Abstract
Synaptotagmins are synaptic vesicle-associated, phospholipid-binding proteins most commonly associated with Ca+2-dependent exocytotic and Ca+2- independent endocytotic events. Synaptotagmin III is a 63.2-kD member of the synaptotagmin homology group; one of its characteristic properties is the ability to bind divalent cations and accessory proteins promiscuously. In the cytosolic portion of this protein, a flexible seven–amino acid linker joins two homologous C2 domains. The C2A domain binds to phospholipid membranes and other accessory proteins in a divalent cation-dependent fashion. The C2B domain promotes binding to other C2B domains, as well as accessory proteins independent of divalent cations. The 3.2 Å crystal structure of synaptotagmin III, residues 295–566, which includes the C2A and C2B domains, exhibits differences in the shape of the Ca+2-binding pocket, the electrostatic surface potential, and the stoichiometry of bound divalent cations for the two domains. These observations may explain the disparate binding properties of the two domains. The C2A and the C2B domains do not interact; synaptotagmin, therefore, covalently links two independent C2 domains, each with potentially different binding partners. A model of synaptotagmin's involvement in Ca+2-dependent regulation of membrane fusion through its interaction with the SNARE complex is presented.
Keywords: SNARE, synaptotagmin, C2 domains, crystallography, calcium-binding protein
Early characterization of synaptic exocytosis demonstrated that Ca+2 floods into the synapse before the propagation of an action potential (Katz and Miledi 1967). This observation implied a receptor for this Ca+2 signal on the presynaptic terminus. Calmodulin, protein kinase C (PKC)1, and the annexins were suggested as possible candidates for the Ca+2 trigger of exocytosis (Pollard et al. 1980; DeLorenzo 1982; Hutton 1986). More recently, however, strong evidence points to the synaptic vesicle protein, synaptotagmin, as the receiver for the Ca+2 signal in the neuron. This protein could potentially transmit the Ca+2 signal through its interactions with the SNARE (soluble N-ethylmaleimide–sensitive factor attachment protein receptor, SNAP receptor) complex at the presynaptic terminal (Brose et al. 1992; Bommert et al. 1993; Reist et al. 1998). Neurotransmitter filled vesicles could then fuse with the presynaptic membrane and release their contents into the synaptic cleft.
Synaptotagmin has been characterized in at least eight different isoforms in neurons. Several synaptotagmin isoforms also localize to nonneuronal cells (Li et al. 1995a). Therefore, this protein appears to be generally important in cellular exocytosis. Synaptotagmin is composed of a short intravesicular NH2-terminal region, a single membrane spanning domain, a lysine- and arginine-rich region, and two homologous C2 domains. The C2 domain was first characterized in conventional protein kinase C isoforms (cPKC; Parker et al. 1986) and implicated in Ca+2-dependent phospholipid binding. The x-ray crystal structure of the C2A domain of synaptotagmin I (SytI) revealed a unique β-sandwich fold with a cluster of three Ca+2-binding loops at the apex of the fold (Sutton et al. 1995). In addition, the x-ray crystal structure of the isolated C2 domain from PKC-β (Sutton and Sprang 1998), the solution NMR structure of synaptotagmin I C2A (Shao et al. 1998), and the crystal structure of PLC-δ1 (Essen et al. 1997) identified up to three Ca+2-binding sites within the Ca+2-binding pockets of their respective C2 domains. Upon Ca+2 binding, few structural changes occur in the divalent cation-binding pocket, apart from changes of the sidechain rotamers of the Ca+2 coordinating aspartate residues (Sutton et al. 1995; Shao et al. 1998). The bound calcium ions quench the negative electrostatic potential within the divalent cation-binding pocket, and the more positive potential from peripheral basic residues dominate the interaction between synaptotagmin and target proteins (Shao et al. 1997). This “electrostatic switch” mechanism may also regulate the interaction of the C2A domain with phospholipid membranes (Davletov et al. 1998).
In vitro, synaptotagmin interacts with several proteins that are essential for exocytotic and endocytotic processes. In its role as a regulator of synaptic vesicle fusion, the C2B domain of synaptotagmin I binds the SNARE protein SNAP-25 (Schiavo et al. 1997) and the C2A domains bind syntaxin-1A (Bennett et al. 1992). The sites of interaction with syntaxin-1A map to a cluster of residues near the Ca+2-binding pocket of the C2A domain (Shao et al. 1997). Interestingly, this cluster site coincides with much of the phospholipid-binding region of the C2A domain of SytI (Chae et al. 1998). Synaptotagmin binds β-SNAP, but not α-SNAP, with a high degree of selectivity (Schiavo et al. 1995); however, the physiological function of this interaction has yet to be established. Synaptotagmin also interacts with the N and P/Q-type Ca+2 channels (Leveque et al. 1994; Martin-Moutot et al. 1996). This interaction implies that exocytosis may occur near the Ca+2 channel where the Ca+2 concentration in the neuron is highest upon depolarization. Synaptotagmin also interacts with the endocytotic adapter protein AP-2 (Zhang et al. 1994). Most recently, synaptotagmin has been implicated as a receptor for clostridial neurotoxins (Nishiki et al. 1994; Li and Singh 1998).
Although most biochemical studies of synaptotagmin have been carried out with the isolated C2A domain of synaptotagmin I, a few studies suggest that the C2B domain may be responsible for homo- and hetero-oligomerization of synaptotagmin isoforms (Chapman et al., 1996). Synaptotagmin I and synaptotagmin II, III can form homo- and heterodimers in a Ca+2-dependent manner (Chapman et al. 1998; Osborne et al. 1999). Likewise, synaptotagmin I and synaptotagmin IV also form heterodimers (Littleton et al. 1999). Therefore, synaptotagmin's ability to associate in various combinations with other isoforms may regulate synaptic diversity. Synaptotagmin can also associate and distinguish between various forms of inositol-polyphosphates through the polybasic region of the C2B domains (Schiavo et al. 1996; Mehrotra et al. 1997). These compounds are known to directly interfere with exocytosis and may be involved in the synaptic vesicle fusion process.
Materials and Methods
Cloning and Expression
The cDNA corresponding to the mouse gene of synaptotagmin III C2A-C2B, residues 295–569 (33,000 kD), was subcloned into pET28A using the NdeI-HinDIII cloning site. The expression plasmid was transformed into BL21(DE3). The bacteria were grown in a BIOFLO3000 using ECPYM1 media (Bernard and Payton 1995) with 50 mg/ml kanamycin sulfate. Heterologous protein expression was initiated at OD600 50.0 with 300 mM IPTG for 3 h at 37°C. Cells were harvested and frozen in liquid nitrogen until needed.
Gel Shift Assay
Native gel electrophoresis was carried out using 10–15% native Phast (Pharmacia) gels with native buffer strips soaked for 2 h in 0.88 M l-alanine, 0.25 M Tris, pH 8.8, and 1 mM EDTA before running the gel. Samples were mixed and allowed to incubate for at least 1 h in 20 mM Hepes, pH 7.8, 150 mM NaCl, 5 mM DTT, and 1 mM EDTA before loading the gel. The concentration of both proteins was determined by amino acid analysis (Keck facility, Yale University). SNARE core complex was expressed in a polycistronic T7 expression vector containing rat synaptobrevin-II, residues 1–96, rat syntaxin-1A, residues 180–262, rat SNAP-25A residues 1–83 and residues 130–206, and purified according to previously published methods (Sutton et al. 1998).
Purification and Crystallization
30 g of frozen bacterial cells were resuspended in 125 ml of extraction buffer (100 mM sodium phosphate, pH 8.0, 200 mM NaCl, and 10% ethylene glycol) with 200 mg of lysozyme and 0.5% Triton X-100. Passing the lysate twice through a pneumatic cell cracker (Microfluidics) at 10,000 lb/in2 facilitated cell lysis. The histidine-tagged protein was incubated in batch with NTA-Ni (Qiagen) resin overnight at 4°C while slowly mixing. The resin was washed in a column with extraction buffer including 50 mM imidazole until the OD280 was below 0.05. The tagged protein was eluted with 300 mM imidazole. At this point, 10% ethylene glycol, and 5 mM CaCl2 were added to the protein solution. Under these conditions, synaptotagmin III precipitated and was removed by centrifugation in a GSA rotor at 10,000 rpm for 20 min. The protein pellet was washed in 25 mM ethanolamine, pH 8.5, 300 mM NaCl, and 10% ethylene glycol. The protein was resuspended in 30 ml of 50 mM sodium phosphate, pH 7.8, and 5 mM EDTA. The histidine tag was removed by cleaving the protein with TEV protease (BRL) overnight at 4°C. The resuspended protein was filtered through a 0.22-μm filter before ion exchange chromatography with a Mono Q 5/5 (Pharmacia) column in 25 mM ethanolamine, pH 8.0. The purified protein eluted from a 0–1-M NaCl gradient at 200 mM NaCl. The protein was concentrated to 40 mg/ml. Synaptotagmin III C2A-C2B crystallized in 1.5 M MgCl2 and 100 mM MES, pH 6.5, at 20°C using the hanging drop method. Large hexagonal crystals grew after ∼1 wk.
Data Collection
Analysis of the systematic absences in the diffraction data narrowed possible space groups choices to either P6122 or P6222: a = 126 Å, b = 126 Å, c = 118 Å. Inspection of the molecular replacement solution of the C2A domain uniquely determined P6222. The Matthews coefficient calculation predicted two molecules in the asymmetric unit given average solvent content; however, only one protein molecule was found resulting in a solvent content of 70%. Native and trimethyl lead acetate (TMLA) data were collected at station BL1-5 at SSRL at a temperature of 20°C. Diffraction data from several crystals were merged to obtain highly redundant and complete native and anomalous derivative data sets. Diffraction data of the derivative were collected at a wavelength of 1.00 Å to take advantage of the lead (Pb) anomalous signal. Diffraction data were integrated and scaled using the DENZO package (Otwinowski and Minor 1998; Table ).
Table 1.
Crystallographic Data
| Crystal | a | b | c | λ | dmin | No. observed reflections | No. unique reflections | Completeness | <I>/<σI> | Rsym |
|---|---|---|---|---|---|---|---|---|---|---|
| Å | % | % | ||||||||
| Native | 125.85 | 125.85 | 118.19 | 1.000 | 3.2 | 61,080 | 9,578 | 99.6 (99.9) | 25.6 | 7.8 (77.3) |
| TMLA | 125.83 | 125.83 | 118.17 | 1.000 | 3.2 | 56,127 | 9,581 | 99.6 (99.9) | 20.5 | 7.6 (72.0) |
Structure Solution
Placement of each C2 domain within the asymmetric unit was attempted by molecular replacement as implemented in CNS (Brunger et al. 1998) using the synaptotagmin I C2A domain (residues 140–265; pdb entry, 1rsy) as the search model. Only the C2A domain of synaptotagmin III was located by this technique. The C2B domain was manually positioned in an electron density map calculated from native amplitudes and phases obtained from the molecular replacement solution combined with the initial SIRAS (single isomorphous replacement, anomalous scattering) phase probability distribution. The C2A and C2B domains were separately refined by rigid body refinement to Rfree and R values of 48 and 46%, respectively. Three heavy atom sites were localized by an automated Patterson search method (Grosse-Kunstleve and Brunger 1999) and confirmed by difference Fourier maps. Two of the heavy atom peaks corresponded to TMLA ions in the Ca+2-binding loops of each domain. The third site mapped to a minor site on the C2A domain. Because of the structural similarity of the two domains, it was difficult to correctly assign each domain to the electron density at this early stage. The two domains were distinguished by identifying a unique cysteine residue (Cys523) in the C2B domain. This cysteine residue is involved in a disulfide linkage between crystallographically related C2B domains. The model was fit to density modified electron density SIRAS maps (Table ), as well as the model-phase combined SIRAS electron density maps.
Table 2.
Cystallographic Phasing
| TMLA | |
|---|---|
| Number of sites | 3 |
| SIR phasing power | 1.80 |
| SIRAS phasing power | 0.05 |
| FOM‡ | 0.82 |
Partial Disorder of the C2B Domain
The program O was used to build the initial model (Jones et al. 1991). Initially, the residues of the C2A domain of synaptotagmin I were changed to the homologous residues for the corresponding synaptotagmin III residues. The electron density of the C2B domain was smeared along the plane of the β-sandwich, especially at the distal end of the domain. This effect is a consequence of the pivoting motion of the C2B domain about the inter-domain linker. This motion would also explain the observed diffuse scatter in the diffraction pattern (not shown). We relied on the primary sequence similarity of the C2B domain with the C2A and PKC-β domains to aid initial model building in the most disordered regions. The seven-residue α-helix between the between strands β7 and β8 of the C2B domain was constructed from the available electron density.
Refinement
The model phases were used as prior phase probability distributions to feed back into the heavy atom model refinement resulting in significantly improved SIRAS phases. The MLHL target function (Pannu et al. 1998) was used with the SIRAS experimental phases. Refinement included several rounds of conjugate gradient minimization, torsion angle simulated annealing at 3,000 K (Rice and Brunger 1994), grouped B-factor refinement for both domains, and individual B-factor refinement for the C2A domain only (Table , A and B). Refinement progress was monitored by a concomitant drop in Rfree (Brunger 1992) and R. All phasing and refinement calculations were carried out with CNS (Brunger et al. 1998). The bulk solvent model used a density level (κsol) of 0.27 e−/Å3 and a B-factor of 25 Å2. The final model had 98.7% of all residues in allowed regions. Three residues in the C2B domain are reported as disallowed (Ala 553, Lys 557, and Ser 475). Due to the disorder in a portion of the C2B domain, several side chain rotamers could not be absolutely determined and were set to those found in common with the PKC-β (1a25) and the synaptotagmin I (1rsy) C2 domains. It should be noted, however, that the side chain rotamers in the Ca+2-binding region of the C2B domain could be unambiguously assigned (see Fig. 3 A). The Mg+2 and the sulfate ions were included in the model after the protein tracing was completed. It should be noted that at this resolution one cannot exclude the possibility that some of the Mg+2 electron density peaks correspond to ordered water molecules. However, the observed Mg+2 sites correspond to known sites in the divalent cation-binding sites of other C2 domains. The last few COOH-terminal residues of synaptotagmin III are predicted to be α-helical by secondary structure prediction (King and Sternberg 1996). Although they are present in the expression construct used for crystallization, they were disordered in the crystal structure.
Table 3.
Crystallographic Refinement
| A | Resolution (Å) | 50–5.47 | 4.34 | 3.79 | 3.45 | 3.2 | 50–3.2 |
|---|---|---|---|---|---|---|---|
| R | 0.307 | 0.235 | 0.314 | 0.302 | 0.344 | 0.292 | |
| RFree | 0.361 | 0.324 | 0.347 | 0.331 | 0.388 | 0.347 | |
| B | Resolution range (Å) | 50–3.2 | |||||
| Number of protein atoms | 2677 | ||||||
| Number of non-protein atoms | 9 | ||||||
| R (%) | 29.2 | ||||||
| RFree (%) | 34.7 | ||||||
| Rmsd lengths (Å) | 0.0084 | ||||||
| Angles (°) | 1.6 | ||||||
| Avg. B (all atoms) (Å)§ | 95.4 | ||||||
| Avg. B (C2A/C2B) (Å)§ | 75.5/ 105.9 |
Figure 3.
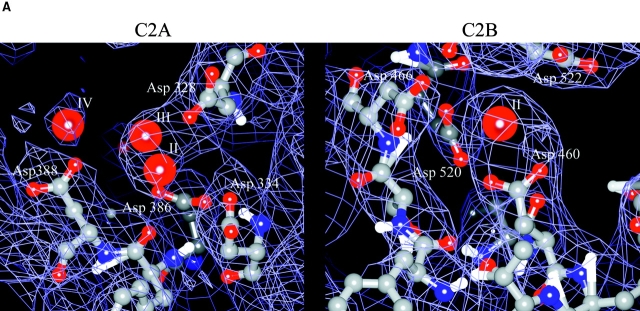
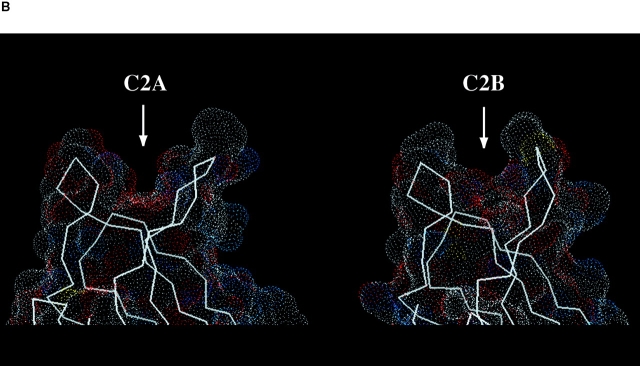
(A) Experimental electron density map using native amplitudes and density modified SIRAS phases of the C2A and C2B Ca+2-binding pockets contoured at 1.5 σ. Some of the coordinating aspartic acid residues are shown as balls-and-sticks. The labels for the Mg+2 sites follow the numbering convention of the C2 domain of PLC-δ1 (Essen et al. 1997). (B) Connelly surface obtained by MidasPlus of synaptotagmin III C2A and C2B using a probe radius of 1.4 Å. Arrows point to the divalent binding pockets in the C2 domains.
Results
Overall Topology
The cytosolic portion of synaptotagmin III contains two tandem C2 domains joined by a short seven-residue linker (Fig. 1). Each of the two homologous C2 domains consists of an eight-stranded Greek key β-sandwich with type I (S-type) C2 topology (Sutton et al. 1995). The Ca+2-binding pocket (CalB) of each domain is located at the apex of the fold and the divalent cations are cradled in between three conserved loops. The fold of each C2 domain contains a series of α-β bulges (Sutton and Sprang 1998), which are unique to C2 domains. This motif determines the overall shape of the domain and may position key residues to surface accessible locations.
Figure 1.
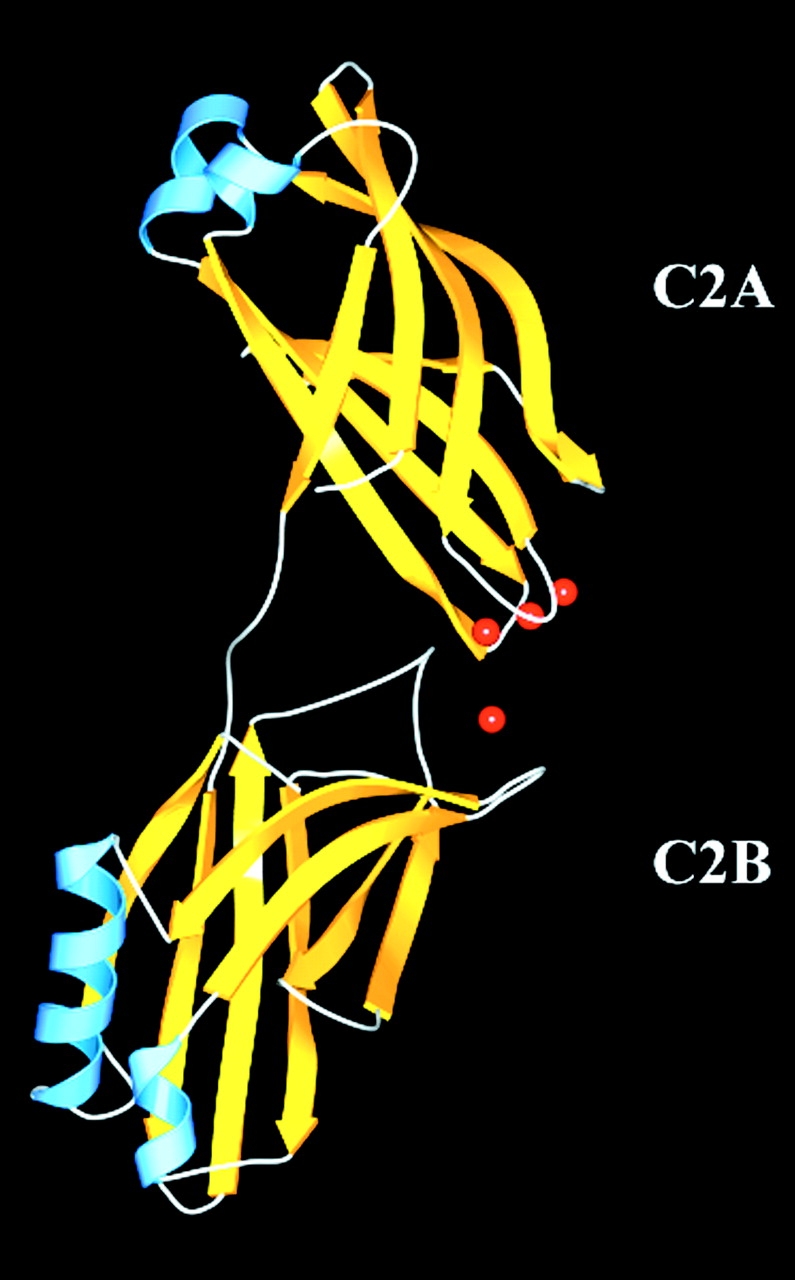
Overall structure of the cytosolic domain of synaptotagmin III. The gold arrows correspond to β-strands, the blue corresponds to α-helices, and the red spheres to Mg+2.
The Ca+2-binding regions of both C2 domains are directed toward each other in the crystal structure. Only ∼300 Å2 of surface area is shared between the two domains, which is likely too small to represent an important interaction in solution. It is therefore unlikely that this contact has physiological function. Thus, we predict that the two C2 domains are largely independent of each other in solution. The Ca+2-binding loop 3 of C2B is disulfide linked to another C2B domain that is related by crystallographic symmetry to another molecule. This covalent linkage does not affect the overall loop conformation since the Ca+2-binding loops of the superimposed C2A and C2B domains are very similar.
The relative orientation of the two C2 domains (Fig. 1) represents the conformation of synaptotagmin III C2A-C2B favored by crystallization. The observed partial disorder of the C2B domain in this crystal form suggests that the relative orientation and position of the two C2 domains is variable in solution. This anisotropic disorder is confined to the plane of the β-sandwich, and the electron density of the β-sheets of the C2B domain is somewhat smeared out. However, the electron density for the C2B domain at the apex of the fold, including the Ca+2-binding pocket, is well defined, allowing sidechain interpretation of the Ca+2-binding region of the C2B domain (see Fig. 3 A).
The C2A and C2B domains are structurally similar with the exception of the α-helix between the strands β7 and β8 of each C2 domain (Fig. 1 and Fig. 2). The seven-residue α-helix of the C2B domain is not present in either the C2A domain of synaptotagmin I, the isolated C2 domain of PKC-β, or the type-II (P-type) C2 domain of PLC-δ1. This conserved α-helical insertion has also been reported in the isolated C2B domain of rabphilin-3A (Ubach et al. 1999) and may have functional importance (Chung et al. 1998). The root-mean-square difference (Rmsd) between the rabphilin C2B and the synaptotagmin III C2B domains computed over all Cα atoms is 1.5Å.
Figure 2.
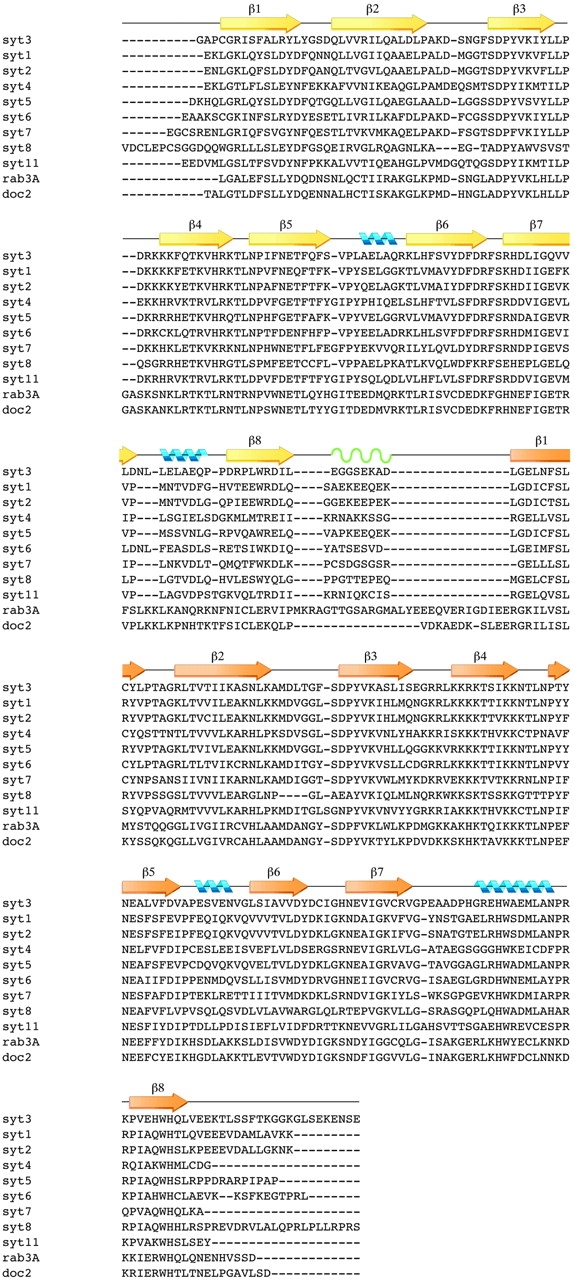
Sequence alignment of the synaptotagmin homology group with ClustalW: yellow arrows correspond to β-strands in the C2A domain; gold arrows correspond to β-strands in the C2B domain; green waves correspond to the flexible linker. Blue helices correspond to α-helices. Syt3, mouse synaptotagmin III (294–588) (Swiss-Prot: SYT3_MOUSE); Syt1, mouse synaptotagmin I (271–421) (Swiss-Prot: SYT1_MOUSE); Syt4, rat synaptotagmin IV (152–425) (Swiss-Prot: SYT4_RAT); Syt5, rat synaptotagmin V (105–386) (Swiss-Prot: SYT3_RAT); Syt6, rat synaptotagmin VI (226–511) (GenBank: U2O105); Syt7, rat synaptotagmin VII (129–403) (GenBank: U20106); Syt8, mouse synaptotagmin VIII (61–355); Syt11, rat synaptotagmin XI (152–430) (GenBank:AF000432); rab3A, rat rabphilin-3a (383–684) (SWISS-PROT: RP3A_RAT); doc2, mouse doc2 (125–412) (PIR: JC4921).
As in the structure of the C2A domain of synaptotagmin I and the isolated C2 domain from protein kinase C, the C2A domain of synaptotagmin III contains a cis-proline (Pro 411) that precedes the β-strand containing the polybasic region. The homologous region in C2B, however, does not contain this proline. Instead, it uses a non-proline β-turn to accommodate the residues leading into the polybasic strand. In the C2B domain, this insertion shifts the polybasic region to a more central location on strand β4.
Ca+2-Binding Pockets
Synaptotagmin III crystallized in the presence of 1.5 M MgSO4. Although Mg+2 could potentially mimic Ca+2 binding, the aspartate residues in the C2A domain that have been shown by solution NMR and x-ray crystallography to pivot upon divalent cation binding, are not in the Ca+2-bound conformation (Fig. 3 A). Under these crystallization conditions, the synaptotagmin III C2A domain is in the unliganded conformation. However, three peaks in electron density difference maps were found in the Ca+2-binding pocket of the C2A domain. These peaks coincide with the positions of the three calcium ions in the PKC-β C2 structure and were interpreted as Mg+2. The Mg+2 could be responding to the negative electrostatic field from the aspartate residues within the divalent cation-binding pocket of the C2A domain without proper coordination of the aspartate residues.
In the C2B domain, significant electron density, consisting of a single 5σ peak, was also found in the vicinity of the Ca+2-binding region (Fig. 3 A). This peak corresponds to the high affinity Ca+2 site observed in the crystal structures of the synaptotagmin I C2A, the PKC-β C2, and the PLC-δ1 C2 domains. Although the divalent cation coordinating aspartate residues are present in the C2B domain, only one Mg+2 binds in the divalent cation-binding pocket, despite the very high Mg+2 concentration used for crystallization. The interpretation of this electron density peak in terms of a divalent cation-binding site is supported by the observed substitution of the site by a trimethyl lead ion in the TMLA derivative.
The Inter-Domain Linker
The linker between C2 domains for most of the synaptotagmin isoforms, including synaptotagmin III, is seven to nine residues in length (Fig. 2). The amino acid composition of synaptotagmin III suggests a rather flexible linkage with two contiguous glycine residues. The primary sequence of the linker is not conserved among the synaptotagmin homology group (Fig. 2). The inter-domain linker may be more rigid for the synaptotagmin isoforms that have fewer glycine residues and more proline residues, such as synaptotagmin VII or synaptotagmin VIII (Fig. 2). The rigidity of the inter-domain linker in some synaptotagmin isoforms may position the C2 domains for docking to vesicle-fusion related protein complexes, or otherwise restrict the range of motion possible between the two C2 domains. Other tandem C2 domain containing proteins such as rabphilin (Shirataki et al. 1993) or rim (Wang et al. 1997) have much longer linkers, thus providing more flexibility between C2 domains.
Discussion
Structural Implications for Biochemically Distinct C2 Domains
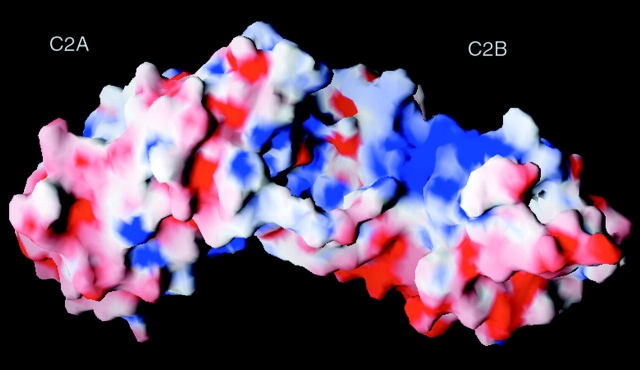
Synaptotagmin III is characterized by promiscuous binding to various accessory proteins and membrane components (Li et al., 1995). The two C2 domains of synaptotagmin have different binding partners and binding affinities. The x-ray crystal structure of the cytosolic domain of synaptotagmin III provides a structural explanation of these disparities between the two C2 domains. Despite the lack of Ca+2 in the crystallization condition, this structure could still mimic some of the Ca+2-binding properties of C2B domains in the synaptotagmin homology group. In the crystal structure, the C2B domain associates with only one Mg+2. The reduced divalent cation-binding capacity of the C2B domain would leave this area of the molecule with a residual negative charge relative to the C2A domain. This difference between the C2A and C2B domains may explain some of the biochemical differences observed in in vitro experiments. Overall, the C2A domain of synaptotagmin III has a more uniform electrostatic surface potential than the C2B domain (Fig. 4). The surface of the C2B domain possesses distinctly basic and acidic areas (Fig. 4). These charged areas may be important for the Ca+2-independent interactions observed in the isolated C2B domain.
Figure 4.
Surface plot showing the electrostatic potential of synaptotagmin. Blue, positive; red, negative charge. Charges were obtained from the OPLS force field. The electrostatic surface was contoured between −10kT/e and +10kT/e. Figure prepared with GRASP (Nicholls et al. 1991).
The Ca+2-binding pocket of the C2B domain is chemically similar to the Ca+2-binding pocket of the C2A domain; however, the shape of the pocket is very different (Fig. 3 B). In principle, this can be explained by either a difference in backbone conformation between the two domains or by a difference in sidechain packing. Superposition of the two domains does not reveal a significant deviation in the backbone position; however, one cannot rule out more subtle backbone differences within the coordinate error of the crystal structure. Although synaptotagmin III has been implicated in Mg+2-dependent phospholipid binding (Fukuda et al. 1997), the crystal structure indicates that 1.5 M Mg+2 is unable to induce rotamers of the aspartic acid residues similar to those found in C2A domains complexed with Ca+2. In the presence of Ca+2, crystallographic and NMR studies indicated that Asp 466 (and homologous residues in other synaptotagmin isoforms) pivot to coordinate the highest affinity Ca+2 in the C2A domain (Sutton et al. 1995; Shao et al. 1998; Sutton and Sprang 1998). Therefore, the Mg+2 in the synaptotagmin C2A-C2B structure are probably responding to the negatively charged surface of the C2A domain rather than coordinating with specific residues. The ionic radius of Mg+2 may also be too small to induce a Ca+2-like coordination.
A sulfate ion from the crystallization medium is located near Lys 356 of the C2A domain of synaptotagmin III. This ion may mimic phospholipid binding to synaptotagmin, since this face of the C2A domain interacts with the phospholipid bilayer (Chae et al. 1998). The crystal structure of the C2A domain of synaptotagmin I also contained a sulfate ion derived from the crystallization (Sutton et al. 1995). The sulfate ion in this structure is in a different position; however, it is associated with the same face of the protein. This indicates that the electrostatic potential is positively charged at this location on the protein.
Model of C2B-mediated Oligomerization
Only one molecule is present in the asymmetric unit of this crystal form. The crystallographic symmetry together with primary sequence analysis of the available synaptotagmin isoforms may provide clues to the mechanism of C2B domain homo- and heterodimerization. Two crystallographically related molecules in this hexagonal crystal form direct their divalent cation-binding pockets toward the sixfold crystallographic axis. The resulting crystal packing contacts include the sequence Asp-Phe-Asp (386–388), which includes two of the divalent cation-binding residues in C2 domains. These three residues are conserved in all C2A domains with the exception of synaptotagmin VII. In the C2B domain, the homologous motif is Asp-Tyr-Asp (520–522), with few exceptions among the other synaptotagmin isoforms. In the crystal structure of the synaptotagmin III C2A-C2B domains, an alternate rotamer of Tyr 521 can be modeled to coordinate Asp 466 of the crystallographically related molecule to form a hydrogen bond. Aspartate 466 anchors the divalent cation-binding chain in the Ca+2-binding pocket of C2 domains. This putative interaction may provide an initial nucleation point for self-association. The sequence variability present in the other isoforms of synaptotagmin, for example, synaptotagmin VII, VIII, and XI, may modulate their individual binding properties to other synaptotagmin isoforms.
Interactions of Synaptotagmin with the SNARE Complex
The core of the SNARE complex, composed of synaptobrevin-II (1–96), syntaxin-1A (180–262), SNAP-25A (1–83), and SNAP-25A (130–206), interacts with the C2A and C2B domains of synaptotagmin III independent of divalent cations (Fig. 5). We have modeled the association between these two moieties using the following arguments: first, the position of the presynaptic membrane restricts the possible contacts between the two C2 domains of synaptotagmin and the SNARE complex. The syntaxin component of the SNARE complex embeds its transmembrane α-helix into the presynaptic membrane; likewise, the synaptobrevin transmembrane α-helix is anchored in the vesicle phospholipid membrane. Our model, therefore, puts the synaptobrevin α-helix on top of the complex and the syntaxin α-helix on the bottom relative to the presynaptic membrane bilayer on the bottom of Fig. 6. The palmitoylated SNAP-25 linker would also contact the presynaptic membrane, so it localizes to the membrane associated with syntaxin. Second, the divalent cation-binding loops (CalB) of the C2 domains of PLA2 contact phospholipid bilayer with some residues embedded in the bilayer itself (Chae et al. 1998; Ball et al. 1999). Ca+2 in the CalB region either directly binds phospholipid headgroups through Ca+2 bridging (Swairjo et al. 1995) or indirectly binds by quenching the electrostatic charge of the CalB domain, in situ (Shao et al. 1997). This restriction would allow the C2A and C2B domains of synaptotagmin to straddle the fusion complex and to interact simultaneously with the presynaptic membrane. Third, the unique electrostatic properties of the SNARE complex and synaptotagmin allow the positively charged, polybasic region of the C2 domains to interact with the negatively charged surface at the center of the SNARE complex (Sutton et al. 1998). Fourth, the cupped shape of the polybasic region of the C2 domains provides a favorable interaction surface with the cylindrical SNARE complex. Fifth, the length and flexibility of the inter-domain linker restrict the possible C2 domain orientation with respect to the SNARE complex. Most isoforms of synaptotagmin have inter-domain linkers of eight to nine residues in length (Fig. 2). The C2 domains presumably act as independent rigid bodies, so the inter-domain linker governs the available flexibility between them. The amino acid composition of this linker is not conserved among synaptotagmin isoforms. The proportion of glycine residues and therefore, the degree of flexibility possible between C2 domains varies by isoform. Some of the linker regions of synaptotagmin contain proline residues; therefore, the C2 domains of these isoforms are probably more restricted in their possible orientations.
Figure 5.
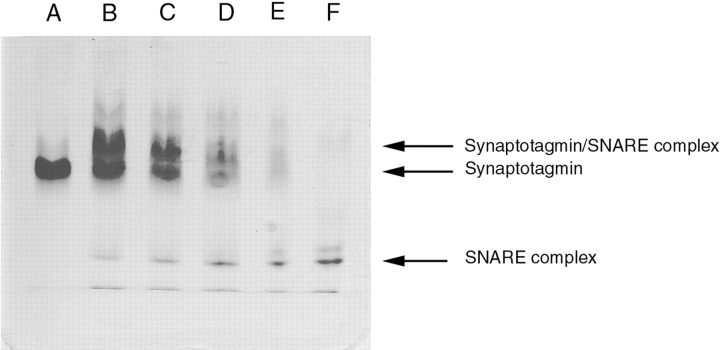
Ca+2-independent synaptotagmin-SNARE complex interaction: 10–15% native Phast gel (Pharmacia) run in the presence of 1 mM EDTA. Lanes B–F contain 1.6 μg of core SNARE complex; lanes A–E contain 3.2, 3.2, 2.4, 1.6, and 0.8 μg of synaptotagmin III C2AB.
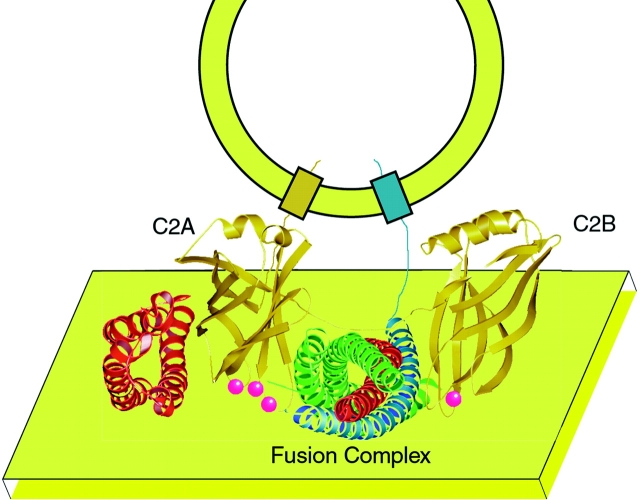
Figure 6.
Cartoon illustrating the proposed association between synaptotagmin (gold colored protein) and the SNARE fusion complex (SNAP25, green helix; syntaxin, red helices; and synaptobrevin, blue helix) and the interaction with the phospholipid membrane (yellow plane). Ca+2 are illustrated as red spheres. The transmembrane anchor of syntaxin and the linker between the H1-H2 domain (red helices to the left of C2A) and the H3 domain of syntaxin (red helices in the SNARE bundle) have been omitted for clarity. In our model, we predict that the Ca+2-binding sites of synaptotagmin interact with the presynaptic membrane. The interactions between the SNARE complex and synaptotagmin probably involve the COOH-terminal (membrane-proximal) portion of the SNARE core complex (Kee and Scheller 1996) and the cup-shaped, polybasic regions of the synaptotagmin C2 domains. The association between the NH2-terminal (H1-H2) domain of syntaxin (red helices to the left of the C2A domain) and the C2A domain of synaptotagmin was modeled according to NMR data (Shao et al. 1997). The peptide distance between the transmembrane domain and the SNARE-binding domain of synaptobrevin (blue helix) is exaggerated in this cartoon.
Specific regions of the SNARE complex are likely to be involved in synaptotagmin C2 domain interactions. The NH2-terminal domain of syntaxin interacts with the C2A domain of synaptotagmin I (Shao et al. 1997). The COOH-terminal 220–240 residues of syntaxin bind in a Ca+2-dependent manner to the cytoplasmic domain of synaptotagmin (Kee and Scheller 1996). The C2B domain of synaptotagmin also binds to SNAP-25. This interaction is not affected by the deletion of the COOH-terminal 26 residues by BoNT/E (Schiavo et al. 1997) suggesting that the NH2-terminal region of SNAP-25 is not involved in binding the C2B domain. Therefore, the C2B domain of synaptotagmin probably interacts with adjacent regions of SNAP-25 and syntaxin on the surface of the synaptic fusion complex in a Ca+2-independent fashion (Fig. 6).
According to the zipper model of SNARE-mediated fusion (Hanson et al. 1997; Lin and Scheller 1997; Fiebig et al. 1999), the SNARE proteins associate from the NH2 to COOH terminus as the neurotransmitter-filled synaptic vesicle and presynaptic membranes approach each other. In the early stages of fusion, synaptotagmin may act as a negative regulator of exocytosis by preventing the completion of the “zippering” action of the SNARE complex by binding to the COOH-terminal, membrane attachment side of the SNARE complex through surface electrostatic interactions on the fusion complex, independent of divalent cations. Indeed, synaptotagmin may bind with higher affinity to the partially assembled SNARE complex than to the fully assembled complex. When an action potential or other fusion signal triggers a Ca+2 flux, as in the case of neuronal exocytosis, synaptotagmin may then bind to the presynaptic phospholipid membrane in a Ca+2-dependent manner in preference to the SNARE complex. Therefore, synaptotagmin binding to the SNARE complex or the presynaptic membrane would be mutually exclusive properties of synaptotagmin. Once synaptotagmin is associated with the membrane, the SNARE complex would then be available to mediate fusion between the vesicle membrane and the presynaptic phospholipid membrane.
Acknowledgments
The authors thank P.D. Adams for stimulating discussions and H. Bellamy for assistance with data collection at SSRL BL1-5. We would like to thank Pat Fleming for help with the packing analysis of synaptotagmin III, Thomas Südhof for the initial clone synaptotagmin III, Josep Rizo for the help with the correct alignment of C2B domains, Mark Bowen for helpful discussion, Mike Reese and Steve Kaiser for preparing the fusion complex, and Stephen Sprang who provided a Welsh grant (I-1229) for early work on synaptotagmin III.
The SSRL Biotechnology Program is supported by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program and the Department of Energy, Office of Biological and Environmental Research. J.A. Ernst is supported by a pre-doctoral fellowship in biophysics from the National Science Foundation. Coordinates will be deposited with the PDB and are also at http://atb.csb. yale.edu.
Footnotes
1.used in this paper: PKC, protein kinase C; SNARE, soluble N-ethylmaleimide–sensitive factor attachment protein receptor; SIRAS, single isomorphous replacement, anomalous scattering; SytI, synaptotagmin I; TMLA, trimethyl lead acetate
References
- Ball A., Nielsen R., Gelb M.H., Robinson B.H. Interfacial membrane docking of cytosolic phospholipase A2 C2 domain using electrostatic potential-modulated spin relaxation magnetic resonance. Proc. Natl. Acad. Sci. USA. 1999;96:6637–6642. doi: 10.1073/pnas.96.12.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M.K., Calakos N., Scheller R.H. Syntaxina synaptic protein implicated in docking of synaptic vesicles at presynaptic active zones. Science. 1992;257:255–259. doi: 10.1126/science.1321498. [DOI] [PubMed] [Google Scholar]
- Bernard A., Payton M. Fermentation and growth of Escherichia coli for optimal protein production. Curr. Prot. Prot. Sci. 1995;5:1–18. doi: 10.1002/0471140864.ps0503s00. [DOI] [PubMed] [Google Scholar]
- Bommert K., Charlton M.P., DeBello W.M., Chin G.J., Betz H., Augustine G.J. Inhibition of neurotransmitter release by C2-domain peptides implicates synaptotagmin in exocytosis. Nature. 1993;363:163. doi: 10.1038/363163a0. [DOI] [PubMed] [Google Scholar]
- Brose N., Petrenko A.G., Südhof T.C., Jahn R. Synaptotagmina calcium sensor on the synaptic vesicle surface. Science. 1992;256:1021–1025. doi: 10.1126/science.1589771. [DOI] [PubMed] [Google Scholar]
- Brunger A.T. The free R valuea novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–474. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- Brunger A.T., Adams P.D., Clore G.M., Gros P., Grosse-Kunstleve R.W., Jiang J.-S., Kuszewski J., Nilges M., Pannu N.S., Read R.J. Crystallography & NMR System (CNS)a new software system for macromolecular structure determination. Acta Cryst. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chae Y.K., Abildgaard F., Chapman E.R., Markley J.L. Lipid binding ridge on loops 2 and 3 of the C2A domain of synaptotagmin I as revealed by NMR spectroscopy. J. Biol. Chem. 1998;273:25659–25663. doi: 10.1074/jbc.273.40.25659. [DOI] [PubMed] [Google Scholar]
- Chapman E.R., Desai R.C., Davis A.F., Tornehl C.K. Delineation of the oligomerization, AP-2 binding, and synprint binding region of the C2B domain of synaptotagmin. J. Biol. Chem. 1998;273:32966–32972. doi: 10.1074/jbc.273.49.32966. [DOI] [PubMed] [Google Scholar]
- Chapman E.R., Hanson P.I., An S., Jahn R. Ca+2 regulates the interaction between synaptotagmin and syntaxin 1. J. Biol. Chem. 1995;270:23667–23671. doi: 10.1074/jbc.270.40.23667. [DOI] [PubMed] [Google Scholar]
- Chung S.-H., Song W.-J., Kim K., Bednarski J., Chen J., Prestwich G.D., Holz R.W. The C2 domains of rabphilin 3A specifically bind phosphatidylinositol 4,5-biphosphate containing vesicles in a Ca+2-dependent manner. J. Biol. Chem. 1998;273:10240–10248. doi: 10.1074/jbc.273.17.10240. [DOI] [PubMed] [Google Scholar]
- Davletov B., Perisic O., Williams R.L. Calcium-dependent membrane penetration is a hallmark of the C2 domain of cytosolic phospholipase A2 whereas the C2A domain of synaptotagmin binds membranes electrostatically. J. Biol. Chem. 1998;273:19093–19096. doi: 10.1074/jbc.273.30.19093. [DOI] [PubMed] [Google Scholar]
- DeLorenzo R.J. Calmodulin in neurotransmitter release and synaptic function. Fed. Proc. 1982;41:2265–2272. [PubMed] [Google Scholar]
- Essen L.O., Perisic O., Lynch D.E., Katan M., Williams R.L. A ternary metal binding site in the C2 domain of phosphoinositide-specific phospholipase C-δ1. Biochemistry. 1997;36:2753–2762. doi: 10.1021/bi962466t. [DOI] [PubMed] [Google Scholar]
- Fiebig K.M., Rice L.M., Pollock E., Brunger A.T. Folding intermediates of SNARE complex assembly. Nat. Struct. Biol. 1999;6:117–123. doi: 10.1038/5803. [DOI] [PubMed] [Google Scholar]
- Fukuda M., Kojima T., Mikoshiba K. Regulation by bivalent cations of phospholipid binding to the C2A domain of synaptotagmin III. Biochem. J. 1997;323:421–425. doi: 10.1042/bj3230421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse-Kunstleve R.W., Brunger A.T. A highly automated heavy-atom search procedure for macromolecular structures. Acta Cryst. D. 1999;In press doi: 10.1107/s0907444999007763. [DOI] [PubMed] [Google Scholar]
- Hanson P.I., Roth R., Morisaki H., Jahn R., Heuser J.E. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- Hutton J.C. Calcium-binding proteins and secretion. Cell Calcium. 1986;7:339–352. doi: 10.1016/0143-4160(86)90037-0. [DOI] [PubMed] [Google Scholar]
- Jones T.A. J.Y., Zou S., Cowan, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Katz B., Miledi R. On the timing of calcium action during neuro-muscular transmission. J. Physiol. 1967;187:535–544. doi: 10.1113/jphysiol.1967.sp008183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y., Scheller R.H. Localization of synaptotagmin-binding domains on syntaxin. J. Neurosci. 1996;16:1975–1981. doi: 10.1523/JNEUROSCI.16-06-01975.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King R.D., Sternberg M.J. Identification and application of the concepts important for accurate and reliable protein secondary structure prediction. Prot. Sci. 1996;5:2298–2310. doi: 10.1002/pro.5560051116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leveque C., el Far O., Martin-Moutot N., Sato K., Kato R., Takahashi M., Seagar M.J. Purification of the N-type calcium channel associated with syntaxin and synaptotagmin. A complex implicated in synaptic vesicle exocytosis. J. Biol. Chem. 1994;269:6306–6312. [PubMed] [Google Scholar]
- Li C., Ullrich B., Zhang J., Anderson R., Brose N., Südhof T. Ca+2-dependent and –independent activities of neural and non-neural synaptotagmins Nature. 375 1995. 594 599a [DOI] [PubMed] [Google Scholar]
- Li C., Davletov B.A., Südhof T.C. Distinct Ca+2 and Sr+2 binding properties of synaptotagmins. Definition of candidate Ca+2 sensors for the fast and slow components of neurotransmitter release J. Biol. Chem. 270 1995. 24898 24902b [DOI] [PubMed] [Google Scholar]
- Li L., Singh B.R. Isolation of synaptotagmin as a receptor for types A and E botulinum neurotoxin and analysis of their comparative binding using a new microtiter plate assay. J. Nat. Toxins. 1998;7:215–226. [PubMed] [Google Scholar]
- Lin R.C., Scheller R.H. Structural organization of the synaptic exocytosis core complex. Neuron. 1997;19:1087–1094. doi: 10.1016/s0896-6273(00)80399-2. [DOI] [PubMed] [Google Scholar]
- Littleton J.T., Serano T.L., Rubin G.M., Ganetzky B., Chapman E.R. Synaptic function modulated by changes in the ratio of synaptotagmin I and IV. Nature. 1999;400:757–760. doi: 10.1038/23462. [DOI] [PubMed] [Google Scholar]
- Martin-Moutot N., Charvin N., Leveque C., Sato K., Nishiki T., Kozaki S., Takahashi M., Seagar M. Interaction of SNARE complexes with P/Q-type calcium channels in rat cerebellar synaptosomes. J. Biol. Chem. 1996;271:6567–6570. doi: 10.1074/jbc.271.12.6567. [DOI] [PubMed] [Google Scholar]
- Mehrotra B., Elliott J.T., Chen J., Olszwqkei J., Profit A., Chaudhary A., Fukuda M., Mikshiba K., Prestwich G. Selective photoaffinity labeling of the inositol polyphosphate binding C2B domains of synaptotagmins. J. Biol. Chem. 1997;272:4237–4244. doi: 10.1074/jbc.272.7.4237. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp K.A., Honig B. Protein folding and associationInsights from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct. Funct. Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Nishiki T., Kamata Y., Nemoto Y., Omori A., Ito T., Takahashi M., Kozaki S. Identification of protein receptor for Clostridium botulinum type B neurotoxin in rat brain synaptosomes. J. Biol. Chem. 1994;269:10498–10503. [PubMed] [Google Scholar]
- Osborne S.L., Herreros J., Bastiaens P.I., Schiavo G. Calcium-dependent oligomerization of synaptotagmins I and II. Synaptotagmins I and II are localized on the same synaptic vesicle and heterodimerize in the presence of calcium. J. Biol. Chem. 1999;274:59–66. doi: 10.1074/jbc.274.1.59. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1998;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pannu N.S., Murshudov G.N., Dodson E.J., Read R.J. Incorporation of prior phase information strengthens maximum likelihood structural refinement. Acta Cryst. D. 1998;54:1285–1294. doi: 10.1107/s0907444998004119. [DOI] [PubMed] [Google Scholar]
- Parker P.J., Coussens L., Totty N., Rhee L., Young S., Chen E., Stabel S., Waterfield M.D., Ullrich A. The complete primary structure of protein kinase C—the major phorbol ester receptor. Science. 1986;233:853–859. doi: 10.1126/science.3755547. [DOI] [PubMed] [Google Scholar]
- Pollard H.D., Pazoles C.J., Creutz C., Zinder O. Role of intracellular proteins in the regulation of calcium action and transmitter release during exocytosis. Monogr. Neural Sci. 1980;7:106–116. doi: 10.1159/000388818. [DOI] [PubMed] [Google Scholar]
- Reist N.E., Buchanan J., Li J., DiAntonio A., Buxton E.M., Schwarz T.L. Morphologically docked synaptic vesicles are reduced in synaptotagmin mutants of Drosophila . J. Neurosci. 1998;18:7662–7673. doi: 10.1523/JNEUROSCI.18-19-07662.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice L.M., Brunger A.T. Torsion angle dynamicsreduced variable conformational sampling enhances crystallographic structure refinement. Proteins Struct. Funct. Genet. 1994;19:277–290. doi: 10.1002/prot.340190403. [DOI] [PubMed] [Google Scholar]
- Schiavo G., Gmachl M.J., Stenbeck G., Sollner T.H., Rothman J.E. A possible docking and fusion particle for synaptic transmission. Nature. 1995;378:733–736. doi: 10.1038/378733a0. [DOI] [PubMed] [Google Scholar]
- Schiavo G., Gu Q.M., Prestwich G.D., Sollner T.H., Rothman J.E. Calcium-dependent switching of the specificity of phosphoinositide binding to synaptotagmin. Proc. Natl. Acad. Sci. USA. 1996;93:13327–13332. doi: 10.1073/pnas.93.23.13327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavo, G., G. Stenbeck, J.E. Rothman, and T.H. Sollner. 1997. Binding of the synaptic vesicle v-SNARE, synaptotagmin, to the plasma membrane t-SNARE, SNAP-25, can explain docked vesicles at neurotoxin-treated synapses. Proc. Natl. Acad. Sci. USA. 997–1001. [DOI] [PMC free article] [PubMed]
- Shao X., Li C., Fernandez I., Zhang X., Südhof T.C., Rizo J. Synaptotagmin-syntaxin interactionthe C2 domain as a Ca+2-dependent electrostatic switch. Neuron. 1997;18:133–142. doi: 10.1016/s0896-6273(01)80052-0. [DOI] [PubMed] [Google Scholar]
- Shao X., Fernandez I., Südhof T.C., Rizo J. Solution structures of the Ca+2-free and Ca+2-bound C2A domain of synaptotagmin Idoes Ca+2induce a conformational change? Biochemistry. 1998;37:16106–16115. doi: 10.1021/bi981789h. [DOI] [PubMed] [Google Scholar]
- Shirataki H., Kaibuchi K., Sakoda T., Kishida S., Yamaguchi T., Wada K., Miyazaki M., Takai Y. Rabphilin-3A, a putative target protein for smg p25A/rab3A p25 small GTP-binding protein related to synaptotagmin. Mol. Cell Biol. 1993;13:2061–2068. doi: 10.1128/mcb.13.4.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton R.B., Davletov B.A., Berghuis A.M., Südhof T.C., Sprang S.R. Structure of the first C2 domain of synaptotagmin Ia novel Ca+2/phospholipid-binding fold. Cell. 1995;80:929–938. doi: 10.1016/0092-8674(95)90296-1. [DOI] [PubMed] [Google Scholar]
- Sutton R.B., Fasshauer D., Jahn R., Brunger A.T. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- Sutton R.B., Sprang S.R. Structure of the protein kinase C-β phospholipid-binding C2 domain complexed with Ca+2 . Structure. 1998;6:1395–1405. doi: 10.1016/s0969-2126(98)00139-7. [DOI] [PubMed] [Google Scholar]
- Swairjo M.A., Concha N.O., Kaetzel M.A., Dedman J.R., Seaton B.A. Ca+2-bridging mechanism and phospholipid head group recognition in the membrane-binding protein annexin V. Nat. Struct. Biol. 1995;2:968–974. doi: 10.1038/nsb1195-968. [DOI] [PubMed] [Google Scholar]
- Ubach J., Garcia J., Nittler M.P., Südhof T.C., Rizo J. The C2B-domain of rabphilinstructural variations in a Janus-faced domain. Nat. Cell Biol. 1999;1:106–112. doi: 10.1038/10076. [DOI] [PubMed] [Google Scholar]
- Wang Y., Okamoto M., Schmitz F., Hofmann K., Südhof T.C. Rim is a putative Rab3 effector in regulating synaptic-vesicle fusion. Nature. 1997;388:593–598. doi: 10.1038/41580. [DOI] [PubMed] [Google Scholar]
- Zhang J.Z., Davletov B.A., Südhof T.C., Anderson R.G. Synaptotagmin I is a high affinity receptor for clathrin AP-2implications for membrane recycling. Cell. 1994;78:751–760. doi: 10.1016/s0092-8674(94)90442-1. [DOI] [PubMed] [Google Scholar]