Abstract
The glycine binding riboswitch forms a unique tandem aptamer structure that binds glycine cooperatively. We employed nucleotide analog interference mapping (NAIM) and mutagenesis to explore the chemical basis of glycine riboswitch cooperativity. Based on the interference pattern, at least two sites appear to facilitate cooperative tertiary interactions, namely, the minor groove of the P1 helix from aptamer 1 and the major groove of the P3a helix from both aptamers. Mutation of these residues altered both the cooperativity and binding affinity of the riboswitch. The data support a model in which the P1 helix of the first aptamer participates in a tertiary interaction important for cooperativity, while nucleotides in the P1 helix of the second aptamer interface with the expression platform. These data have direct analogy to well-characterized mutations in hemoglobin, which provides a framework for considering cooperativity in this RNA-based system.
Keywords: riboswitch, glycine, NAIM, cooperativity
INTRODUCTION
Riboswitches are folded RNA domains that bind specific metabolites and control gene expression by modulation of transcription termination, translation initiation or RNA degradation (Winkler and Breaker 2005). These highly structured RNAs reside in the 5′ untranslated region (UTR) of mRNAs that encode genes related to the biosynthesis or degradation of the corresponding metabolite. Regulation by riboswitches typically occurs through conformational changes in the RNA induced by binding of the small effector molecule (Mandal and Breaker 2004). In most cases, metabolite binding induces the formation of the P1 helix, which effects the conformation of the downstream expression platform (Schwalbe et al. 2007).
Among 11 classes of riboswitches reported to date, the glycine riboswitch has been identified upstream of some genes that encode glycine-related proteins (Barrick et al. 2004). Unlike most riboswitches, activation by glycine up-regulates gene expression in Bacillus subtilis (Mandal et al. 2004), where the gcvT gene encodes three enzymes involved in the glycine cleavage system. Glycine is among the smallest ligands to be recognized by an RNA aptamer, second only to magnesium (Cromie et al. 2006). Despite its small size, glycine is recognized by the riboswitch with high specificity.
The secondary structure of the glycine riboswitch comprises two similar aptamer domains joined by a linker region (Fig. 1). Surprisingly, these two aptamers each can bind glycine independently, but in the tandem arrangement, the binding takes place cooperatively. Recently, tandem riboswitch architectures that act as a “genetic logic gates” were reported (Sudarsan et al. 2006). However, most of these tandem configurations are composed of two complete riboswitches including the expression platform, so they function in an independent rather than a cooperative manner. The glycine riboswitch has an interrelated double aptamer domain followed by an expression platform, which is the only known riboswitch example that uses a cooperative binary system. This special feature makes the glycine riboswitch a digital RNA sensor that is particularly sensitive to the concentration of glycine.
FIGURE 1.
Secondary structure model of the glycine riboswitch. Consensus sequences were identified from 104 different glycine riboswitch candidate by bioinformatics. Conserved nucleotides in more than 75%, 90%, and 97% of the sequences are shown as black, blue, and red, respectively. Each aptamer has two conserved paired regions (P1 and P3) and two stem–loops (P2 and P4) whose sequence is not conserved. The conserved sequences are located in the P3 stem–loop, the upper region of the P1 stem, and the J1/2 and J3/1 joiner regions on top of helix P1. The P2 and P4 stem loops are variable in length and sequence.
Cooperative ligand binding is an important mechanism of biological systems (Perutz 1989). Since the early study of oxygen binding to hemoglobin (Bohr et al. 1904), various proteins that have cooperative activity have been identified in multisubunit enzymes and receptor proteins (Koshland and Hamadani 2002), and cooperative events typically occur through allosteric interactions. Ligand binding induces a conformational change at one site that affects the ligand affinity at another site or sites. It is an economical system for the cell, because cooperativity makes it possible to sense and efficiently respond to a ligand within a narrow range of concentrations (Forsen and Linse 1995).
We set out to investigate the chemical basis of glycine riboswitch cooperativity using nucleotide analog interference mapping (NAIM) (Ryder and Strobel 1999). NAIM is a chemogenetic technique that makes it possible to rapidly identify the functional groups throughout an RNA that are important for activity. Using phosphorothioate-tagged nucleotide analogs to alter, delete, or add functional groups, every occurrence of a particular functional group in an RNA can be simultaneously characterized. NAIM has been applied to a variety of functional RNAs including ribozymes (Boudvillain and Pyle 1998; Strauss-Soukup and Strobel 2000), RNA–RNA interactions (Heide et al. 2001), and RNA–protein binding interactions (Rox et al. 2002; Szewczak et al. 2002). However, NAIM requires a method to separate active and inactive forms of the RNA based on their activity, which can pose a challenge for its application to some RNAs. Here we report the use of polyacrylamide coelectrophoresis (PACE) to distinguish functional glycine riboswitches, and we apply this method for the analysis of two versions of the glycine riboswitch. The results provide information regarding the chemical basis of ligand binding and cooperativity between the two individual glycine binding domains.
RESULTS
Characterization of the Fusobacterium nucleatum glycine riboswitch
Our first experimental goal was to identify a minimal RNA sequence that retains the ability to function as a cooperative glycine riboswitch. This objective is important both for ease of experimental analysis and to facilitate future structural investigation (Strauss-Soukup and Strobel 2000; Adams et al. 2004). The initial report of the glycine riboswitch was performed on the sequences from Vibrio cholerae (VC) and Bacillus subtilis (BS) that are 226 and 206 nucleotides (nt), respectively. From phylogenetic sequence alignment (Mandal et al. 2004), we selected a putative glycine riboswitch motif in Fusobacterium nucleatum (FN) that is only 158 nt in size but retains the conserved features of the glycine riboswitch. We tested if the FN RNA shares the biochemical properties reported for the larger glycine riboswitches. Glycine binding affinity and cooperativity were measured by the in-line probing assay (Soukup and Breaker 1999). Patterns of spontaneous cleavage, primarily in the unpaired loop regions, occurred in a glycine-dependent manner similar to those reported for the VC riboswitch (Fig. 2A). In the presence of glycine, the core conserved central loop was protected from cleavage, while the linker region between the two aptamers showed enhanced cleavage. The FN RNA also showed molecular discrimination characteristics similar to those of VC (Fig. 2B). It bound glycine, exhibited reduced affinity for the glycine methyl ester, ethyl ester, and t-butyl ester, and showed no affinity for all other analogs. The FN RNA binds glycine with an apparent dissociation constant of 20 μM (Fig. 2C), which is similar to the 30 μM Kd reported for the VC RNA. The Hill coefficient for the FN RNA is 1.4, indicating positive cooperativity. This is comparable to the values of 1.6 and 1.4 for the VC and BS riboswitches, respectively (Mandal et al. 2004). Together these results demonstrate that, despite its smaller size, the FN RNA has biochemical properties consistent with a functional glycine riboswitch. It provides a good minimal system to explore glycine riboswitch function by NAIM and other approaches.
FIGURE 2.
Characterization of a Fusobacterium nucleatum (FN) glycine riboswitch. (A) Secondary structure and in-line probing cleavage pattern of the VC and FN glycine riboswitches. The glycine-mediated changes in spontaneous cleavage are labeled. In-line probing of the VC was reproduced by the authors in this study and gave results identical to those previously reported (Mandal et al. 2004). (B) In-line probing gel of FN glycine riboswitch. NR, OH, and T1 represent no reaction, partial digestion with alkali, and partial digestion with RNase T1, respectively. Ala represents reactions performed in the presence of 1 mM alanine. (C) Plot of the normalized fraction of the RNA bound to ligand versus the logarithm of glycine concentration. Binding affinity and cooperativity of the FN riboswitch were calculated by the best fit to the plot as described (Mandal et al. 2004). (D) Ligand specificity of the FN riboswitch. Fraction cleaved in the presence of glycine (2, 3) and various ligands. Ligand concentration was 100 μM for reaction 2 and 1 mM for reactions 3–10. No ligand reaction (1) was used for control. The site of modulation used for quantitation was G61 in aptamer 1.
Native gel analysis of glycine riboswitch
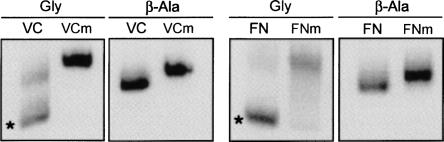
We next set out to develop an efficient method to separate the riboswitch RNA based on glycine binding activity. We attempted to achieve separation using a glycine affinity column, but were unsuccessful due to the poor binding affinity for the derivatized glycine analog. We next attempted native gel electrophoresis with glycine based on the PACE method (Cilley and Williamson 1999). Glycine binds with relatively weak affinity and causes essentially no change in molecular weight, which makes traditional native gel shift methods ineffective. However, riboswitches are predicted to adopt at least two stable conformations, one in the presence and one in the absence of ligand. This structural change can be detected by native gel electrophoresis. The problem of low binding affinity can be overcome by inclusion of glycine within the gel matrix. We used this approach to successfully separate the active and inactive forms of the glycine riboswitch (Fig. 3). The electrophoretic mobility of the VC and FN riboswitches in a gel containing 5 mM glycine was substantially faster than that in a gel containing an equivalent amount of β-alanine. As an additional control, we introduced the G-to-C point mutation into the central loop segment of both RNAs. This point mutation reduces binding affinity of the VC RNA by 1000-fold as assayed by in-line probing (Mandal et al. 2004). No change in the electrophoretic mobility of these control RNAs was observed in the presence of either glycine or β-alanine. Taken together, these results show that the glycine-bound RNA migrates more rapidly through the gel than unbound RNA, which suggests that the RNA adopts a more compact structure when bound to glycine. The result is consistent with a previous report that found, using small-angle X-ray scattering, that the VC RNA undergoes significant compaction upon glycine binding (Lipfert et al. 2007). The native gel provides a simple physical method to separate RNAs capable of binding glycine from those that are not.
FIGURE 3.
Native gel shift analysis of the (A) VC and (B) FN glycine riboswitches with 5 mM Mg(OAc)2, 5 mM glycine or β-alanine. Mutant glycine riboswitches (VCm, FNm) that have ∼1,000-fold loss of affinity were used as a negative control. The G-to-C mutation was introduced into the J1/2 loop region of the aptamer 2 (G146 in VC, G85 in FN) (Mandal et al. 2004). Bands of active riboswitches are indicated by an asterisk.
NAIM analysis of the glycine riboswitch
We performed NAIM using three sets of analogs: four phosphorothioate-tagged parental nucleotides (AαS, GαS, CαS, UαS), four 2′-deoxynucleotides (dAαS, dGαS, dCαS, dUαS), and four purine nucleotide analogs (7-deazapurine phosphorothioate nucleotides; 7dAαS, 7dGαS, N-methylpurine phosphorothioate nucleotides; NMeAαS, NMeGαS). We used two RNA constructs, the originally characterized VC and the shorter FN riboswitch, to compare data sets from RNAs with different phylogenetic origins. Functional RNAs were physically separated by PACE, the RNAs cleaved at the phosphorothioate linkage with iodine, and the products resolved on a denaturing polyacrylamide gel. Sites of analog interference were detected as weak bands in the bound fraction and enhanced bands in the unbound fraction (Fig. 4).
FIGURE 4.
Representative autoradiogram of the NAIM gel. AαS and dAαS NAIM gel of the VC glycine riboswitch. (U) unselected; (+) active; (−) inactive RNA. Sites of interference are indicated to the left of the gel and denoted with an asterisk to the left of each corresponding band. No-iodine-treated controls are shown as a control for RNA degradation.
The glycine riboswitch contains two glycine aptamers that can each bind glycine independently but bind glycine cooperatively as a tandem configuration. Thus, equivalent interferences are expected at those sites responsible for common chemical features important for ligand recognition by a single aptamer or at sites important for cooperativity that are common to both aptamers.
Consistent with this expectation, many of the interferences were symmetrical within the tandem aptamer and appeared in identical sequences or functional groups between the two sequences tested (Fig. 5A). Interferences were observed within the core P1 and P3 stems and the J1/2 and J3/1 loop regions of the riboswitch. No interference was observed in the P2 and P4 stem–loop. Seventeen symmetric positions were observed in each aptamer. The data suggest that these core elements are essential for riboswitch function, while stem–loops P2 and P4 are not directly involved in glycine binding or cooperative interactions. Symmetrical interferences include 2′-deoxy interferences and 7-deaza interferences at sites where the functional group, but not the sequence, is conserved. For each of the individual aptamer domains, four equivalent phosphorothioate interference sites were observed in the J3/3a and P3a stem, and three equivalent 2′-deoxy interference sites were observed within the upper stem of P1 and J3/3a. Ten purine nucleotide interference sites were identified within P1 (top of the P1 and junction region connected to P1), P3, P3a, and J3/3a. Previous biochemical work showed that mutation in one of the symmetric interference sites reduced glycine binding affinity and cooperativity (Mandal et al. 2004). Mutation in G17 to C within the VC aptamer 1 (G17C) showed complete loss of glycine binding in aptamer 1 and weaker binding (100-fold) to aptamer 2. This site displayed N-methyl G interference, indicating possible interaction with the N2 exocyclic amine of G, a contact that would be disrupted by a G-to-C mutation. An equivalent correlation was observed in aptamer 2 at G147 (G147C). In this case, the mutation of the second aptamer eliminated all binding, while the unaltered first aptamer showed a 1000-fold reduction in binding affinity (Mandal et al. 2004). The observation of interference in both of the tandem aptamers suggests that these functional groups are essential for glycine riboswitch function, either for RNA folding, effector binding, or a symmetrical site of cooperativity.
FIGURE 5.
Sites of nucleotide analog interference. (A) Symmetric interference sites between the tandem aptamers in each constructs. Only κ values greater than 2.0 are depicted. (B) Asymmetric interference sites observed between the tandem aptamers in FN and VC constructs. Mutations used for the subsequent study of VC riboswitch cooperativity are indicated with boxes. To measure interference sites near the 3′ end, extended RNA constructs were used for dAαS, dGαS, and NMeGαS NAIM. See Materials and Methods section.
We also identified several interference sites that are not symmetric within the tandem aptamer configuration. A map of the interferences that were asymmetrically distributed between aptamers 1 and 2 is shown in Figure 5B. These asymmetric sites are good candidates for positions involved in cooperative interactions, because the potential interface between the two aptamers is likely to be at least partially asymmetric, using some functional groups on one aptamer that are not used on the second. The asymmetric interferences clustered into two regions in aptamer 1, including the top of P1 and within the P3a hairpin. Within aptamer 2, only the J3a/b loop showed selective interferences. The overall asymmetrical interference patterns between the two riboswitch constructs tested were similar but not identical.
The P1 helix of aptamer 1 displayed asymmetric dAαS (VC A13, FN A7) and NMeGαS (VC G127, FN G6) interferences in both riboswitch constructs. Both of these analogs probe functional groups in the helical minor groove. The 2′-OH can act as both a hydrogen donor and acceptor, and it is often involved in the tertiary interactions with a variety of functional group partners (Strobel and Doudna 1997). N-methyl guanosine replaces one proton of the N2 exocyclic amine with a methyl group, which disrupts minor groove hydrogen bonds while retaining the same duplex stability (Rife et al. 1998). We further investigated the minor groove surface of VC G127 using inosine phosphorothioate analog (IαS), which deletes the exocyclic amine (Fig. 6). G127 showed weak (κ = 1.9) but clear interference with IαS. G127 also showed weak dGαS interference (κ = 1.9; data not shown). Interference with IαS, NMeGαS, and dGαS is a pattern consistent with a minor groove tertiary interaction (Ortoleva-Donnelly et al. 1998a). This result suggests the C12–G127 base pair within the P1 helix of the first aptamer is involved in a tertiary interaction important for VC riboswitch cooperativity.
FIGURE 6.
NAIM analysis of the selected asymmetric sites (G127, A67, A68) in P1 stem and P3a stem of aptamer 1 in VC glycine riboswitch. Interference values (κ) are indicated to the right of each band.
The second region of asymmetric interference is within the P3a hairpin of the first aptamer. Two consecutive N-methyl A interference sites (VC A67 and A68, FN A39 and A40) suggest a tertiary contact is made in the major groove of P3a (Ortoleva-Donnelly et al. 1998b). To explore this possibility further, we performed NAIM using the purine phosphorothioate analog (PurαS), which lacks the N6 exocyclic amine (Ryder and Strobel 1999). A67 and A68 showed different patterns in the purine NAIM assay (Fig. 6). A67 showed weak PurαS interference (1.8), while A68 had no effect (1.0). Such data suggest that A67 uses its exocyclic amine for tertiary hydrogen bond formation within the glycine riboswitch structure. A hydrogen bonding contact may also be made with A68, but the pattern is more consistent with close approach to the major groove at A68 that does not involve a direct tertiary interaction to the amine.
Testing cooperative interactions in the P1 minor groove
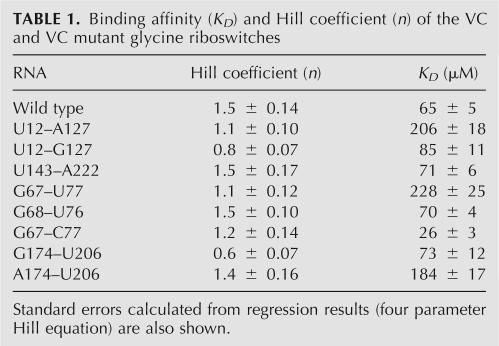
The asymmetric interference data suggest that the minor groove of P1 and the major groove of P3 might be sites of tertiary interaction that facilitate cooperativity between the glycine aptamers. To further explore the functional contribution of these two regions, we introduced several mutations and measured their functional activity and cooperativity. Unlike backbone nucleotide functional groups, the importance of base functional groups can be readily explored by mutation. For these studies we focused on the VC glycine riboswitch because it has been well characterized and it has a higher cooperativity (n = 1.6) than the FN riboswitch (n = 1.4). Glycine binding affinity and cooperativity of the wild-type and mutant VC were measured by in-line probing. To compare mutant affinity and cooperativity with wild-type VC, we used the same curve fitting as reported for wild-type VC. Three regions (aptamer 1, linker, aptamer 2) of RNA were quantitated and calculated. For each of the VC variants, these three regions of the riboswitch showed the same glycine affinity. Due to clearer resolution and higher band intensity, data from the aptamer 1 region (G122–G123) gave the best curve fit and minimal standard error, so we used this plot to calculate the affinity and cooperativity (Table 1).
TABLE 1.
Binding affinity (KD) and Hill coefficient (n) of the VC and VC mutant glycine riboswitches
We first explored minor groove interactions at G127 in the P1 helix (Fig. 7). The C12–G127 base pair in aptamer 1 was mutated to a U–A (U12–A127), which removed the minor groove exocyclic amine, and to a U–G (U12–G127) wobble pair, which changed the conformation of the G. For the wild-type VC riboswitch, the Kd and Hill coefficient were measured to be 65 μM and 1.5, respectively, similar to the values previously reported (Kd = 30 μM, n = 1.6). The U12–A127 mutant showed loss of cooperativity (n = 1.1) and decreased binding affinity (Kd = 206 μM). The U12–G127 mutant showed complete loss of cooperativity (n = 0.8) but retained binding affinity (Kd = 85 μM) (Fig. 7). All three segments of the riboswitch (aptamer 1, linker, and aptamer 2) showed similar glycine affinity. If the mutation affected a single aptamer, the Kd values of each aptamer would be expected to differ, as observed for the G17C and G147C mutants (Mandal et al. 2004). This suggests that activity changes by the mutation came from overall riboswitch modulation, not from the altered function of a single aptamer. To determine if the identity of this base pair is specific to aptamer 1, we introduced the same C–G-to-U–A mutation in the corresponding site in the P1 helix of aptamer 2. Consistent with the model, the U143–A222 mutation did not affect glycine affinity (Kd = 71 μM) or cooperativity (n = 1.5) (Fig. 7). The results support the hypothesis that the minor groove of the aptamer 1 P1 helix participates in a tertiary interaction that is important for glycine riboswitch cooperativity.
FIGURE 7.
In-line probing plot of VC mutants indicated in Figure 5B. Binding plot of VC wild-type (circle) and VC variants (triangle). Plots of the normalized fraction of the RNA bound to ligand versus the concentration of glycine were derived from the in-line probing assay. Depicted is the best fit curve as calculated from the quantitated data of the bands G122 and G123. Calculated values of the binding affinity (Kd) and cooperativity (Hill coefficient, n) are shown in Table 1.
Testing cooperative interactions in the P3a major groove
The asymmetric interference pattern suggested that the P3a major groove might also be involved in cooperative tertiary interactions between the two aptamers. The NAIM data focused primarily upon the exocyclic amino groups of the consecutive adenosines A67 and A68. To determine the possible contribution of the P3a major groove, we individually mutated A67 and A68 to G, changing the AU pairs to GU wobble pairs. The A67G mutation (G67–U77) reduced cooperativity (n = 1.1) and binding affinity (Kd = 228 μM), while the A68G mutation (G68–U76) showed no change in cooperativity (n = 1.5) and binding affinity (Kd = 70 μM) (Fig. 7). These results are consistent with PurαS NAIM data, suggesting the exocyclic amine of A67 acts as a possible hydrogen bond donor for tertiary structure of glycine riboswitch.
Both A67 and A68 participate in A–U base pairs in aptamer 1. Since the A67–U77 base pair is 97% conserved as a purine–pyrimidine base pair (Fig. 1), some fraction of the aptamers have a G–C pair at this site. A G–C pair also has a potential major groove hydrogen bond donor at the N4 exocyclic amine group of C, which could potentially serve the same role as the N6 exocyclic amine of A. The A67G–U77C double mutant (G67–C77) showed loss of cooperativity (n = 1.2) compared to wild type (Fig. 7), indicating that the G–C pair cannot fully replace the A–U pair in the VC glycine riboswitch sequence context. Interestingly, the glycine binding affinity of G67–C77 was Kd = 26 μM, 2.5-fold tighter than the wild type.
The asymmetric nature of the A67 NAIM result was possibly related to the fact that the sequence identity was different between the two aptamers. The corresponding site in aptamer 2 is G174, thus sequence differences preclude detection of NMeAαS interference in P3a of aptamer 2. However, both A67 and G174 have 7-deaza interference. Therefore, the major groove of G174–C206 may also take part in a tertiary interaction. To further test the importance of P3a helix of aptamer 2, we performed mutagenesis at the corresponding base pair within the aptamer 2 (G174–C206). We mutated C to U (G174–U206), which creates a wobble pair and eliminates the N4 exocyclic amine in C. G174–U206 showed not only complete loss of cooperativity but actually showed negative cooperativity (n = 0.6), yet the binding affinity of G174–U206 for glycine was unaffected (Kd = 73 μM) (Fig. 7). We also mutated the G–C to an A–U (A174–U206). The A174–U206 significantly recovered cooperativity (n = 1.4), but the binding affinity was decreased (Kd = 184 μM) compared to that of wild type (Fig. 7). The results suggest that the major grooves of P3a in both aptamers 1 and 2 are involved in cooperative glycine binding.
DISCUSSION
The glycine riboswitch is the first reported natural RNA with a tandem aptamer structure and homotrophic cooperative binding. In this study, we determined possible sites for glycine binding and tertiary cooperativity using NAIM and mutagenesis analysis. NAIM showed several symmetric and asymmetric interference sites in the tandem aptamer configuration. This study focused on the asymmetric interference sites as possible sites of tertiary interaction between the aptamers. The resulting mutagenesis data suggest that the minor groove of the P1 helix in aptamer 1 and the major groove of P3a in both aptamers provide tertiary interactions important for glycine riboswitch cooperativity.
Chemical basis of P1 helix minor groove interaction site
If there is a cooperative interaction with the aptamer 1 P1 helical minor groove, what is the nature of this interaction and what is the interaction partner? The helical minor groove is a common site of helix docking (Strobel and Doudna 1997; Doherty et al. 2001). One motif commonly observed for RNA helix packing is the A-minor motif, which involves close contact between the minor groove edge of a donor adenosine and the minor groove edge of the acceptor duplex (Nissen et al. 2001; Battle and Doudna 2002). In the type I A-minor motif, N1 and N3 imino groups of the adenosine form hydrogen bonds with the 2′-OH and N2 amine of the G in the G–C acceptor, respectively. The adenosine 2′-OH hydrogen bonds with the O2 carbonyl and 2′-OH of the cytidine. The interaction is specific to Watson–Crick base pairs and discriminates strongly against wobble pairs (Battle and Doudna 2002). The interference and mutagenesis results at the C12–G127 base pair are suggestive of a type I A minor motif. Most notably, the C–G-to-U–G mutation showed complete loss of cooperativity without loss of glycine binding affinity.
If an A-minor interaction is responsible for a cooperative interaction to the P1 helix, which A in the sequence serves as the donor adenosine? One possibility is the A-rich loop in J3a/b within aptamer 2. In this region, three adenosine residues (A178, A201, and A202) showed asymmetric 2′-deoxy interference (Fig. 5B), as would be expected for a type I or type II A-minor motif. However, these residues did not display diaminopurine interference, as would also be expected (data not shown). In an effort to identify the tertiary contact, a nucleotide analog interference suppression experiment (NAIS) was performed to look for changes in the overall adenosine interference pattern specific to the noncooperative U12–G127 mutant, but no interference suppression was observed. This may have occurred because (1) the donor adenosine is uninformative due to strong phosphorothioate interference (such as A73, A171, and A209), (2) the PACE gel shift assay is incompatible with a successful NAIS experiment, or (3) the interaction with C12–G127 is not through an A-minor contact. Thus, the data support a model in which a tertiary contact to the aptamer 1 P1 minor groove is a key contact for glycine riboswitch cooperativity, but the tertiary interaction partner remains to be identified.
Functional role of P1 helix in riboswitches
The observation that the P1 helix hosts an interaction essential for riboswitch cooperativity provides an interesting parallel to the general role of the P1 helix in riboswitch function. In most riboswitches, modulation of P1 helix formation is the key conformational switch that affects gene expression. The 3′ strand of the P1 helix forms an alternative interaction with the 5′ end of the downstream expression platform, which results in transcriptional termination or alters translational initiation. Obviously, there are two P1 helices in the tandem glycine riboswitch, one for each aptamer. At first glance these appear to be equivalent, but the phylogenentic and biochemical data suggest that they are playing different roles in riboswitch function. The P1 stem of aptamer 2 appears to play the role of a typical riboswitch P1 helix. Nucleotides on the 3′ side of the helix are complementary with sequence in the downstream expression platform. There does not appear to be an equivalent region of complementarity to strands in the aptamer 1 P1 helix. The sequence and length of the helix also display greater variability than seen for aptamer 2. The biochemical data presented here indicate that the aptamer 1 P1 helix contributes to a tertiary interaction important for cooperativity, which implies that the P1 helix in each aptamer has evolved to serve different roles within the glycine riboswitch.
Comparison with cooperativity in hemoglobin
To understand cooperativity in the glycine riboswitch RNA it is useful to make comparison to hemoglobin, the best-studied cooperative system in biology (Royer et al. 2001). Hemoglobin is an allosteric tetrameric protein comprised of two αβ dimers (α1β1–α2β2). The dimers form a quaternary structure that adopt a T (deoxy, tense) or R (oxy, relaxed) state, according to the definitions in the classical Monod–Wyman–Changeux (MWC) model for cooperativity (Monod et al. 1965). The T to R transition (unbound to bound) through ligand binding changes the overall structure of the tetramer and induces a shift of the α1/β2 intersubunit contacts. The interaction of the contacts along the α1/β2 interface provides the important chemical connection for cooperativity (Turner et al. 1992). Mutations in the interfacial contact region change the overall structure and mechanism of cooperativity in oxygen binding (Turner et al. 1992).
This study reports the effects on cooperativity and affinity that resulted from several glycine riboswitch variants mutated at possible interdomain contact sites. There were independent effects on cooperativity and binding affinity, in a pattern similar to several classic hemoglobin mutations. For example, removal of the exocyclic amine in the minor groove of P1 (U12–A127) and the major groove of P3a stem (G67–U77) in aptamer 1 caused decreased affinity and cooperativity. The hemoglobin mutant Bassett (αAsp94 → Ala) showed similar results with reduced binding affinity and cooperativity (Abdulmalik et al. 2004). Crystallographic structures of hemoglobin Bassett revealed that disruption of a hydrogen bond in the α1/β2 interface of the R state is responsible for this effect (Safo et al. 2005). By analogy to hemoglobin Bassett, the exocyclic amines in P1 and P3a are likely crucial functional groups for formation of the fully bound RNA. A second interesting example is P3a mutant G67–C77, which showed reduced cooperativity but increased binding affinity. This unanticipated result has been reported in many hemoglobin mutants (Turner et al. 1992). One example is hemoglobin Roanne (αAsp94 → Glu), which has a destabilized unbound structure (T state), higher oxygen affinity, and reduced cooperativity (Kister et al. 1995). Still another phenotype was observed upon mutation in the P3a stem of aptamer 2 (A174–U206), in that it retained cooperativity but displayed decreased binding affinity. This behavior is similar to hemoglobin mutant (αVal96 → Trp), which is known to stabilize the T state by formation of an additional water-mediated hydrogen bond (Puius et al. 1998), resulting in no change in cooperativity but reduced binding affinity (Tsai and Ho 2002). While explanations for the effects observed in the glycine riboswitch await detailed molecular explanation, these precedents for independent effects on affinity and cooperativity observed in hemoglobin lead to analogous predictions in this RNA-based cooperative system.
MATERIALS AND METHODS
DNA oligonucleotides and chemicals
DNA oligonucleotides were synthesized by W.M Keck Foundation Biotechnology Resource Laboratory at Yale University and used without further purification. Phosphorothioate nucleotide analogs were synthesized or purchased from Glen Research. Glycine and other chemicals were obtained from Sigma.
DNA constructs
The Vibrio cholera VC1422 glycine riboswitch was generated by PCR amplification of Vibrio cholera genome. Fusobacterium nucleatum FN 0328 riboswitch was prepared by the annealing of synthetic oligonucleotides and amplified by PCR. In both cases, the PCR DNA was cloned into pCR2.1-TOPO (Invitrogen) plasmid. The sequence of the cloned plasmid DNA was confirmed by DNA sequencing. The resulting plasmid DNA was used as a template for PCR amplification or used as a template for site-directed mutagenesis. Preparation of the plasmid DNA for mutant VC riboswitch was achieved by PCR reaction using corresponding primers.
In vitro transcription
Plasmid DNA encoding the glycine riboswitch was amplified by PCR with a forward primer containing the T7 promoter and an appropriate reverse primer. PCR DNA containing the double glycine aptamer region was used as a template. RNAs were transcribed in 40 mM Tris-HCl (pH 7.5), 4 mM spermidine, 10 mM DTT, 15 mM MgCl2, 0.05% Triton X-100, and 1 mM each 5′-nucleotide triphosphate for 3 h at 37°C. All the RNAs were purified by 6% polyacrylamide gel electrophoresis, eluted into 0.3 mM NaOAc (pH 5.2), precipitated with ethanol, and resuspended in water. RNA concentrations were determined by UV absorbance at 260 nm wavelength.
Preparation of analog incorporated RNA
The PCR DNA was used as a template for in vitro transcription using either the wild-type or Y639F mutant form of T7 RNA polymerase and the analog and NTP concentrations reported previously (Ryder et al. 2000). RNA was synthesized with 5% incorporation of each of the phosphorothioate nucleotide analogs. Transcription, purification, and quantitation of the analog incorporated RNA was performed by the same method as described above.
In-line probing assay and Hill plot fitting
RNA transcripts were dephosphorylated and 5′-32P-labeled as described elsewhere (Ryder and Strobel 1999). For each in-line probing reaction, ∼20 nM of end-labeled RNA was incubated at 23°C for 40 h in 50 mM Tris-HCl (pH 8.3), 20 mM MgCl2, and 100 mM KCl in the presence or absence of ligand. Spontaneously cleaved RNA was separated by 6% denaturing PAGE and visualized by PhosphorImager (Molecular Dynamics). Individual band intensities were quantitated using ImageQuaNT software. For mutant VC constructs, three different regions in the aptamers were quantitated; G122–G123 (aptamer 1), A135–G137 (linker), C177–A178 (aptamer 2). To control for loading differences, the band intensities were normalized to a nonvariable band (U94). Apparent Kd values were determined by plotting each normalized fraction cleaved versus the logarithm of ligand concentration. The Hill coefficient was calculated by fitting each plot using the four parameter Hill function in SigmaPlot 9.0 software.
Native gel analysis
Nondenaturing gel electrophoresis was performed by a modified PACE method (Cilley and Williamson 1999). End-labeled RNA was incubated in a TB buffer (90 mM Tris-borate at pH 8.3) containing 5 mM glycine, 5 mM MgCl2 for 20 min at 23°C. One-fifth volume of glycerol loading dye (50% glycerol, 0.1% xylene cyanol FF) was added to the RNA mixture. Glycine bound and unbound forms were separated on a 6% native acrylamide gel in TB buffer containing 5 mM glycine and 5 mM MgCl2. Electrophoresis was carried out at 10°C for 2 h with TB buffer containing 5 mM MgCl2. The separated RNA was visualized by PhosphorImager or autoradiography.
Nucleotide analog interference mapping
5′-End 32P-labeled and analog incorporated RNAs (∼5 pmol) were incubated in a TB buffer containing 5 mM glycine, 5 mM MgCl2 for 20 min at 23°C. The glycine-bound RNA and -unbound RNA were eluted into 0.3 mM NaOAc. RNAs were precipitated by ethanol and resuspended in water. Radioactivity of the RNA was measured by scintillation counting and adjusted by dilution to have the same activity per unit volume. Typically, 50,000–100,000 cpm of RNA were used for each lane. One-tenth volume of 100 mM iodine in ethanol was added to the RNA to cleave the phosphorothioate linkages. Cleavage products were resolved by denaturing 6% PAGE. Reactions containing no iodine were run in parallel to confirm that the cleavage pattern was specific to the iodine treatment and did not result from nonspecific degradation. For dAαS, dGαS, and NMeGαS NAIM, RNA constructs with a 15-nt overhang on the 3′ end were used to resolve interference sites in the P1 stem of aptamer 2. The sequences included are VC, 5′-AGUGAAAGGCCAAUC-3′, and FN, 5′-UAAUUGUGCAAUUUAU-3′.
Interference quantitation
Peak intensities for both glycine-bound RNA and -unbound RNA were quantitated by PhosphorImager. In this study, the extent of interference (κ) at each position was calculated by dividing band intensity in the unbound fraction by band intensity in the bound fraction. This calculation method is different from the previously reported method. Each value was normalized to the average value of a noninterfering band to adjust for possible loading differences. In this study, the sites that have κ values of 2 or greater were highlighted in Figure 5.
ACKNOWLEDGMENTS
We thank Ian Suydam for critical comments on manuscript, Ron Breaker and Jeff Barrick for helpful discussions, and Rüdiger Welz for gel shift analysis. This work was supported by NSF grant MCB0544255.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.771608.
REFERENCES
- Abdulmalik, O., Safo, M.K., Lerner, N.B., Ochotorena, J., Daikhin, E., Lakka, V., Santacroce, R., Abraham, D.J., Asakura, T. Characterization of hemoglobin bassett (α94Asp → Ala), a variant with very low oxygen affinity. Am. J. Hematol. 2004;77:268–276. doi: 10.1002/ajh.20184. [DOI] [PubMed] [Google Scholar]
- Adams, P.L., Stahley, M.R., Kosek, A.B., Wang, J., Strobel, S.A. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004;430:45–50. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- Barrick, J.E., Corbino, K.A., Winkler, W.C., Nahvi, A., Mandal, M., Collins, J., Lee, M., Roth, A., Sudarsan, N., Jona, I., et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl. Acad. Sci. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle, D.J., Doudna, J.A. Specificity of RNA–RNA helix recognition. Proc. Natl. Acad. Sci. 2002;99:11676–11681. doi: 10.1073/pnas.182221799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohr, C., Hasselbach, K.A., Krogh, A. Uber einen in biologischen beziehung wichtigen einfluss, den die kohlen-sauerspannung des blutes auf dessen sauerstoffbindung ubt. Skand. Arch. Physiol. 1904;15:401–412. [Google Scholar]
- Boudvillain, M., Pyle, A.M. Defining functional groups, core structural features and inter-domain tertiary contacts essential for group II intron self-splicing: A NAIM analysis. EMBO J. 1998;17:7091–7104. doi: 10.1093/emboj/17.23.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cilley, C.D., Williamson, J.R. PACE analysis of RNA–peptide interactions. Methods Mol. Biol. 1999;118:129–141. doi: 10.1385/1-59259-676-2:129. [DOI] [PubMed] [Google Scholar]
- Cromie, M.J., Shi, Y., Latifi, T., Groisman, E.A. An RNA sensor for intracellular Mg2+ . Cell. 2006;125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- Doherty, E.A., Batey, R.T., Masquida, B., Doudna, J.A. A universal mode of helix packing in RNA. Nat. Struct. Biol. 2001;8:339–343. doi: 10.1038/86221. [DOI] [PubMed] [Google Scholar]
- Forsen, S., Linse, S. Cooperativity: Over the Hill. Trends Biochem. Sci. 1995;20:495–497. doi: 10.1016/s0968-0004(00)89115-x. [DOI] [PubMed] [Google Scholar]
- Heide, C., Busch, S., Feltens, R., Hartmann, R.K. Distinct modes of mature and precursor tRNA binding to Escherichia coli RNase P RNA revealed by NAIM analyses. RNA. 2001;7:553–564. doi: 10.1017/s1355838201001765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kister, J., Kiger, L., Francina, A., Hanny, P., Szymanowicz, A., Blouquit, Y., Prome, D., Galacteros, F., Delaunay, J., Wajcman, H. Hemoglobin Roanne [α94 (G1) Asp → Glu]: A variant of the α 1 β 2 interface with an unexpected high oxygen affinity. Biochim. Biophys. Acta. 1995;1246:34–38. doi: 10.1016/0167-4838(94)00190-r. [DOI] [PubMed] [Google Scholar]
- Koshland D.E., Jr, Hamadani, K. Proteomics and models for enzyme cooperativity. J. Biol. Chem. 2002;277:46841–46844. doi: 10.1074/jbc.R200014200. [DOI] [PubMed] [Google Scholar]
- Lipfert, J., Das, R., Chu, V.B., Kudaravalli, M., Boyd, N., Herschlag, D., Doniach, S. Structural transitions and thermodynamics of a glycine-dependent riboswitch from Vibrio cholerae . J. Mol. Biol. 2007;365:1393–1406. doi: 10.1016/j.jmb.2006.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal, M., Breaker, R.R. Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- Mandal, M., Lee, M., Barrick, J.E., Weinberg, Z., Emilsson, G.M., Ruzzo, W.L., Breaker, R.R. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- Monod, J., Wyman, J., Changeux, J.P. On the nature of allosteric transitions: A plausible model. J. Mol. Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- Nissen, P., Ippolito, J.A., Ban, N., Moore, P.B., Steitz, T.A. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc. Natl. Acad. Sci. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortoleva-Donnelly, L., Kronman, M., Strobel, S.A. Identifying RNA minor groove tertiary contacts by nucleotide analogue interference mapping with N2-methylguanosine. Biochemistry. 1998a;37:12933–12942. doi: 10.1021/bi980723j. [DOI] [PubMed] [Google Scholar]
- Ortoleva-Donnelly, L., Szewczak, A.A., Gutell, R.R., Strobel, S.A. The chemical basis of adenosine conservation throughout the Tetrahymena ribozyme. RNA. 1998b;4:498–519. doi: 10.1017/s1355838298980086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perutz, M.F. Mechanisms of cooperativity and allosteric regulation in proteins. Q. Rev. Biophys. 1989;22:139–237. doi: 10.1017/s0033583500003826. [DOI] [PubMed] [Google Scholar]
- Puius, Y.A., Zou, M., Ho, N.T., Ho, C., Almo, S.C. Novel water-mediated hydrogen bonds as the structural basis for the low oxygen affinity of the blood substitute candidate rHb(α96Val → Trp) Biochemistry. 1998;37:9258–9265. doi: 10.1021/bi9727287. [DOI] [PubMed] [Google Scholar]
- Rife, J.P., Cheng, C.S., Moore, P.B., Strobel, S.A. N 2-methylguanosine is iso-energetic with guanosine in RNA duplexes and GNRA tetraloops. Nucleic Acids Res. 1998;26:3640–3644. doi: 10.1093/nar/26.16.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rox, C., Feltens, R., Pfeiffer, T., Hartmann, R.K. Potential contact sites between the protein and RNA subunit in the Bacillus subtilis RNase P holoenzyme. J. Mol. Biol. 2002;315:551–560. doi: 10.1006/jmbi.2001.5261. [DOI] [PubMed] [Google Scholar]
- Royer W.E., Jr, Knapp, J.E., Strand, K., Heaslet, H.A. Cooperative hemoglobins: Conserved fold, diverse quaternary assemblies and allosteric mechanisms. Trends Biochem. Sci. 2001;26:297–304. doi: 10.1016/s0968-0004(01)01811-4. [DOI] [PubMed] [Google Scholar]
- Ryder, S.P., Strobel, S.A. Nucleotide analog interference mapping. Methods. 1999;18:38–50. doi: 10.1006/meth.1999.0755. [DOI] [PubMed] [Google Scholar]
- Ryder, S.P., Ortoleva-Donnelly, L., Kosek, A.B., Strobel, S.A. Chemical probing of RNA by nucleotide analog interference mapping. Methods Enzymol. 2000;317:92–109. doi: 10.1016/s0076-6879(00)17008-9. [DOI] [PubMed] [Google Scholar]
- Safo, M.K., Abdulmalik, O., Lin, H.R., Asakura, T., Abraham, D.J. Structures of R- and T-state hemoglobin Bassett: Elucidating the structural basis for the low oxygen affinity of a mutant hemoglobin. Acta Crystallogr. D Biol. Crystallogr. 2005;61:156–162. doi: 10.1107/S0907444904030501. [DOI] [PubMed] [Google Scholar]
- Schwalbe, H., Buck, J., Furtig, B., Noeske, J., Wohnert, J. Structures of RNA switches: Insight into molecular recognition and tertiary structure. Angew. Chem. Int. Ed. Engl. 2007;46:1212–1219. doi: 10.1002/anie.200604163. [DOI] [PubMed] [Google Scholar]
- Soukup, G.A., Breaker, R.R. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss-Soukup, J.K., Strobel, S.A. A chemical phylogeny of group I introns based upon interference mapping of a bacterial ribozyme. J. Mol. Biol. 2000;302:339–358. doi: 10.1006/jmbi.2000.4056. [DOI] [PubMed] [Google Scholar]
- Strobel, S.A., Doudna, J.A. RNA seeing double: Close packing of helices in RNA tertiary structure. Trends Biochem. Sci. 1997;22:262–266. doi: 10.1016/s0968-0004(97)01056-6. [DOI] [PubMed] [Google Scholar]
- Sudarsan, N., Hammond, M.C., Block, K.F., Welz, R., Barrick, J.E., Roth, A., Breaker, R.R. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006;314:300–304. doi: 10.1126/science.1130716. [DOI] [PubMed] [Google Scholar]
- Szewczak, L.B., DeGregorio, S.J., Strobel, S.A., Steitz, J.A. Exclusive interaction of the 15.5 kD protein with the terminal box C/D motif of a methylation guide snoRNP. Chem. Biol. 2002;9:1095–1107. doi: 10.1016/s1074-5521(02)00239-9. [DOI] [PubMed] [Google Scholar]
- Tsai, C.H., Ho, C. Recombinant hemoglobins with low oxygen affinity and high cooperativity. Biophys. Chem. 2002;98:15–25. doi: 10.1016/s0301-4622(02)00081-9. [DOI] [PubMed] [Google Scholar]
- Turner, G.J., Galacteros, F., Doyle, M.L., Hedlund, B., Pettigrew, D.W., Turner, B.W., Smith, F.R., Moo-Penn, W., Rucknagel, D.L., Ackers, G.K. Mutagenic dissection of hemoglobin cooperativity: Effects of amino acid alteration on subunit assembly of oxy and deoxy tetramers. Proteins. 1992;14:333–350. doi: 10.1002/prot.340140303. [DOI] [PubMed] [Google Scholar]
- Winkler, W.C., Breaker, R.R. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]