNA. For simplicity, only the factors relevant to this report are shown. eIF4F (4G + 4E) binds the BTE structure in the 3′ UTR. The BTE and eIF4E bind eIF4G directly, while eIF4E and unidentified pulled-down proteins (indicated by question mark) may or may not bind the BTE directly. Long-distance base-pairing (parallel lines connecting a stem–loop in the BTE with the stem–loop adjacent to the AUG) juxtaposes the BTE-bound factors near the 5′ end. This delivers eIF4F and possibly other factors to the 43S ribosomal complex at the 5′ end (dashed arrow). eIF4G and possibly other factors interact with the ribosome to facilitate ribosomal scanning (horizontal arrow). See text for details. Reproduced with permission from Miller et al. (2007); ©2007 the Biochemical Society.
MATERIALS AND METHODS
Plasmid construction
Reporter plasmids were constructed as described previously (Guo et al. 2000). Plasmid pET28a_4G86 was constructed for expression of the truncated version of eIF4G (4G86), lacking the N-terminal 765 amino acids. A 2221-nt long fragment was generated by PCR using pET3d harboring the eIF4G ORF as a template. Forward primer (BamHI_TEV_4G-p86) includes a BamHI site in the 5′ end, followed by nucleotides coding for the tobacco etch virus (TEV) proteinase cleavage site, and sequence corresponding to nucleotides 2296–2314 on eIF4G cDNA: (in bold): AAGGATCCGAAAACCTGTATTTTCAGTCTATGAGACCAACATCTCGCGGTG. Reverse primer XhoI-4G-3′r contains at its 5′-end and XhoI site followed by sequence complementary to nucleotides 4464–4447 in the eIF4G ORF (in bold): AACTCGAGTTATTAAGTCAACATGAAGGCATC. The PCR product was cut with BamHI and XhoI and cloned into plasmid pET28a that had been digested with the same enzymes.
RNA transcription
Plasmid templates were linearized by restriction digestion or amplified by PCR to ensure correct RNA length. The RNAs were synthesized by in vitro transcription with T7 or SP6 polymerase using Megascript (for uncapped RNAs) or mMessage mMachine (for capped RNAs) kits (Ambion). RNAs used as probes in filter-binding assays were synthesized according to the Promega small-scale transcription protocol using [α-32P]CTP as a label. Unincorporated nucleotides were removed on a BioRad P30 spin column. RNA integrity was verified by 1% agarose gel electrophoresis.
Isolation of BTE-interacting proteins (BTEIPs)
Bait RNAs were biotin labeled at the 3′-terminus by a modification of the method of von Ahsen and Noller (1995). RNAs were oxidized by adding an equal volume of RNA (3 nmol total) to fresh 100 mM NaIO3, to a total volume of 100 μL, followed by a 1-h incubation in the dark at room temperature. An equal volume of 50% ethylene glycol was added and incubation continued for 15 min in the dark to destroy any remaining periodate. Oxidized RNA was precipitated with ethanol and dissolved in 80 μL of H2O. Biotin amidocaproyl hydrazide (Sigma) in DMSO (20 μL) was added to the RNA to a final concentration of 10 mM each and incubated for 2 h at 37°C. A total of 100 μL of 0.2 M sodium borohydride and 200 μL of 1 M Tris-HCl (pH 8.2) were added and incubated 30 min on ice in the dark. The RNA was precipitated with ethanol, redissolved in ddH2O, and purified on a BioRad P30 spin column. Magnetic beads (Promega) conjugated to streptavidin were washed three times in 0.5X SSC. One nanomole of biotinylated RNA was added to the beads in 0.5X SSC, and incubated for 10 min at room temperature. Beads were captured using a magnetic stand and washed four times with 0.1X SSC. A total of 500 μL of wheat germ extract plus 500 μL of 2X binding buffer (40 mM Tris-HCl at pH 7.5, 100 mM potassium acetate, 4 mM DTT, 4 mM MgCl2, 2 mM EDTA, 10% glycerol) were added to the beads and incubated 10 min at room temperature. Unbound proteins were removed by three to seven washes with 1X binding buffer containing 5 μg/mL tRNA (Sigma). Smaller volume washes were used in Figure 1D than in Figure 1B; thus, in Figure 1D the binding buffer was not sufficiently diluted by wash buffer in the first wash to elute much of the nonspecifically bound protein. By the second wash, the column was equilibrated with the wash buffer, thus removing the majority of nonspecifically bound protein Bound protein (Fig. 1B, lanes 7–11) was eluted by high salt or by heating to 95°C in 1X SDS-PAGE loading buffer (50 mM Tris-HCl at pH 6.5, 2% SDS, 15% glycerol, 0.72 M 2-mercaptoethanol, 0.01% bromophenol blue) for 5 min.
Protein expression and purification
His-tagged eIF4E and eIFiso4E in pET23d vectors were introduced into E. coli (BL21 cells) and expression was induced, at ODλ=600 ∼ 0.8, with 100 mM IPTG. Four hours after induction, cells were harvested from 1 L of culture by centrifugation at 10,000g for 10 min. The cells were frozen in −80°C for at least 1 h and sonicated 12 times for 30 sec each with 2 min cooling on ice in binding buffer (25 mM HEPES-KOH at pH 7.6, 100 mM KCl, 2 mM MgCl2, 10% glycerol plus 0.1 mM phenylmethyl-sulphonyl fluoride, 0.1% Soybean trypsin inhibitor, and 1 tablet/10 mL of Complete protease inhibitor cocktail, EDTA-free [La Roche]). The homogenate from 1 L of cells was centrifuged at 38,000g for 20 min at 4°C and supernatant was applied to 1 mL of Ni-NTA Superflow Cartridge (Qiagen). The cartridge was washed with 10 vol of binding buffer plus 10 mM imidazole and then with 10 vol of binding buffer plus 20 mM imidazole. The his-tagged proteins were eluted with 250 mM imidazol in the same buffer.
Recombinant (wild-type) wheat eIF4F and eIFiso4F were expressed from dicistronic constructs in a pET3D vector harboring eIF4G and eIF4E or eIFiso4G and eIFiso4E from wheat, respectively, and purified as described (Mayberry et al. 2007). The dicistronic plasmids were introduced into E.coli (BL21 cells) and induced with 0.1 M IPTG. Four hours post-induction, cells were harvested by centrifugation and sonicated prior to purification. The lysates were loaded onto a phosphocellulose column, followed by a m7GTP sepharose affinity column, and lastly, the protein was concentrated on a second phosphocellulose column. The proteins were dialyzed against N′-100 (25 mM Hepes-KOH at pH 7.6, 100 mM KCl, 1 mM MgCl2, 1 mM DTT) to remove excess m7GTP, and concentrated on Microcon YM-10 (Amicon) with three changes of N′-100. Recombinant scaffold proteins were expressed from pET3d harboring wheat eIF4G (Mayberry et al. 2007) and eIFiso4G (van Heerden and Browning 1994) and purified on a phosphocellulose column and centrifuged through Microcon YM-100 (eIF4G) or Microcon YM-50 (eIFiso4G). Expression and purification of p86 was performed as for eIF4G, followed by an additional step on 1 mL of Ni-NTA Superflow Cartridge (Qiagen) as described for his-tagged eIF4E. The purity of all proteins was verified by SDS-PAGE and Coomassie Brilliant Blue staining and concentration determined by Bradford assay (BioRad Protein Assay).
In vitro translation
In vitro translation reactions with cap analog, m7G(5′)ppp(5′)G, added in trans, were set up using wheat germ extract from Promega as described previously (Guo et al. 2000) or with S30 extracts prepared as described previously (Lax et al. 1986). A total of 3.2 mM MgCl2 was added to samples containing cap analog. To supplement depleted extracts, protein was diluted with N′-100 to a total volume of 5 μL. The protein was then added to wheat germ translation mix to a total volume of 50 μL. Luciferase assays were performed using the Luciferase Assay Reporter system from Promega Corporation.
Depleted extracts were prepared as described by Gallie and Browning (2001). A wheat germ extract (Promega) was loaded onto an m7GTP-Sepharose affinity column equilibrated in N′-100, and unbound fractions showing the highest protein concentration at 280 nm were harvested and pooled, and then realiquoted for storage at −80°C prior to use.
Western blotting
BTEIPs were blotted onto PVDF membrane and probed using antibodies to known initiation factors eIFiso4E, eIFiso4G, eIF4A, and eIF4B. Antibody reactive proteins were visualized using the ECF detection kit (Amersham-Pharmacia). For Western blots to detect eIF4E and eIF4G, proteins were electrophoresed in NuPage 4%–12% Bis-Tris gels (Invitrogen) and transferred to Hybond P PVDF membranes (Amersham Bioscience). Membranes were blocked in 3% milk in 1X PBS 0.1% Tween overnight, then probed with primary antibody at a dilution of 1:3000. Blots were washed and incubated with Goat Anti-Rabbit IgG (H+L) HRP conjugate (Bio-Rad) at 1:20000. Chemiluminescent detection was performed with SuperSignal West Pico substrate (Pierce). Protein ladders were included on each gel, and after transfer to the membrane, the positions of each size on the protein ladder were marked on the membrane. Following chemiluminescent detection, the mobilities and sizes of BTEIPs were confirmed.
UV cross-linking assay
UV cross-linking and immunoprecipitation were carried out as described (Gazo et al. 2004). Briefly, 200 μL of wheat germ S30 extract was incubated with 24 mM Hepes-KOH (pH 7.6), 2.9 mM MgAc2, 100 mM KAc, 30 mM KCl, 2.4 mM DTT, 0.1 mM spermine, 1 mM ATP, 0.2 mM GTP, 50 μM amino acids, 7.8 μM creatine phosphate, 3 μg of creatine kinase, 0.75 A260 unit of yeast tRNA, and 50 μL of [32P]BTE or 50 μL of [32P]BTEBF. The reaction mixture was incubated for 20 min at 27°C, followed by irradiation in a StrataLinker (Stratagene) for 4 min. The cross-linking reactions were then incubated with 5000 U RNase T1 for 15 min at 37°C. The reaction mixture was added to 2.5 mg of protein A Sepharose (Pharmacia) containing either 10 μL of rabbit preimmune, anti-eIF4F, or anti-eIFiso4F serum and incubated at room temperature for 2 h with mixing. After washing three times with 10 mM Tris•HCl (pH 8), 500 mM NaCl, and 0.1% NP-40, the beads were collected by heating in 50 μL of Laemmli sample buffer for 2 min at 90°C. The proteins were separated by 12.5% PAGE and detected by autoradiography.
Filter-binding assay
The binding assay was performed essentially as described by Wong and Lohman (1993). [32P]-labeled RNAs such as BTE or a 200-nt long pGEMLuc vector-derived transcript (0.4 nM) were incubated with the indicated proteins in a final volume of 50 μL of binding buffer, 25 mM HEPES-KOH (pH 7.6), 100 mM potassium acetate, 30 mM KCl, 2 mM MgCl2, 1 mM DTT, 0.1 mg/mL BSA, 50 μg/mL tRNA, 50 μg/mL poly(dI-dC), and 2.5% glycerol. Samples were filtered through nitrocellulose and hybond N++ membranes (Amersham Bioscience) in a 96-well manifold (Schleicher and Schuell) connected to a vacuum aspirator. RNA–protein complexes were retained on nitrocellulose and all free radiolabeled RNA was retained on hybond N++ (nylon) membrane. Both membranes were exposed to a PhosphorImager screen (Amersham Bioscience) and intensity of obtained spots was quantified using ImageQuant software (Molecular Dynamics). The percent of bound RNA was determined by dividing the value on the nitrocellulose membrane (RNA bound to protein) by the sum of the values on the nitrocellulose (RNA bound to protein) and nylon (unbound RNA). Data were fitted using GraphPad software (GraphPad Software, Inc.) and Kd values were calculated using the best-fitted curve. Each protein level was measured in triplicate and averaged. Each experiment was repeated at least three times.
ACKNOWLEDGMENTS
We thank Liang Guo for advice and plasmids and the Doudna lab for advice on filter-binding assays. We also thank Aurélie Rakotondrafara for assistance with Figure 7, and we thank her and Andrew Firth for critical reading of the manuscript and inspiring comments. This research was funded by grants from NIH (GM067104) to W.A.M., from NSF (MCB0214996) and The Welch Foundation (F1339) to K.S.B., and by USDA National Needs Fellowships to E.M.A. and E.L.P.K., and an Iowa State University Plant Sciences Institute Fellowship to E.L.P.K.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.777308.
REFERENCES
- Ali, I.K., McKendrick, L., Morley, S.J., Jackson, R.J. Truncated initiation factor eIF4G lacking an eIF4E binding site can support capped mRNA translation. EMBO J. 2001;20:4233–4242. doi: 10.1093/emboj/20.15.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, M.L., Metz, A.M., Timmer, R.T., Rhoads, R.E., Browning, K.S. Isolation and sequence of the cDNAs encoding the subunits of the isozyme form of wheat protein synthesis initiation factor 4F. J. Biol. Chem. 1992;267:23232–23236. [PubMed] [Google Scholar]
- Allen, E., Wang, S., Miller, W.A. Barley yellow dwarf virus RNA requires a cap-independent translation sequence because it lacks a 5′ cap. Virology. 1999;253:139–144. doi: 10.1006/viro.1998.9507. [DOI] [PubMed] [Google Scholar]
- Basso, J., Dallaire, P., Charest, P.J., Devantier, Y., Laliberte, J.F. Evidence for an internal ribosome entry site within the 5′ nontranslated region of turnip mosaic potyvirus RNA. J. Gen. Virol. 1994;75:3157–3165. doi: 10.1099/0022-1317-75-11-3157. [DOI] [PubMed] [Google Scholar]
- Batten, J.S., Desvoyes, B., Yamamura, Y., Scholthof, K.B. A translational enhancer element on the 3′-proximal end of the Panicum mosaic virus genome. FEBS Lett. 2006;580:2591–2597. doi: 10.1016/j.febslet.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Browning, K.S. The plant translational apparatus. Plant Mol. Biol. 1996;32:107–144. doi: 10.1007/BF00039380. [DOI] [PubMed] [Google Scholar]
- Browning, K.S. Plant translation initiation factors: It is not easy to be green. Biochem. Soc. Trans. 2004;32:589–591. doi: 10.1042/BST0320589. [DOI] [PubMed] [Google Scholar]
- Browning, K.S., Webster, C., Roberts, J.K., Ravel, J.M. Identification of an isozyme form of protein synthesis initiation factor 4F in plants. J. Biol. Chem. 1992;267:10096–10100. [PubMed] [Google Scholar]
- Danthinne, X., Seurinck, J., Meulewaeter, F., Van Montagu, M., Cornelissen, M. The 3′ untranslated region of satellite tobacco necrosis virus RNA stimulates translation in vitro. Mol. Cell. Biol. 1993;13:3340–3349. doi: 10.1128/mcb.13.6.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregorio, E., Preiss, T., Hentze, M.W. Translational activation of uncapped mRNAs by the central part of human eIF4G is 5′ end dependent. RNA. 1998;4:828–836. doi: 10.1017/s1355838298980372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz, J.A., Nieto, C., Moriones, E., Truniger, V., Aranda, M.A. Molecular characterization of a Melon necrotic spot virus strain that overcomes the resistance in melon and nonhost plants. Mol. Plant Microbe Interact. 2004;17:668–675. doi: 10.1094/MPMI.2004.17.6.668. [DOI] [PubMed] [Google Scholar]
- Diaz-Pendon, J.A., Truniger, V., Nieto, C., Garcia-Mas, J., Bendahmane, A., Aranda, M.A. Advances in understanding recessive resistance to plant viruses. Mol. Plant Pathol. 2004;5:223–233. doi: 10.1111/j.1364-3703.2004.00223.x. [DOI] [PubMed] [Google Scholar]
- Doudna, J.A., Sarnow, P. Translation initiation by viral internal ribosome entry sites. In: Mathews M.B., et al., editors. Translational control in biology and medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 129–153. [Google Scholar]
- Fabian, M.R., White, K.A. 5′–3′ RNA–RNA interaction facilitates cap- and poly(A) tail-independent translation of tomato bushy stunt virus mRNA: A potential common mechanism for tombusviridae. J. Biol. Chem. 2004;279:28862–28872. doi: 10.1074/jbc.M401272200. [DOI] [PubMed] [Google Scholar]
- Fabian, M.R., White, K.A. Analysis of a 3′-translation enhancer in a tombusvirus: A dynamic model for RNA–RNA interactions of mRNA termini. RNA. 2006;12:1304–1314. doi: 10.1261/rna.69506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher, L., Corbin, S.D., Browning, K.S., Ravel, J.M. The absence of a m7G cap on β-globin mRNA and alfalfa mosaic virus RNA 4 increases the amounts of initiation factor 4F required for translation. J. Biol. Chem. 1990;265:19582–19587. [PubMed] [Google Scholar]
- Gallie, D.R. Cap-independent translation conferred by the 5′ leader of tobacco etch virus is eukaryotic initiation factor 4G dependent. J. Virol. 2001;75:12141–12152. doi: 10.1128/JVI.75.24.12141-12152.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie, D.R. Translational control in plants and chloroplasts. In: Mathews M.B., et al., editors. Translational control in biology and medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 747–774. [Google Scholar]
- Gallie, D.R., Browning, K.S. eIF4G functionally differs from eIFiso4G in promoting internal initiation, cap-independent translation, and translation of structured mRNAs. J. Biol. Chem. 2001;276:36951–36960. doi: 10.1074/jbc.M103869200. [DOI] [PubMed] [Google Scholar]
- Gazo, B.M., Murphy, P., Gatchel, J.R., Browning, K.S. A novel interaction of Cap-binding protein complexes eukaryotic initiation factor (eIF) 4F and eIF(iso)4F with a region in the 3′-untranslated region of satellite tobacco necrosis virus. J. Biol. Chem. 2004;279:13584–13592. doi: 10.1074/jbc.M311361200. [DOI] [PubMed] [Google Scholar]
- Gingras, A.C., Raught, B., Sonenberg, N. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Gross, J.D., Moerke, N.J., von der Haar, T., Lugovskoy, A.A., Sachs, A.B., McCarthy, J.E., Wagner, G. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell. 2003;115:739–750. doi: 10.1016/s0092-8674(03)00975-9. [DOI] [PubMed] [Google Scholar]
- Guo, L., Allen, E., Miller, W.A. Structure and function of a cap-independent translation element that functions in either the 3′ or the 5′ untranslated region. RNA. 2000;6:1808–1820. doi: 10.1017/s1355838200001539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, L., Allen, E.M., Miller, W.A. Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol. Cell. 2001;7:1103–1109. doi: 10.1016/s1097-2765(01)00252-0. [DOI] [PubMed] [Google Scholar]
- Herbreteau, C.H., Weill, L., Decimo, D., Prevot, D., Darlix, J.L., Sargueil, B., Ohlmann, T. HIV-2 genomic RNA contains a novel type of IRES located downstream of its initiation codon. Nat. Struct. Mol. Biol. 2005;12:1001–1007. doi: 10.1038/nsmb1011. [DOI] [PubMed] [Google Scholar]
- Hershey, J.W.B., Merrick, W.C. The pathway and mechanism of initiation of protein synthesis. In: Sonenberg N., et al., editors. Translational control of gene expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 33–88. [Google Scholar]
- Hinnebusch, A.G. eIF3: A versatile scaffold for translation initiation complexes. Trends Biochem. Sci. 2006;31:553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Hinnebusch, A.G., Dever, T.E., Asano, K. Mechanism of translation initiation in the yeast. In: Mathews M.B., et al., editors. Translational control in biology and medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 225–268. [Google Scholar]
- Hinton, T.M., Coldwell, M.J., Carpenter, G.A., Morley, S.J., Pain, V.M. Functional analysis of individual binding activities of the scaffold protein eIF4G. J. Biol. Chem. 2007;282:1695–1708. doi: 10.1074/jbc.M602780200. [DOI] [PubMed] [Google Scholar]
- Imataka, H., Gradi, A., Sonenberg, N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaag, H.M., Kawchuk, L., Rohde, W., Fischer, R., Emans, N., Prufer, D. An unusual internal ribosomal entry site of inverted symmetry directs expression of a potato leafroll polerovirus replication-associated protein. Proc. Natl. Acad. Sci. 2003;100:8939–8944. doi: 10.1073/pnas.1332697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, R.J. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem. Soc. Trans. 2005;33:1231–1241. doi: 10.1042/BST0331231. [DOI] [PubMed] [Google Scholar]
- Jan, E. Divergent IRES elements in invertebrates. Virus Res. 2006;119:16–28. doi: 10.1016/j.virusres.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Jang, S.K., Krausslich, H.G., Nicklin, M.J., Duke, G.M., Palmenberg, A.C., Wimmer, E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junemann, C., Song, Y., Bassili, G., Goergen, D., Henke, J., Niepmann, M. Picornavirus internal ribosome entry site elements can stimulate translation of upstream genes. J. Biol. Chem. 2007;282:132–141. doi: 10.1074/jbc.M608750200. [DOI] [PubMed] [Google Scholar]
- Kapp, L.D., Lorsch, J.R. The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- Karetnikov, A., Lehto, K. The RNA2 5′ leader of Blackcurrant reversion virus mediates efficient in vivo translation through an internal ribosomal entry site mechanism. J. Gen. Virol. 2007;88:286–297. doi: 10.1099/vir.0.82307-0. [DOI] [PubMed] [Google Scholar]
- Karetnikov, A., Keranen, M., Lehto, K. Role of the RNA2 3′ nontranslated region of Blackcurrant reversion nepovirus in translational regulation. Virology. 2006;354:178–191. doi: 10.1016/j.virol.2006.06.028. [DOI] [PubMed] [Google Scholar]
- Katsamba, P.S., Park, S., Laird-Offringa, I.A. Kinetic studies of RNA–protein interactions using surface plasmon resonance. Methods. 2002;26:95–104. doi: 10.1016/S1046-2023(02)00012-9. [DOI] [PubMed] [Google Scholar]
- Kim, C.Y., Takahashi, K., Nguyen, T.B., Roberts, J.K., Webster, C. Identification of a nucleic acid binding domain in eukaryotic initiation factor eIFiso4G from wheat. J. Biol. Chem. 1999;274:10603–10608. doi: 10.1074/jbc.274.15.10603. [DOI] [PubMed] [Google Scholar]
- Kneller, E.L., Rakotondrafara, A.M., Miller, W.A. Cap-independent translation of plant viral RNAs. Virus Res. 2006;119:63–75. doi: 10.1016/j.virusres.2005.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh, D.C., Liu, D.X., Wong, S.M. A six-nucleotide segment within the 3′ untranslated region of hibiscus chlorotic ringspot virus plays an essential role in translational enhancement. J. Virol. 2002;76:1144–1153. doi: 10.1128/JVI.76.3.1144-1153.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh, D.C., Wong, S.M., Liu, D.X. Synergism of the 3′-untranslated region and an internal ribosome entry site differentially enhances the translation of a plant virus coat protein. J. Biol. Chem. 2003;278:20565–20573. doi: 10.1074/jbc.M210212200. [DOI] [PubMed] [Google Scholar]
- Kozak, M. Influence of mRNA secondary structure on binding and migration of 40S ribosomal subunits. Cell. 1980;19:79–90. doi: 10.1016/0092-8674(80)90390-6. [DOI] [PubMed] [Google Scholar]
- Lax, S.R., Lauer, S.J., Browning, K.S., Ravel, J.M. Purification and properties of protein synthesis initiation and elongation factors from wheat germ. Methods Enzymol. 1986;118:109–128. doi: 10.1016/0076-6879(86)18068-2. [DOI] [PubMed] [Google Scholar]
- Le, H., Tanguay, R.L., Balasta, M.L., Wei, C.C., Browning, K.S., Metz, A.M., Goss, D.J., Gallie, D.R. Translation initiation factors eIF-iso4G and eIF-4B interact with the poly(A)-binding protein and increase its RNA binding activity. J. Biol. Chem. 1997;272:16247–16255. doi: 10.1074/jbc.272.26.16247. [DOI] [PubMed] [Google Scholar]
- Legault, P., Li, J., Mogridge, J., Kay, L.E., Greenblatt, J. NMR structure of the bacteriophage λ N peptide/boxB RNA complex: Recognition of a GNRA fold by an arginine-rich motif. Cell. 1998;93:289–299. doi: 10.1016/s0092-8674(00)81579-2. [DOI] [PubMed] [Google Scholar]
- Levis, C., Astier-Manifacier, S. The 5′ untranslated region of PVY RNA, even located in an internal position, enables initiation of translation. Virus Genes. 1993;7:367–379. doi: 10.1007/BF01703392. [DOI] [PubMed] [Google Scholar]
- Lomakin, I.B., Hellen, C.U., Pestova, T.V. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 2000;20:6019–6029. doi: 10.1128/mcb.20.16.6019-6029.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano, J., Gingras, A.C., Sonenberg, N., Burley, S.K. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997;89:951–961. doi: 10.1016/s0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- Marcotrigiano, J., Lomakin, I.B., Sonenberg, N., Pestova, T.V., Hellen, C.U., Burley, S.K. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol. Cell. 2001;7:193–203. doi: 10.1016/s1097-2765(01)00167-8. [DOI] [PubMed] [Google Scholar]
- Marintchev, A., Wagner, G. Translation initiation: Structures, mechanisms, and evolution. Q. Rev. Biophys. 2004;37:197–284. doi: 10.1017/S0033583505004026. [DOI] [PubMed] [Google Scholar]
- Mathews, M.B., Sonenberg, N., Hershey, J.W.B. Origins and principles of translational control. In: Mathews M.B., et al., editors. Translational control in biology and medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 1–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo, H., Li, H., McGuire, A.M., Fletcher, C.M., Gingras, A.C., Sonenberg, N., Wagner, G. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat. Struct. Biol. 1997;4:717–724. doi: 10.1038/nsb0997-717. [DOI] [PubMed] [Google Scholar]
- Mayberry, L.K., Dennis, M.D., Allen, M.L., Ruud Nitka, K., Murphy, P.A., Campbell, L., Browning, K.S. Expression and purification of recombinant wheat translation initiation factors eIF1, eIF1A, eIF4A, eIF4B, eIF4F, eIF(iso)4F, and eIF5. Methods Enzymol. 2007;430:397–408. doi: 10.1016/S0076-6879(07)30015-3. [DOI] [PubMed] [Google Scholar]
- Merrick, W.C. Cap-dependent and cap-independent translation in eukaryotic systems. Gene. 2004;332:1–11. doi: 10.1016/j.gene.2004.02.051. [DOI] [PubMed] [Google Scholar]
- Meulewaeter, F., Danthinne, X., Van Montagu, M., Cornelissen, M. 5′- and 3′-sequences of satellite tobacco necrosis virus RNA promoting translation in tobacco. Plant J. 1998;14:169–176. doi: 10.1046/j.1365-313x.1998.00104.x. [DOI] [PubMed] [Google Scholar]
- Miller, W.A., Rasochova, L. Barley yellow dwarf viruses. Annu. Rev. Phytopathol. 1997;35:167–190. doi: 10.1146/annurev.phyto.35.1.167. [DOI] [PubMed] [Google Scholar]
- Miller, W.A., White, K.A. Long-distance RNA–RNA interactions in plant virus gene expression and replication. Annu. Rev. Phytopathol. 2006;44:447–467. doi: 10.1146/annurev.phyto.44.070505.143353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, W.A., Waterhouse, P.M., Gerlach, W.L. Sequence and organization of barley yellow dwarf virus genomic RNA. Nucleic Acids Res. 1988;16:6097–6111. doi: 10.1093/nar/16.13.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, W.A., Beckett, R., Liu, S. Barley yellow dwarf virus: Luteoviridae or Tombusviridae? Mol. Plant Pathol. 2002;3:177–183. doi: 10.1046/j.1364-3703.2002.00112.x. [DOI] [PubMed] [Google Scholar]
- Miller, W.A., Wang, Z., Treder, K. The amazing diversity of cap-independent translation elements in the 3′-untranslated regions of plant viral RNAs. Biochem. Soc. Trans. 2007;35:1629–1633. doi: 10.1042/BST0351629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa, S., Oguro, A., Ohtsu, T., Imataka, H., Sonenberg, N., Nakamura, Y. RNA aptamers to mammalian initiation factor 4G inhibit cap-dependent translation by blocking the formation of initiation factor complexes. RNA. 2006;12:1825–1834. doi: 10.1261/rna.2169406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzingo, A.F., Dhaliwal, S., Dutt-Chaudhuri, A., Lyon, A., Sadow, J.H., Hoffman, D.W., Robertus, J.D., Browning, K.S. The structure of eukaryotic translation initiation factor-4E from wheat reveals a novel disulfide bond. Plant Physiol. 2007;143:1504–1518. doi: 10.1104/pp.106.093146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao, I., Obokata, J. In vitro selection of translational regulatory elements. Anal. Biochem. 2006;354:1–7. doi: 10.1016/j.ab.2006.03.051. [DOI] [PubMed] [Google Scholar]
- Nicholson, M.G., Rue, S.M., Clements, J.E., Barber, S.A. An internal ribosome entry site promotes translation of a novel SIV Pr55Gag isoform. Virology. 2006;349:325–334. doi: 10.1016/j.virol.2006.01.034. [DOI] [PubMed] [Google Scholar]
- Niepel, M., Gallie, D.R. Identification and characterization of the functional elements within the tobacco etch virus 5′ leader required for cap-independent translation. J. Virol. 1999;73:9080–9088. doi: 10.1128/jvi.73.11.9080-9088.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto, C., Morales, M., Orjeda, G., Clepet, C., Monfort, A., Sturbois, B., Puigdomenech, P., Pitrat, M., Caboche, M., Dogimont, C., et al. An eIF4E allele confers resistance to an uncapped and nonpolyadenylated RNA virus in melon. Plant J. 2006;48:452–462. doi: 10.1111/j.1365-313X.2006.02885.x. [DOI] [PubMed] [Google Scholar]
- Pause, A., Methot, N., Svitkin, Y., Merrick, W.C., Sonenberg, N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 1994;13:1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier, J., Sonenberg, N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- Pestova, T.V., Kolupaeva, V.G. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes & Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Hellen, C.U., Shatsky, I.N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Lorsch, J.R., Hellen, C.U. The mechanism of translation initiation in eukaryotes. In: Mathews M.B., et al., editors. Translational control in biology and medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 87–128. [Google Scholar]
- Poyry, T.A., Kaminski, A., Jackson, R.J. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes & Dev. 2004;18:62–75. doi: 10.1101/gad.276504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevot, D., Darlix, J.L., Ohlmann, T. Conducting the initiation of protein synthesis: The role of eIF4G. Biol. Cell. 2003a;95:141–156. doi: 10.1016/s0248-4900(03)00031-5. [DOI] [PubMed] [Google Scholar]
- Prevot, D., Decimo, D., Herbreteau, C.H., Roux, F., Garin, J., Darlix, J.L., Ohlmann, T. Characterization of a novel RNA-binding region of eIF4GI critical for ribosomal scanning. EMBO J. 2003b;22:1909–1921. doi: 10.1093/emboj/cdg175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, F., Morris, T.J. Cap-independent translational enhancement of turnip crinkle virus genomic and subgenomic RNAs. J. Virol. 2000;74:1085–1093. doi: 10.1128/jvi.74.3.1085-1093.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakotondrafara, A.M., Polacek, C., Harris, E., Miller, W.A. Oscillating kissing stem–loop interactions mediate 5′ scanning-dependent translation by a viral 3′ cap-independent translation element. RNA. 2006;12:1893–1906. doi: 10.1261/rna.115606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, S., Yumak, H., Domashevskiy, A., Khan, M.A., Gallie, D.R., Goss, D.J. Tobacco etch virus mRNA preferentially binds wheat germ eukaryotic initiation factor (eIF) 4G rather than eIFiso4G. J. Biol. Chem. 2006;281:35826–35834. doi: 10.1074/jbc.M605762200. [DOI] [PubMed] [Google Scholar]
- Robaglia, C., Caranta, C. Translation initiation factors: A weak link in plant RNA virus infection. Trends Plant Sci. 2006;11:40–45. doi: 10.1016/j.tplants.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Sasaki, J., Nakashima, N. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J. Virol. 1999;73:1219–1226. doi: 10.1128/jvi.73.2.1219-1226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheets, K., Redinbaugh, M.G. Infectious cDNA transcripts of Maize necrotic streak virus: Infectivity and translational characteristics. Virology. 2006;350:171–183. doi: 10.1016/j.virol.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Shen, R.Z., Rakotondrafara, A.M., Miller, W.A. Trans-regulation of cap-independent translation by a viral subgenomic RNA. J. Virol. 2006;80:10045–10054. doi: 10.1128/JVI.00991-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, R.E., Clark J.M., Jr Effect of capping upon the mRNA properties of satellite tobacco necrosis virus ribonucleic acid. Biochemistry. 1979;18:1366–1371. doi: 10.1021/bi00574a037. [DOI] [PubMed] [Google Scholar]
- Tarun S.Z., Jr, Sachs, A.B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Tarun S.Z., Jr, Wells, S.E., Deardorff, J.A., Sachs, A.B. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc. Natl. Acad. Sci. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmer, R.T., Benkowski, L.A., Schodin, D., Lax, S.R., Metz, A.M., Ravel, J.M., Browning, K.S. The 5′ and 3′ untranslated regions of satellite tobacco necrosis virus RNA affect translational efficiency and dependence on a 5′ cap structure. J. Biol. Chem. 1993;268:9504–9510. [PubMed] [Google Scholar]
- van Heerden, A., Browning, K.S. Expression in Escherichia coli of the two subunits of the isozyme form of wheat germ protein synthesis initiation factor 4F. Purification of the subunits and formation of an enzymatically active complex. J. Biol. Chem. 1994;269:17454–17457. [PubMed] [Google Scholar]
- van Lipzig, R., Gultyaev, A.P., Pleij, C.W., van Montagu, M., Cornelissen, M., Meulewaeter, F. The 5′ and 3′ extremities of the satellite tobacco necrosis virus translational enhancer domain contribute differentially to stimulation of translation. RNA. 2002;8:229–236. doi: 10.1017/s1355838202018071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Ahsen, U., Noller, H.F. Identification of bases in 16S rRNA essential for tRNA binding at the 30S ribosomal P site. Science. 1995;267:234–237. doi: 10.1126/science.7528943. [DOI] [PubMed] [Google Scholar]
- von der Haar, T., Ball, P.D., McCarthy, J.E. Stabilization of eukaryotic initiation factor 4E binding to the mRNA 5′ cap by domains of eIF4G. J. Biol. Chem. 2000;275:30551–30555. doi: 10.1074/jbc.M004565200. [DOI] [PubMed] [Google Scholar]
- von der Haar, T., Gross, J.D., Wagner, G., McCarthy, J.E. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat. Struct. Mol. Biol. 2004;11:503–511. doi: 10.1038/nsmb779. [DOI] [PubMed] [Google Scholar]
- von der Haar, T., Oku, Y., Ptushkina, M., Moerke, N., Wagner, G., Gross, J.D., McCarthy, J.E. Folding transitions during assembly of the eukaryotic mRNA cap-binding complex. J. Mol. Biol. 2006;356:982–992. doi: 10.1016/j.jmb.2005.12.034. [DOI] [PubMed] [Google Scholar]
- Wang, M., Hudak, K.A. A novel interaction of pokeweed antiviral protein with translation initiation factors 4G and iso4G: A potential indirect mechanism to access viral RNAs. Nucleic Acids Res. 2006;34:1174–1181. doi: 10.1093/nar/gkj520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., Miller, W.A. A sequence located 4.5 to 5 kilobases from the 5′ end of the barley yellow dwarf virus (PAV) genome strongly stimulates translation of uncapped mRNA. J. Biol. Chem. 1995;270:13446–13452. doi: 10.1074/jbc.270.22.13446. [DOI] [PubMed] [Google Scholar]
- Wang, S., Browning, K.S., Miller, W.A. A viral sequence in the 3′-untranslated region mimics a 5′ cap in facilitating translation of uncapped mRNA. EMBO J. 1997;16:4107–4116. doi: 10.1093/emboj/16.13.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., Guo, L., Allen, E., Miller, W.A. A potential mechanism for selective control of cap-independent translation by a viral RNA sequence in cis and in trans . RNA. 1999;5:728–738. doi: 10.1017/s1355838299981979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells, S.E., Hillner, P.E., Vale, R.D., Sachs, A.B. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- Wilson, J.E., Powell, M.J., Hoover, S.E., Sarnow, P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol. Cell. Biol. 2000;20:4990–4999. doi: 10.1128/mcb.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, I., Lohman, T.M. A double-filter method for nitrocellulose-filter binding: Application to protein–nucleic acid interactions. Proc. Natl. Acad. Sci. 1993;90:5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii, M., Nishikiori, M., Tomita, K., Yoshioka, N., Kozuka, R., Naito, S., Ishikawa, M. The Arabidopsis cucumovirus multiplication 1 and 2 loci encode translation initiation factors 4E and 4G. J. Virol. 2004;78:6102–6111. doi: 10.1128/JVI.78.12.6102-6111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]