Abstract
Recent studies of prokaryotic ribosomes have dramatically increased our knowledge of ribosomal RNA (rRNA) structure, functional centers, and their interactions with antibiotics. However, much less is known about how rRNA function differs between prokaryotic and eukaryotic ribosomes. The core decoding sites are identical in yeast and human 18S rRNAs, suggesting that insights obtained in studies with yeast rRNA mutants can provide information about ribosome function in both species. In this study, we examined the importance of key nucleotides of the 18S rRNA decoding site on ribosome function and aminoglycoside susceptibility in Saccharomyces cerevisiae cells expressing homogeneous populations of mutant ribosomes. We found that residues G577, A1755, and A1756 (corresponding to Escherichia coli residues G530, A1492, and A1493, respectively) are essential for cell viability. We also found that residue G1645 (A1408 in E. coli) and A1754 (G1491 in E. coli) both make significant and distinct contributions to aminoglycoside resistance. Furthermore, we found that mutations at these residues do not alter the basal level of translational accuracy, but influence both paromomycin-induced misreading of sense codons and readthrough of stop codons. This study represents the most comprehensive mutational analysis of the eukaryotic decoding site to date, and suggests that many fundamental features of decoding site function are conserved between prokaryotes and eukaryotes.
Keywords: 18S rRNA, decoding site, translation termination, aminoglycosides, elongation fidelity
INTRODUCTION
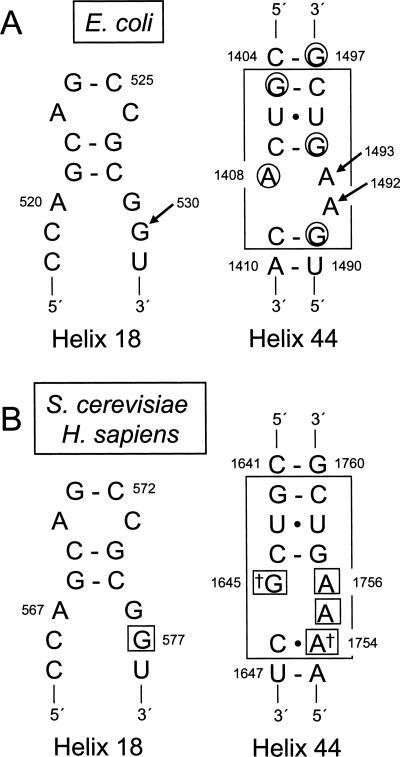
Ribosome function has been studied by biochemical and genetic approaches for decades. These studies have shown that ribosomal RNA (rRNA), as the main component of ribosomes, plays a crucial role in essentially all aspects of translation (Moore and Steitz 2002). More recently, X-ray crystallographic and cryo-EM studies of prokaryotic ribosomes have provided a wealth of structural information about various steps in translation. For example, tRNA selection is a multistep process that occurs when the anticodon of an aminoacyl-tRNA base pairs with a mRNA codon located in the ribosomal A site. Crystallographic studies have shown that three universally conserved nucleotides of Escherichia coli 16S rRNA—G530 from helix 18 and A1492 and A1493 from helix 44—probe the codon–anticodon helix to ensure the accuracy of the codon–anticodon match during translation elongation (Fig. 1A). When a cognate codon–anticodon interaction occurs, G530 transitions from a syn to an anti conformation, while A1492 and A1493 flip out into the minor groove of the codon–anticodon helix (Carter et al. 2000; Ogle et al. 2001). These conformational changes allow the decoding site to monitor the proper codon–anticodon interaction by hydrogen bonding to 2′ hydroxyl groups on either side of the cognate codon–anticodon helix, providing a sensitive test of codon–anticodon pairing.
FIGURE 1.
Decoding site residues from E. coli 16S rRNA and S. cerevisiae and H. sapiens 18S rRNAs. (A) Secondary structures of the portions of helices 18 and 44 of E. coli 16S rRNA that contribute to the decoding site. The portion of helix 18 containing residue G530 is shown on the left. Residues of the core decoding stem in helix 44 are boxed on the right (Pfister et al. 2003a). Bases protected from DMS modification by paromomycin are circled (Yoshizawa et al. 1998). Arrows indicate the decoding site residues G530, A1492, and A1493. (B) Secondary structures of corresponding portions of helices 18 and 44 in S. cerevisiae and H. sapiens 18S rRNAs. Residues corresponding to the core decoding stem in E. coli helix 44 are enclosed by the large box. Daggers mark the two bases of the decoding site not conserved between prokaryotes and eukaryotes. Small boxes indicate the residues mutated in this study.
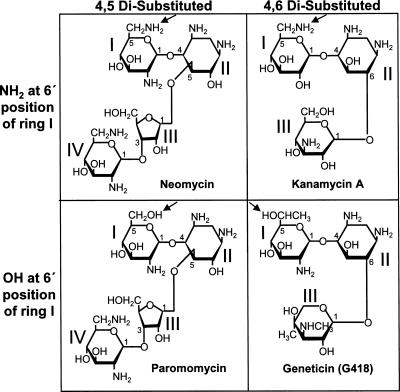
In previous studies, comparison of ribosome structures in the presence and absence of antibiotics has helped us understand the mechanism of action of compounds that inhibit protein synthesis, including the aminoglycosides (Carter et al. 2000). Aminoglycosides are clinically useful antibiotics because they inhibit prokaryotic protein synthesis at significantly lower concentrations than eukaryotic protein synthesis (Keeling and Bedwell 2005). This family of antibiotics contains a common 2-deoxystreptamine core (ring II) glycosidically linked to a glucopyranosyl (ring I) at position 4 (Fig. 2; Hobbie et al. 2006a). Based on how additional sugars are attached to ring II, aminoglycosides can be structurally divided into 4,5- and 4,6-disubstituted classes. Structural and chemical protection studies have shown that aminoglycosides bind to specific nucleotides in the decoding site of 16S rRNA in bacteria (Fig. 1A), where they induce a conformational change similar to the transition that occurs upon cognate tRNA binding (Purohit and Stern 1994; Ogle et al. 2001; Ogle and Ramakrishnan 2005). As a result, aminoglycosides interfere with bacterial protein synthesis by facilitating amino acid misincorporation.
FIGURE 2.
Structures of aminoglycosides used in this study. Structures of different aminoglycoside groups are based on linkage to ring II (4,5 disubstituted versus 4,6 disubstituted) and constituent group at 6′ position of ring I (NH2 versus OH). The relevant constituent group at the 6′ position of ring I in each structure is marked with an arrow.
rRNA genes (rDNA) are present in multiple copies in most bacterial genomes (for example, the E. coli genome contains seven rRNA operons). Since mutations that confer aminoglycoside resistance are recessive to the corresponding wild-type rDNA alleles, mutational studies to confirm the proposed model of aminoglycoside action have been difficult. For many years, the functional analysis of rRNA mutations was limited to studies of cells containing mixed populations of wild-type and mutant ribosomes (De Stasio et al. 1989; De Stasio and Dahlberg, 1990; Recht et al. 1999b). Due to the dominant nature of drug sensitivity, wild-type ribosomes in these mixed populations frequently masked the resistant phenotype of various mutations and prevented unambiguous determination of drug susceptibilities (Hobbie et al. 2006a).
Bacterial strains that express a single rDNA unit have recently been constructed that enable the analysis of mutations that confer drug resistance in homogenous populations of mutant ribosomes (Sander et al. 1996; Asai et al. 1999). This has led to several studies of the decoding site of bacterial ribosomes (Pfister et al. 2003a,b; 2005a,b). The A1408G mutation, among all decoding site mutations, conferred the most significant resistance to aminoglycosides (Sander et al. 1996; Recht et al. 1999a,b; Pfister et al. 2003a; Gregory et al. 2005; Hobbie et al. 2005). Given that a guanine lies at the corresponding position in eukaryotes, A1408 was concluded to be the main determinant of the prokaryotic specificity of aminoglycosides (Hobbie et al. 2006a). However, the A1408G mutation in E. coli 16S rRNA conferred only an intermediate level of resistance to aminoglycosides with a hydroxyl group at the 6′ position of ring I, such as paromomycin and geneticin (G418) (Recht et al. 1999b; Gregory et al. 2005). In contrast, mutations at G1491 conferred much greater resistance to these compounds (Pfister et al. 2005b). These results suggested that G1491 also plays an important role in determining bacterial sensitivity to aminoglycosides.
rDNA genes are much more highly repeated in eukaryotic organisms (Wai et al. 2000). For example, Saccharomyces cerevisiae contains ∼150 copies of a 9.1-kb repeat at the RDN1 locus on chromosome 12 (Raue and Planta 1991). Each repeat unit contains the genes for 5.8S, 18S, and 25S rRNAs that are transcribed by RNA polymerase I, as well as the 5S rRNA gene transcribed by RNA polymerase III. Previous studies have used a yeast strain in which the majority of the chromosomal rDNA repeats were deleted (Chernoff et al. 1994; Velichutina et al. 2001), with cell viability maintained by the expression of an rDNA repeat from a plasmid. However, a small number of copies of rDNA repeats remained in the genome of that strain, again resulting in mixed populations of wild-type and mutant ribosomes. More recently, Nomura and co-workers constructed an improved yeast strain in which the chromosomal rDNA repeats were completely deleted (designated rdnΔΔ) by sequential deletion steps (Wai et al. 2000). This rdnΔΔ strain allowed specific rRNA mutations to be expressed from an rDNA repeat on a high copy plasmid, resulting in a strain carrying a homogeneous population of mutant ribosomes.
In the current study, we used this rdnΔΔ yeast strain to investigate the importance of key nucleotides of the 18S rRNA decoding site on ribosome function and aminoglycoside susceptibility in eukaryotic cells. We found that residues G577, A1755, and A1756 (corresponding to E. coli residues G530, A1492, and A1493) are essential for cell viability. We also found that residues G1645 (A1408 in E. coli) and A1754 (G1491 in E. coli) both contribute to aminoglycoside resistance and influence both paromomycin-induced misreading of sense codons and readthrough of stop codons.
RESULTS
Mutagenesis of nucleotides in the yeast 18S rRNA decoding site
The decoding site within helix 44 of 18S rRNA is located in the ribosomal A site, where the anticodon of an aminoacyl-tRNA base pairs with a mRNA codon. Three universally conserved nucleotides in the E. coli decoding site—G530, A1492, and A1493—interact with the codon–anticodon helix to monitor the accuracy of base pairing (Cate et al. 1999; Ogle et al. 2001; Yusupov et al. 2001). The prokaryotic decoding site is also the target for many aminoglycosides antibiotics, which interfere with translational fidelity (Carter et al. 2000). The nucleotides of the central portion of the decoding site in helix 44 of 16S rRNA (Fig. 1A) are highly conserved among bacteria (Vicens and Westhof 2001; Pfister et al. 2003a). Nucleotides beyond this central region are more variable, and these latter differences do not influence aminoglycoside binding or sensitivity (Lynch and Puglisi 2001b; Pfister et al. 2003a).
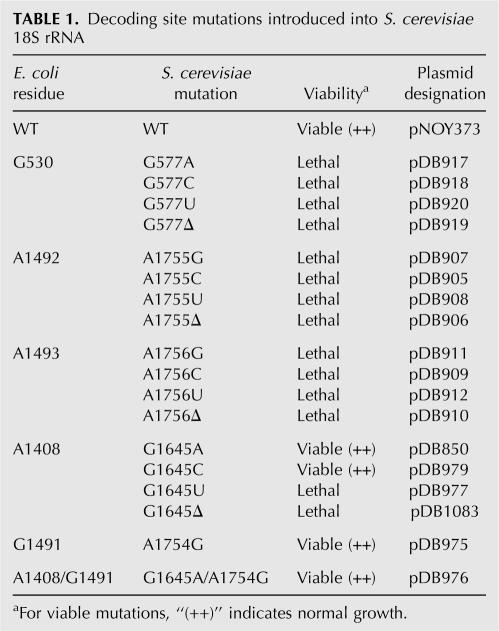
The decoding site in the 18S rRNA of the yeast S. cerevisiae shares significant homology with its prokaryotic counterpart (Fig. 1B), suggesting that essential functions of the decoding process may be conserved. In fact, only two nucleotides, G1645 and A1754 (S. cerevisiae numbering), diverge within this region between E. coli 16S rRNA and either yeast or human 18S rRNA. We initially investigated the role of conserved residues of the eukaryotic decoding site in translational fidelity. To do this, site-directed mutagenesis was first used to introduce all possible base substitutions and single base deletions of the critical nucleotides that monitor codon–anticodon pairing: yeast 18S rRNA residues G577 (E. coli G530), A1755 (E. coli A1492), and A1756 (E. coli A1493) (Fig. 1B). Plasmids expressing these mutant rRNAs under rRDN promoter control were introduced into the rdnΔΔ yeast strain NOY891 and plasmid shuffling was used to test the viability of strains expressing only these mutant rRNAs (Table 1). We found that all possible base substitutions or the deletion of each of these three nucleotides were lethal. Since these residues are also essential in E. coli 16S rRNA (Powers and Noller 1990; Yoshizawa et al. 1999), these results suggest that the function of these residues in translational fidelity is evolutionarily conserved in eukaryotes.
TABLE 1.
Decoding site mutations introduced into S. cerevisiae 18S rRNA
Genetic and structure studies have suggested that the variable 16S decoding site residues A1408 and G1491 are responsible for the sensitivity of prokaryotic ribosomes to distinct subclasses of aminoglycosides (Sander et al. 1996; Recht et al. 1999a,b; Carter et al. 2000; Vicens and Westhof 2001; Pfister et al. 2003a, 2005b). Since the functional importance of the corresponding residues in the eukaryotic decoding site has not been systematically examined, we next mutagenized the two variable bases of the yeast decoding site, G1645 and A1754. First, we introduced all possible base substitutions or a single base deletion at G1645. While the G1645U and the G1645 deletion (G1645Δ) were lethal, strains expressing 18S rRNA with the G1645A and G1645C mutations were viable and the mutant strains grew with normal growth rates (Table 1). The G1645A mutation recreated the asymmetric internal loop found in prokaryotic species (consisting of the A1408•A1493 pair and the unpaired A1492) without any detectable growth defect, indicating that this mutation does not impair the function of eukaryotic ribosomes. Consistent with this observation, the reciprocal mutation A1408G introduced into E. coli 16S rRNA also had no effect on bacterial growth (Pfister et al. 2003a). The E. coli residue G1491 (yeast A1754) has also been extensively mutagenized (Hobbie et al. 2005, 2006b; Pfister et al. 2005b). We found that the A1754G mutation also had no effect on cell growth (Table 1). Since the single G1645A and A1754G substitutions were viable, we next constructed the double mutant that converted the yeast decoding site into the bacterial version of this structure (Fig. 1). The strain with both the G1645A and A1754G mutations was viable and also did not exhibit any growth defect, suggesting that the combination of the two mutations did not interfere with normal eukaryotic ribosome function. Furthermore, the level of 18S rRNA in each of the viable mutant rRNA strains was similar to the wild-type strain (data not shown).
Decoding site mutations alter aminoglycoside resistance in yeast
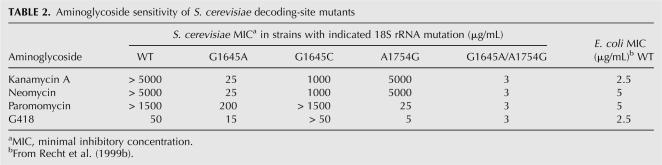
The two nonconserved nucleotides in the core of the prokaryotic decoding site, A1408 and G1491, have been shown to be involved in aminoglycoside binding (Recht et al. 1999b; Carter et al. 2000). To determine whether the corresponding nucleotides in yeast 18S rRNA, G1645 and A1754, are responsible for the aminoglycoside resistance of yeast ribosomes, we used a minimum inhibitory concentration (MIC) assay to determine the affect of mutations at these positions on aminoglycosides susceptibility. Aminoglycosides are generally categorized into 4,5 and 4,6 disubstituted classes (Fig. 2). However, recent studies have shown that the 16S rRNA of prokaryotes exhibit distinct interactions with aminoglycosides carrying a hydroxyl group versus an amino group at the 6′ position of ring I (Recht et al. 1999b; Pfister et al. 2003a, 2005b; Hobbie et al. 2005, 2006b). Because of these differences, we chose to examine the response to four aminoglycosides (Fig. 2): neomycin (4,5 disubstituted with an amino group at the 6′ position of ring I); kanamycin (4,6 disubstituted with an amino group at the 6′ position of ring I); paromomycin (4,5 disubstituted with a hydroxyl group at the 6′ position of ring I); and G418 (4,6 disubstituted with a hydroxyl group at the 6′ position of ring I). A strain expressing wild-type 18S rRNA and strains expressing 18S rRNA with the G1645A, G1645C, and A1754G single mutations or the G1645A/A1754G double mutation were tested to determine the effect of these rRNA mutations on the sensitivity to these aminoglycosides (Table 2).
TABLE 2.
Aminoglycoside sensitivity of S. cerevisiae decoding-site mutants
We found that the G1645A mutation resulted in a ∼200-fold increase in sensitivity to kanamycin A and neomycin, which both have an amino group at the 6′ position of ring I. In contrast, the G1645A mutation increased the sensitivity to paromomycin and G418 (which both have a hydroxyl group at the 6′ position of ring I) by only 7.5- and 3.3-fold, respectively. These results were consistent with the reciprocal A1408G mutation in E. coli, which conferred a high level of resistance to aminoglycosides carrying an amino group at the 6′ position of ring I but only modest resistance to aminoglycosides carrying a hydroxyl group at that position (Recht et al. 1999b). In contrast, the G1645C mutation increased sensitivity to kanamycin A and neomycin by about fivefold, did not alter sensitivity to paromomycin, and modestly reduced sensitivity to G418 (Table 2).
At the second nonconserved nucleotide of the decoding site, the A1754G mutation conferred ∼60-fold greater sensitivity to paromomycin and 10-fold greater sensitivity to G418. In contrast, the strain with the A1754G mutation exhibited only a slight increase in sensitivity to kanamycin A and neomycin. This was also consistent with the effect of the reciprocal G1491A mutation in bacteria, which conferred greater resistance to aminoglycosides carrying a hydroxyl group at the 6′ position of ring I than to aminoglycosides with an amino group at that position. The most striking changes in aminoglycoside sensitivity were observed with a strain carrying both the G1645A and A1754G mutations, which conferred sensitivity to 3 μg/mL for each of the four aminoglycosides tested. Notably, this level of sensitivity was similar to that previously reported for a wild-type E. coli strain (Table 2; Recht et al. 1999b). These results indicate that nucleotide divergence at both G1645 and A1754 is responsible for the differential sensitivity to aminoglycosides observed between prokaryotes and eukaryotes.
The effect of decoding site mutations on paromomycin-induced readthrough correlates with drug susceptibility
The mechanism by which the decoding site monitors codon–anticodon interactions during translation elongation in prokaryotes is well understood (Ogle et al. 2001). The decoding site is also thought to influence how release factors mediate translation termination, although the mechanism by which this occurs is currently unknown. We previously used a dual luciferase readthrough reporter system to monitor the efficiency of stop-codon recognition in yeast cells (Fig. 3A; Keeling et al. 2004; Salas-Marco and Bedwell 2004, 2005; Kallmeyer et al. 2006; Salas-Marco et al. 2006). To more directly assess whether mutations made at the nonconserved decoding site residues affect the efficiency of translation termination, we carried out dual luciferase readthrough assays with strains expressing rRNA carrying the G1645A, G1645C, A1754G, or G1645A/A1754G mutations in the presence or absence of paromomycin.
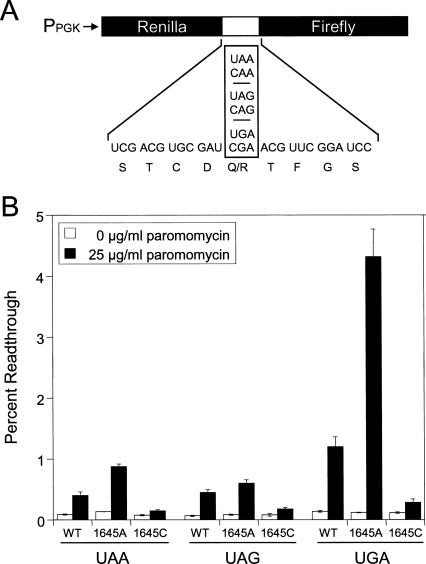
FIGURE 3.
Translational readthrough in yeast strains carrying the G1645A and G1645C mutations. (A) Schematic of the dual luciferase readthrough reporter plasmids. (B) Readthrough measured in yeast strains with the G1645A and G1645C mutations. Cells were grown in SM glucose medium at 30°C with and without 25 μg/mL paromomycin. Bars represent mean ± standard deviation.
Paromomycin is a 4,5 disubstituted aminoglycoside that increases misincorporation and suppresses nonsense mutations in yeast (Palmer et al. 1979; Singh et al. 1979; Chernoff et al. 1994; Salas-Marco and Bedwell 2005). Due to the dramatic differences in the MIC of paromomycin for strains carrying these different rRNA mutations, we first grew strains expressing wild-type, G1645A, or G1645C 18S rRNAs in the presence of 25 μg/mL paromomycin. In the absence of paromomycin, we observed minimal differences in the readthrough measured in these mutant strains (Fig. 3B). Since an increased rate of stop-codon suppression can be caused by defects in either translation elongation or termination (Salas-Marco and Bedwell 2005), these results suggest that these mutations do not significantly impair either elongation fidelity or termination efficiency. In the presence of 25 μg/mL paromomycin, the wild-type strain exhibited 4.5- to 8.6-fold increases in readthrough at the three stop codons. The G1645A mutation resulted in somewhat higher readthrough than wild type at the UAA and UAG codons (6.4- and 7.2-fold increases, respectively), but a much higher level of readthrough (36.3-fold) at the UGA stop codon. Interestingly, the G1645C mutation resulted in a much lower increase in readthrough than wild type at all three stop codons (only 1.9- to 2.4-fold). The relative readthrough observed in strains carrying the G1645A or G1645C mutations generally correlated with their respective sensitivities to paromomycin in the MIC assay.
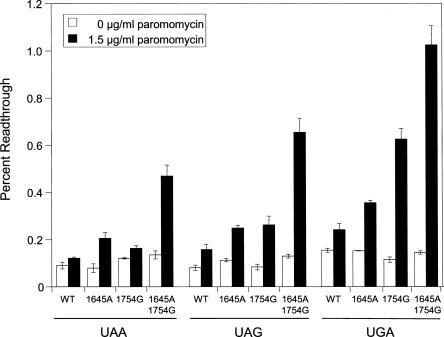
Because of the much higher paromomycin sensitivity associated with the A1754G mutation, we next grew strains expressing wild-type, G1645A, A1754G, or G1645A/A1754G 18S rRNAs in the presence of 1.5 μg/mL paromomycin (Fig. 4). In the absence of paromomycin, we again observed little or no differences in the readthrough measured in these mutant strains, suggesting that these mutations do not significantly impair elongation fidelity or termination efficiency. The wild-type strain exhibited only a slight (1.3- to 1.9-fold) increase in readthrough in the presence of this low paromomycin concentration. The strain carrying the G1645A mutation had a somewhat higher (2.2- to 2.6-fold) increase in readthrough, while the A1754G strain exhibited a 1.4- to 5.4-fold increase in readthrough. The strain carrying both the G1645A and A1754G mutations exhibited the highest readthrough at all three stop codons (3.5- to 7.1-fold) in the presence of 1.5 μg/mL paromomycin. The fact that the strain carrying both the G1645A and A1754G mutations exhibited the highest levels of paromomycin-induced readthrough was consistent with it also having the highest paromomycin sensitivity in the MIC assay.
FIGURE 4.
Translational readthrough in yeast strains carrying the G1645A, A1754G, and G1645A/A1754G mutations. Cells were grown in SM glucose medium at 30°C with or without 1.5 μg/mL paromomycin. Bars represent mean ± standard deviation.
Some decoding site mutations affect paromomycin-induced misreading
We previously developed a luciferase misincorporation reporter system that relies on an H245R mis-sense mutation to inactivate firefly luciferase activity (Fig. 5A) (Salas-Marco and Bedwell 2005). The luciferase activity of this misincorporation reporter can only be restored if a histidine is misincorporated during translation in place of the encoded arginine residue. This system provides a measure of the relative level of misreading during translation elongation and thus indicates whether elongation fidelity errors contribute to translational readthrough.
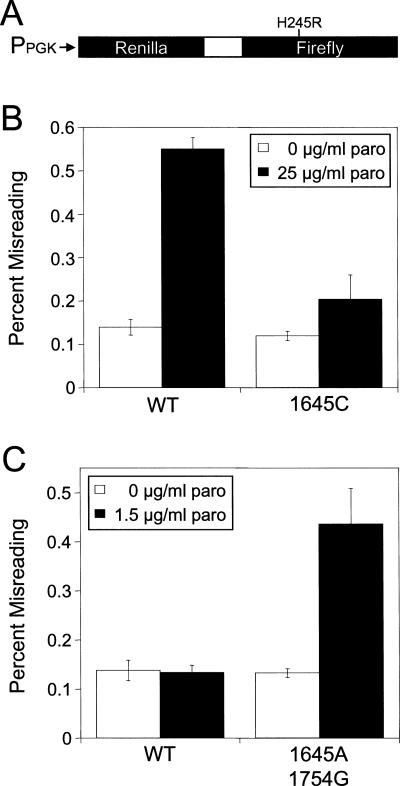
FIGURE 5.
Misincorporation associated with mutations in the yeast decoding site. (A) Schematic of the dual luciferase misincorporation reporter system. (B) Misincorporation in a yeast strain carrying the G1645C mutation. Cells were grown in SM glucose medium at 30°C with and without 25 μg/mL paromomycin. (C) Misincorporation in a yeast strain carrying the G1645A and A1754G mutations. Cells were grown in SM glucose medium at 30°C with and without 1.5 μg/mL paromomycin. Bars represent mean ± standard deviation.
We used the misincorporation assay to examine elongation fidelity in two strains expressing rRNA mutations: a strain expressing the G1645C mutation that decreased paromomycin-induced readthrough, and the G1645A/A1754G strain that was associated with the largest increase in paromomycin-induced readthrough. In the absence of paromomycin, neither mutation significantly altered the level of misreading relative to the wild-type strain, suggesting that these mutations themselves do not affect elongation fidelity (Fig. 5B,C). However, treatment with 25 μg/mL paromomycin caused a 3.9-fold increase in misreading in the wild-type strain, while the G1645C strain exhibited a much smaller (1.7-fold) increase in misreading (Fig. 5B). This indicated that the G1645C mutation suppresses paromomycin-induced misincorporation.
Because of the much higher paromomycin sensitivity observed in the strain carrying both the G1645A and A1754G mutations, we examined misreading of this strain in the presence of 1.5 μg/mL paromomycin (Fig. 5C). Growth in the presence of this low level of paromomycin did alter the level of misreading in the wild-type strain, while the strain expressing 18S rRNA with the G1645A and A1754G mutations exhibited a 3.3-fold increase in misreading. These data suggest that these mutations together enhance paromomycin-induced misreading. The altered misincorporation in these two mutant strains again correlates with their respective levels of drug sensitivity.
DISCUSSION
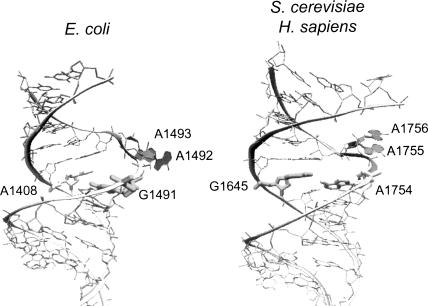
Previous studies have shown that conformational changes associated with the 16S rRNA decoding site residues G530, A1492, and A1493, are essential for prokaryotic ribosome function (Powers and Noller 1990; Yoshizawa et al. 1999; Carter et al. 2000; Ogle et al. 2001; Abdi and Fredrick 2005). In particular, cognate aminoacyl-tRNA binding causes 16S rRNA residues A1492 and A1493 to flip out from the core of helix 44 into a conformation that allows them to interact with the minor groove of the codon–anticodon helix (Fig. 6, left; Carter et al. 2000; Ogle et al. 2001). An accumulating body of evidence suggests that the corresponding region in eukaryotic 18S rRNA functions like its prokaryotic counterpart. First, most rRNA residues in the core decoding site are conserved between prokaryotes and eukaryotes. A recent structural analysis of the human decoding stem found that the residues corresponding to A1492 and A1493 also have the potential to flip out like their bacterial counterparts (Fig. 6, right; Kondo et al. 2006). Finally, in the current study we examined all possible mutations at three residues of the yeast 18S rRNA (G570, A1755, and A1756) that correspond to the critical residues of the E. coli decoding site (G530, A1492, and A1493, respectively). Notably, we found that all possible mutations at these three residues were lethal. When taken together, these findings suggest that the basic features of decoding site function are conserved between prokaryotes and eukaryotes.
FIGURE 6.
Comparison of prokaryotic and eukaryotic decoding sites. Structures of RNA fragments containing the E. coli (left) or S. cerevisiae and H. sapiens (right) decoding sites are shown. The two nonconserved residues (A1408 and G1491 in E. coli; G1645 and A1754 in S. cerevisiae) are highlighted. The conserved adenines (A1492 and A1493 in E. coli; A1755 and A1756 in S. cerevisiae) are filled and shown in the flipped-out conformation (see text for details). PDB files used to model these structures are 1J7T (left) and 2FQN (right).
A number of studies have investigated how aminoglycoside binding to the decoding stem of 16S rRNA influences ribosome function. Crystal structures of aminoglycosides bound to the prokaryotic 30S ribosomal subunit (Carter et al. 2000; Ogle et al. 2001, 2002) and to oligonucleotides containing a minimal 16S rRNA decoding site (Vicens and Westhof 2001, 2002, 2003) have shown that aminoglycoside binding (particularly the contacts made by rings I and II) induces A1492 and A1493 to flip out of helix 44 in the absence of cognate aminoacyl-tRNA binding, thus making the ribosome more prone to errors in tRNA selection. Both structural and functional studies indicate that the degree of aminoglycoside binding to the prokaryotic decoding site is greatly influenced by two nonconserved nucleotides, A1408 and G1491. A1408 hydrogen bonds with the amino or hydroxyl group of the 6′ position of ring I of aminoglycosides to form a pseudo-base-pair interaction (Vicens and Westhof 2001, 2002, 2003). In contrast, a guanine at 1408 cannot form a hydrogen bond with an amino group but can hydrogen bond with a hydroxyl group at the 6′ position of ring I. This model explains why the A1408G mutation was previously shown to cause greater bacterial resistance to aminoglycosides with an amino group as opposed to a hydroxyl group at the 6′ position of ring I (Recht et al. 1999b). In contrast, G1491 provides a stacking interaction with ring I to stabilize the pseudo-base-pair interaction between ring I and A1408, while an adenine at position 1491 weakens this interaction. It was proposed that a pseudo-base pair between a hydroxyl at the 6′ position of ring I and A1408 is too weak to allow proper drug binding to bacterial ribosomes in the context of a G1491A mutation, thus explaining why the G1491A mutation confers high-level resistance to aminoglycosides with a hydroxyl group at that position (Pfister et al. 2005b). Thus, A1408 and G1491 facilitate the proper insertion of ring I carrying either an amino or hydroxyl group into the binding site, whereas G1408 and A1491 cause the binding pocket to be shallower and less stable, thus preventing proper insertion and binding of ring I (Recht et al. 1999b; Lynch and Puglisi 2001a; Vicens and Westhof, 2001, 2003). Consistent with these findings, a recent study demonstrated that the introduction of the eukaryotic decoding site into bacteria 16S rRNA conferred aminoglycoside resistance comparable to eukaryotic ribosomes (Hobbie et al. 2007).
Our mutational analysis suggests that the eukaryotic decoding site interacts with aminoglycosides in a similar manner. The yeast 18S rRNA residues G1645 and A1754 correspond to 16S rRNA residues A1408 and G1491, respectively. Our results indicate that naturally occurring differences at these two nucleotides are responsible for the greater resistance of eukaryotes to aminoglycosides. The G1645A mutation increased sensitivity to all four types of aminoglycosides tested but caused a more pronounced susceptibility to kanamycin A and neomycin (which both carry an amino group at the 6′ position of ring I) (Table 2). These results define G1645 of yeast 18S rRNA as an important determinant of eukaryotic aminoglycoside resistance. The A1754G mutation in yeast 18S rRNA increased sensitivity to both paromomycin and G418, both of which contain a hydroxyl at the 6′ position of ring I (Table 2). Finally, the G1645A/A1754G double mutant conferred a high level of sensitivity to aminoglycosides with either an amino or hydroxyl group at the 6′ position of ring I, resulting in yeast cells that are as susceptible to these compounds as wild-type E. coli cells. Together, these results confirm that the major determinants of the differential aminoglycoside sensitivity observed between prokaryotes and eukaryotes are the yeast 18S rRNA residues G1645 and A1754 (corresponding to E. coli residues A1408 and G1491, respectively).
Unexpectedly, we also found that the G1645C mutation resulted in a unique pattern of sensitivity to various aminoglycosides. It caused much smaller decreases in resistance to kanamycin A or neomycin than the G1645A mutation, did not significantly alter paromomycin resistance, and conferred slightly greater G418 resistance than the wild-type G1645 residue. This suggests that the G1645C mutation is somewhat less favorable for the insertion of aminoglycosides with a 6′ hydroxyl on ring I than the original G1645 residue. While it is more conducive than G1645 to the insertion of aminoglycosides with a 6′ amino group on ring I, it is less prone to the binding of these compounds than the G1645A mutation. The other mutations examined at residue 1645, G1645U and G1645Δ, resulted in a lethal phenotype. The G1645U mutation may be lethal because of its ability to form a Watson–Crick base pair with A1756 (see Fig. 1B). In contrast, the G1645Δ mutation will enlarge the internal bulge in the opposite strand of the decoding stem. Both of these mutations may inhibit the ability of A1755 and/or A1756 to flip out into the minor groove of the codon–anticodon helix during tRNA selection.
Finally, our results suggest that the two nonconserved nucleotides of the eukaryotic decoding site do not participate directly in tRNA selection or stop-codon recognition, since mutations at these residues do not have significant effects on these processes under normal conditions. However, the G1645A and A1754G mutations both stimulate the misreading and readthrough induced by aminoglycosides, in accordance with their conferred drug susceptibility. Together, these results suggest that the growth inhibition of these compounds correlates with their binding affinity for the decoding site and consequently their effects on translation accuracy. This is consistent with the observation that clinically relevant aminoglycosides can be used to successfully treat bacterial infections in patients (Keeling and Bedwell 2005). Interestingly, the rdn15 mutation (identified as an A1754G mutation in 18S rRNA) was previously isolated as a suppressor of the conditional lethality associated with a yeast eRF1 mutation (sup45-R2ts) (Velichutina et al. 2001). Thus, while this nonconserved nucleotide does not appear to contribute directly to the decoding of sense codons, it clearly influences the recognition and/or function of this mutant release factor. Further genetic studies with this novel rdnΔΔ strain should allow other hypotheses of eukaryotic rRNA function to be tested.
MATERIALS AND METHODS
Strains, plasmids, and genetic methods
The S. cerevisiae strain NOY891 (MATa ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rdnΔΔ∷HIS3) was used to generate strains with homogeneous populations of ribosomes carrying the indicated rRNA mutations. In this strain, chromosomal rDNA repeats are completely deleted and cell growth is supported by the high copy plasmid pNOY353 (TRP1 selectable marker) carrying a single rDNA repeat under GAL7 promoter control (Wai et al. 2000). The multicopy plasmid pNOY373 (LEU2 selectable marker), which carries a single rDNA repeat under Pol I promoter control, was used as template for mutagenesis. The ∼3.9-kb PstI–MluI fragment containing 18S rRNA gene was subcloned into pBluescript II KS+ (HindIII→MluI) (pDB840) prior to mutagenesis. All rDNA mutations were introduced using a QuikChange site-directed mutagenesis protocol (Stratagene). After confirming the mutation by sequencing the entire PstI–MluI fragment, the fragment was cloned back into pNOY373. Mutated plasmids were introduced into NOY891 using a plasmid shuffle technique on plates containing glucose as carbon source to prevent rRNA transcription of wild-type rRNA from pNOY353. Following the loss of pNOY353, the remaining plasmid was recovered from viable strains and sequenced to confirm that the desired rRNA mutation was present. By avoiding the simultaneous transcription from both rRNA plasmids, transcription-stimulated recombination was avoided (Keil and Roeder 1984; Voelkel-Meiman et al. 1987). Others have also observed frequent recombination between two transcriptionally active rRNA plasmids using this rdnΔΔ system (Rakauskaite and Dinman 2006).
Antibiotics and determination of MIC
Antibiotics kanamycin (K-1377), neomycin (N-6386), paromomycin (P-5057), and hygromycin B (H-7772) were obtained from Sigma. G418 (Geneticin) (345,810) was obtained from Calbiochem. To determine the MIC of each aminoglycoside, cultures were grown to mid-log phase in YPD medium (Burke et al. 2000) at 30°C, and equal cell numbers (determined by A 600 units) were streaked at 30°C on YPD plates with different concentrations of aminoglycosides. The MIC of each aminoglycoside was defined as the lowest drug concentration at which growth is completely inhibited after incubation for 5 days at 30°C.
Dual luciferase readthrough assays
Dual luciferase assays were performed using the Dual Luciferase Assay System (Promega) as previously described (Keeling et al. 2004). Briefly, yeast strains were transformed with the indicated readthrough reporter plasmids. To measure readthrough of a UAAA tetranucleotide termination signal, plasmids pDB688 (CAA sense codon control) and pDB689 (UAA stop codon) were used. For readthrough of UAGA tetranucleotide termination signal, plasmids pDB728 (CAG sense codon control) and pDB730 (UAG stop codon) were used. For readthrough of UGAA tetranucleotide termination signal, plasmids pDB711 (CGA sense codon control) and pDB712 (UGA stop codon) were used. Cells were grown in SM glucose medium (Burke et al. 2000) at 30°C and ∼104 cells from each strain were assayed for luminescence with a Lumat LB 9507 luminometer (Berthold). Assays were done in quadruplicate, and the percent readthrough in each strain was expressed as the Firefly/Renilla luciferase activity ratio (nonsense) divided by the Firefly/Renilla luciferase activity ratio (sense) multiplied by 100. The measurement of readthrough efficiency under paromomycin treatment was performed in the same manner except strains with readthrough reporter plasmids were cultured with the indicated drug concentration for at least three generations prior to the assay.
Misincorporation assays
The misincorporation assay was performed using a previously described reporter system (Salas-Marco and Bedwell 2005). The misincorporation reporter constructs contain dual luciferase genes with an in-frame linker between the upstream Renilla and downstream firefly genes containing the wild-type H245 (CAC) residue (pDB688) or the H245R (CGC) mutation (pDB868). After transforming yeast strains with the misincorporation reporters, assays were performed as described above for the readthrough assay. The percent misreading was expressed as Firefly (H245R)/Renilla luciferase activity divided by the Firefly (wild type)/Renilla luciferase activity multiplied by 100. The measurement of misreading under paromomycin treatment was performed in a similar manner except the strains were cultured with the indicated drug concentration for at least three generations prior to the assay.
ACKNOWLEDGMENTS
We thank Masayasu Nomura for providing the rdnΔΔ yeast strain and rRNA plasmids and Kim Keeling for providing comments on the manuscript. This work was supported by NIH grant RO1 GM 68854 (to D.M.B.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.805208.
REFERENCES
- Abdi, N.M., Fredrick, K. Contribution of 16S rRNA nucleotides forming the 30S subunit A and P sites to translation in Escherichia coli . RNA. 2005;11:1624–1632. doi: 10.1261/rna.2118105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai, T., Zaporojets, D., Squires, C., Squires, C.L. An Escherichia coli strain with all chromosomal rRNA operons inactivated: Complete exchange of rRNA genes between bacteria. Proc. Natl. Acad. Sci. 1999;96:1971–1976. doi: 10.1073/pnas.96.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, D., Dawson, D.C., Stearns, T. Methods in yeast genetics: A Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. [Google Scholar]
- Carter, A.P., Clemons, W.M., Brodersen, D.E., Morgan-Warren, R.J., Wimberly, B.T., Ramakrishnan, V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- Cate, J.H., Yusupov, M.M., Yusupova, G.Z., Earnest, T.N., Noller, H.F. X-ray crystal structures of 70S ribosome functional complexes. Science. 1999;285:2095–2104. doi: 10.1126/science.285.5436.2095. [DOI] [PubMed] [Google Scholar]
- Chernoff, Y.O., Vincent, A., Liebman, S.W. Mutations in eukaryotic 18S ribosomal RNA affect translational fidelity and resistance to aminoglycoside antibiotics. EMBO J. 1994;13:906–913. doi: 10.1002/j.1460-2075.1994.tb06334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Stasio, E.A., Dahlberg, A.E. Effects of mutagenesis of a conserved base-paired site near the decoding region of Escherichia coli 16 S ribosomal RNA. J. Mol. Biol. 1990;212:127–133. doi: 10.1016/0022-2836(90)90309-A. [DOI] [PubMed] [Google Scholar]
- De Stasio, E.A., Moazed, D., Noller, H.F., Dahlberg, A.E. Mutations in 16S ribosomal RNA disrupt antibiotic–RNA interactions. EMBO J. 1989;8:1213–1216. doi: 10.1002/j.1460-2075.1989.tb03494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory, S.T., Carr, J.F., Rodriguez-Correa, D., Dahlberg, A.E. Mutational analysis of 16S and 23S rRNA genes of Thermus thermophilus . J. Bacteriol. 2005;187:4804–4812. doi: 10.1128/JB.187.14.4804-4812.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie, S.N., Pfister, P., Brull, C., Westhof, E., Bottger, E.C. Analysis of the contribution of individual substituents in 4,6-aminoglycoside–ribosome interaction. Antimicrob. Agents Chemother. 2005;49:5112–5118. doi: 10.1128/AAC.49.12.5112-5118.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie, S.N., Bruell, C., Kalapala, S., Akshay, S., Schmidt, S., Pfister, P., Bottger, E.C. A genetic model to investigate drug–target interactions at the ribosomal decoding site. Biochimie. 2006a;88:1033–1043. doi: 10.1016/j.biochi.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Hobbie, S.N., Pfister, P., Bruell, C., Sander, P., Francois, B., Westhof, E., Bottger, E.C. Binding of neomycin-class aminoglycoside antibiotics to mutant ribosomes with alterations in the A site of 16S rRNA. Antimicrob. Agents Chemother. 2006b;50:1489–1496. doi: 10.1128/AAC.50.4.1489-1496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie, S.N., Kalapala, S.K., Akshay, S., Bruell, C., Schmidt, S., Dabow, S., Vasella, A., Sander, P., Bottger, E.C. Engineering the rRNA decoding site of eukaryotic cytosolic ribosomes in bacteria. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallmeyer, A.K., Keeling, K.M., Bedwell, D.M. Eukaryotic release factor 1 phosphorylation by CK2 protein kinase is dynamic but has little effect on the efficiency of translation termination in Saccharomyces cerevisiae . Eukaryot. Cell. 2006;5:1378–1387. doi: 10.1128/EC.00073-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling, K.M., Bedwell, D.M. Pharmacological suppression of premature stop mutations that cause genetic diseases. Curr. Pharmacogenomics. 2005;3:259–269. [Google Scholar]
- Keeling, K.M., Lanier, J., Du, M., Salas-Marco, J., Gao, L., Kaenjak-Angeletti, A., Bedwell, D.M. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae . RNA. 2004;10:691–703. doi: 10.1261/rna.5147804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil, R.L., Roeder, G.S. Cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae . Cell. 1984;39:377–386. doi: 10.1016/0092-8674(84)90016-3. [DOI] [PubMed] [Google Scholar]
- Kondo, J., Urzhumtsev, A., Westhof, E. Two conformational states in the crystal structure of the Homo sapiens cytoplasmic ribosomal decoding A site. Nucleic Acids Res. 2006;34:676–685. doi: 10.1093/nar/gkj467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, S.R., Puglisi, J.D. Structural origins of aminoglycoside specificity for prokaryotic ribosomes. J. Mol. Biol. 2001a;306:1037–1058. doi: 10.1006/jmbi.2000.4420. [DOI] [PubMed] [Google Scholar]
- Lynch, S.R., Puglisi, J.D. Structure of a eukaryotic decoding region A-site RNA. J. Mol. Biol. 2001b;306:1023–1035. doi: 10.1006/jmbi.2000.4419. [DOI] [PubMed] [Google Scholar]
- Moore, P.B., Steitz, T.A. The involvement of RNA in ribosome function. Nature. 2002;418:229–235. doi: 10.1038/418229a. [DOI] [PubMed] [Google Scholar]
- Ogle, J.M., Ramakrishnan, V. Structural insights into translational fidelity. Annu. Rev. Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- Ogle, J.M., Brodersen, D.E., Clemons W.M., Jr, Tarry, M.J., Carter, A.P., Ramakrishnan, V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- Ogle, J.M., Murphy, F.V., Tarry, M.J., Ramakrishnan, V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- Palmer, E., Wilhelm, J.M., Sherman, F. Phenotypic suppression of nonsense mutants in yeast by aminoglycoside antibiotics. Nature. 1979;277:148–150. doi: 10.1038/277148a0. [DOI] [PubMed] [Google Scholar]
- Pfister, P., Hobbie, S., Vicens, Q., Bottger, E.C., Westhof, E. The molecular basis for A-site mutations conferring aminoglycoside resistance: Relationship between ribosomal susceptibility and X-ray crystal structures. ChemBioChem. 2003a;4:1078–1088. doi: 10.1002/cbic.200300657. [DOI] [PubMed] [Google Scholar]
- Pfister, P., Risch, M., Brodersen, D.E., Bottger, E.C. Role of 16S rRNA helix 44 in ribosomal resistance to Hygromycin B. Antimicrob. Agents Chemother. 2003b;47:1496–1502. doi: 10.1128/AAC.47.5.1496-1502.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister, P., Corti, N., Hobbie, S., Bruell, C., Zarivach, R., Yonath, A., Bottger, E.C. 23S rRNA base pair 2057–2611 determines ketolide susceptibility and fitness cost of the macrolide resistance mutation 2058A→G. Proc. Natl. Acad. Sci. 2005a;102:5180–5185. doi: 10.1073/pnas.0501598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister, P., Hobbie, S., Brull, C., Corti, N., Vasella, A., Westhof, E., Bottger, E.C. Mutagenesis of 16S rRNA C1409–G1491 base-pair differentiates between 6′OH and 6′NH3+ aminoglycosides. J. Mol. Biol. 2005b;346:467–475. doi: 10.1016/j.jmb.2004.11.073. [DOI] [PubMed] [Google Scholar]
- Powers, T., Noller, H.F. Dominant lethal mutations in a conserved loop in 16S rRNA. Proc. Natl. Acad. Sci. 1990;87:1042–1046. doi: 10.1073/pnas.87.3.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purohit, P., Stern, S. Interactions of a small RNA with antibiotic and RNA ligands of the 30S subunit. Nature. 1994;370:659–662. doi: 10.1038/370659a0. [DOI] [PubMed] [Google Scholar]
- Rakauskaite, R., Dinman, J.D. An arc of unpaired “hinge bases” facilitates information exchange among functional centers of the ribosome. Mol. Cell. Biol. 2006;26:8992–9002. doi: 10.1128/MCB.01311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raue, H.A., Planta, R.J. Ribosome biogenesis in yeast. Prog. Nucleic Acid Res. Mol. Biol. 1991;41:89–129. doi: 10.1016/s0079-6603(08)60007-0. [DOI] [PubMed] [Google Scholar]
- Recht, M.I., Douthwaite, S., Dahlquist, K.D., Puglisi, J.D. Effect of mutations in the A site of 16 S rRNA on aminoglycoside antibiotic–ribosome interaction. J. Mol. Biol. 1999a;286:33–43. doi: 10.1006/jmbi.1998.2446. [DOI] [PubMed] [Google Scholar]
- Recht, M.I., Douthwaite, S., Puglisi, J.D. Basis for prokaryotic specificity of action of aminoglycoside antibiotics. EMBO J. 1999b;18:3133–3138. doi: 10.1093/emboj/18.11.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Marco, J., Bedwell, D.M. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol. Cell. Biol. 2004;24:7769–7778. doi: 10.1128/MCB.24.17.7769-7778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Marco, J., Bedwell, D.M. Discrimination between defects in elongation fidelity and termination efficiency provides mechanistic insights into translational readthrough. J. Mol. Biol. 2005;348:801–815. doi: 10.1016/j.jmb.2005.03.025. [DOI] [PubMed] [Google Scholar]
- Salas-Marco, J., Fan-Minogue, H., Kallmeyer, A.K., Klobutcher, L.A., Farabaugh, P.J., Bedwell, D.M. Distinct paths to stop codon reassignment by the variant-code organisms Tetrahymena and Euplotes . Mol. Cell. Biol. 2006;26:438–447. doi: 10.1128/MCB.26.2.438-447.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander, P., Prammananan, T., Bottger, E.C. Introducing mutations into a chromosomal rRNA gene using a genetically modified eubacterial host with a single rRNA operon. Mol. Microbiol. 1996;22:841–848. doi: 10.1046/j.1365-2958.1996.01532.x. [DOI] [PubMed] [Google Scholar]
- Singh, A., Ursic, D., Davies, J. Phenotypic suppression and misreading Saccharomyces cerevisiae . Nature. 1979;277:146–148. doi: 10.1038/277146a0. [DOI] [PubMed] [Google Scholar]
- Velichutina, I.V., Hong, J.Y., Mesecar, A.D., Chernoff, Y.O., Liebman, S.W. Genetic interaction between yeast Saccharomyces cerevisiae release factors and the decoding region of 18 S rRNA. J. Mol. Biol. 2001;305:715–727. doi: 10.1006/jmbi.2000.4329. [DOI] [PubMed] [Google Scholar]
- Vicens, Q., Westhof, E. Crystal structure of paromomycin docked into the eubacterial ribosomal decoding A site. Structure. 2001;9:647–658. doi: 10.1016/s0969-2126(01)00629-3. [DOI] [PubMed] [Google Scholar]
- Vicens, Q., Westhof, E. Crystal structure of a complex between the aminoglycoside tobramycin and an oligonucleotide containing the ribosomal decoding a site. Chem. Biol. 2002;9:747–755. doi: 10.1016/s1074-5521(02)00153-9. [DOI] [PubMed] [Google Scholar]
- Vicens, Q., Westhof, E. Crystal structure of geneticin bound to a bacterial 16S ribosomal RNA A site oligonucleotide. J. Mol. Biol. 2003;326:1175–1188. doi: 10.1016/s0022-2836(02)01435-3. [DOI] [PubMed] [Google Scholar]
- Voelkel-Meiman, K., Keil, R.L., Roeder, G.S. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell. 1987;48:1071–1079. doi: 10.1016/0092-8674(87)90714-8. [DOI] [PubMed] [Google Scholar]
- Wai, H.H., Vu, L., Oakes, M., Nomura, M. Complete deletion of yeast chromosomal rDNA repeats and integration of a new rDNA repeat: Use of rDNA deletion strains for functional analysis of rDNA promoter elements in vivo. Nucleic Acids Res. 2000;28:3524–3534. doi: 10.1093/nar/28.18.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa, S., Fourmy, D., Puglisi, J.D. Structural origins of gentamicin antibiotic action. EMBO J. 1998;17:6437–6448. doi: 10.1093/emboj/17.22.6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa, S., Fourmy, D., Puglisi, J.D. Recognition of the codon–anticodon helix by ribosomal RNA. Science. 1999;285:1722–1725. doi: 10.1126/science.285.5434.1722. [DOI] [PubMed] [Google Scholar]
- Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H., Noller, H.F. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]