Abstract
To learn more about holocentric chromosome structure and function, we generated a monoclonal antibody (mAb), 6C4, that recognizes the poleward face of mitotic chromosomes in Caenorhabditis elegans. Early in mitosis, mAb 6C4 stains dots throughout the nucleoplasm. Later in prophase, mAb 6C4 stains structures on opposing faces of chromosomes which orient towards the centrosomes at metaphase. Colocalization with an antibody against a centromeric histone H3–like protein and the MPM-2 antibody, which identifies a kinetochore-associated phosphoepitope present in a variety of organisms, shows that the mAb 6C4 staining is present adjacent to the centromere.
Expression screening using mAb 6C4 identified a protein in C. elegans that we named HCP-1 (for holocentric protein 1). We also identified a second protein from the C. elegans genome sequence database, HCP-2, that is 54% similar to HCP-1. When expression of HCP-1 is reduced by RNA interference (RNAi), staining with mAb 6C4 is eliminated, indicating that hcp-1 encodes the major mAb 6C4 antigen. RNAi with hcp-1 and hcp-2 together results in aberrant anaphases and embryonic arrest at ∼100 cells with different amounts of DNA in individual nuclei. These results suggest that HCP-1 is a centromere-associated protein that is involved in the fidelity of chromosome segregation.
Keywords: Caenorhabditis elegans, chromosome, mitosis, centromere, kinetochore
During anaphase of mitosis, sister chromatids are moved to opposite sides of the dividing daughter cells. In insects of the hemipteran order, various plants, and nematodes, the separating sister chromatids orient parallel to the poles and move as a whole. This type of movement was defined as holokinetic and reflects that these chromosomes attach to the mitotic spindle all along their length (Schrader 1935). Subsequent EM of these chromosomes revealed that this holokinetic movement was due to a diffuse or nonlocalized kinetochore structure that extends over the entire poleward surface of each chromosome (Buck 1967; Comings and Okada 1972; Albertson and Thomson 1982; Goday et al. 1985, Goday et al. 1992). The kinetochore is an ultrastructure located on the chromosome that is responsible for attachment of the chromosome to the mitotic spindles and regulating the progression of mitosis (for reviews see Pluta et al. 1995; Allshire 1997). Further indications that a diffuse kinetochore is present on holokinetic chromosomes was obtained when chromosome fragments generated in these organisms by irradiation were shown to be capable of proper segregation during mitosis, indicating that each chromosomal fragment had a functioning kinetochore associated with it (Hughs-Schrader and Ris 1941; Ris 1942; Hughs-Schrader and Schrader 1961). These chromosomes are generally referred to as holocentric to denote that the kinetochore structure extends over most of the chromosome (Reider 1982). By contrast, monocentric chromosomes like those of mammals have a discrete or localized kinetochore structure, and chromosome fragments generated from monocentric chromosomes lack the ability to segregate properly when separated from the centromere (Mather and Stone 1933).
Consistent with their function, the kinetochores of both holocentric and monocentric chromosomes resemble one another, at least at the ultrastructural level. Monocentric chromosomes are described with the site of spindle attachment localized to a distinct region of the chromosome, the primary constriction which is observed as a narrowing of the chromosome. EM studies of the primary constriction revealed that the kinetochore is composed of a trilaminar disk structure adjacent to each sister chromatid during metaphase (Jokelaninen 1967; Comings and Okada 1971; Roos 1973; Ris and Witt 1981; Reider 1982). However, holocentric chromosomes lack a primary constriction, and EM revealed that they also have a trilaminar kinetochore which extends nearly the entire length of the chromosome (Buck 1967; Comings and Okada 1972; Albertson and Thomson 1982; Goday et al. 1985, Goday et al. 1992). The similarity in kinetochore ultrastructure between monocentric and holocentric chromosomes suggests that the kinetochore is a conserved organelle and that differences attributed to each chromosome organization are due to the amount of the chromosome encompassed by the kinetochore (Albertson and Thomson 1982).
The molecular composition of the centromere is beginning to be elucidated. Antibodies have been useful in probing the structure of, and identifying gene products localized to the centromere. Antisera from patients with certain autoimmune diseases recognize the primary constriction of monocentric chromosomes, and by immunoelectron microscopy, identify the kinetochore (Moroi et al. 1980; Brenner et al. 1981). Further support that these antibodies recognize components of the centromere comes from the injection of purified antibodies from anticentromere serum into HeLa cells (Bernat et al. 1990, Bernat et al. 1991). In these experiments, kinetochore structure is disrupted and chromosome segregation is blocked. Several centromeric proteins (CENPs)1 have been identified using these autoantibodies and have been subsequently shown to be important for kinetochore structure and function (for reviews see Earnshaw and Mackay 1994; Pluta et al. 1995).
We have used a similar immunocytological approach to study the structure and function of holocentric chromosomes in the nematode Caenorhabditis elegans (Herman et al. 1976, Herman et al. 1979; Albertson and Thomson 1982). We generated a mAb, 6C4, which recognizes the centromere region of mitotic chromosomes in C. elegans. Using mAb 6C4, we have identified a protein, holocentric protein (HCP)-1, that together with a related protein, HCP-2, is important for proper chromosome segregation.
Materials and Methods
Immunostaining Procedures
mAb 6C4 was isolated in a screen of hybridoma cell lines which produce antibodies that recognize C. elegans embryos. Hybridoma cell lines were generated and ascites fluid was produced as described (Harlow and Lane 1988). Bristol strain N2 was cultured as described by Brenner 1974. Embryos were prepared by hypochlorite treatment (Albertson 1984) and attached to coverslips (type 1.5) pretreated with 3-aminopropyltriethoxysilane (Sigma Chemical Co.), and overlaid with an untreated coverslip. The coverslips were quick-frozen on dry ice and the overlaying coverslip rapidly removed. Fixation was in N,N-dimethylformamide or methanol at −20°C for 10 min. Slides were blocked for 30 min in PBS with 2 mM MgCl2, 0.1% Tween 20, and 3% BSA. Immunostaining was performed with mAb 6C4 ascites fluid at 1:500 dilution in blocking solution. Primary antibody was incubated for 1 h at room temperature. Coverslips were then washed three times in block for 5 min each before incubation with 10 μg/ml rhodamine-conjugated goat anti–mouse IgG (Jackson ImmunoResearch Laboratories, Inc.) for 30 min. Coverslips were then washed three times for 5 min each, with 0.05 mg/ml 4′,6-diamidino-3-phenylindole dihydrochloride (DAPI) included in the first wash. Coverslips were mounted on slides with 100% glycerol. Double labeling experiments were performed under similar conditions, except either an anti–β-tubulin antibody (Amersham Pharmacia Biotech), an anti–HCP-3 antibody (Buchwitz et al. 1999), or MPM-2 antibody (DAKO) at 10 μg/ml was included with mAb 6C4. Detection was by rhodamine-conjugated goat anti–rabbit IgG, goat anti–mouse IgG, or fluorescein-conjugated goat anti–mouse IgM.
High Resolution Microscopy
Samples were examined by three-dimensional multiple wavelength fluorescence microscopy using Deltavision (Applied Precision). Images were collected at the indicated wavelengths for 30 0.2-μm optical sections per nucleus and were subsequently deconvolved mathematically (Hiraoka et al. 1991; Carrington et al. 1995). The limit of detection (resolution between two points) was 90 nm. Data were examined as either optical sections or as a projection of the entire stack.
Expression Library Screening
The mixed stage C. elegans cDNA library in λgt11 was a gift from Dr. Pete Okema (University of Illinois at Chicago, Chicago, IL). Immunoscreening of the cDNA library was carried out by the method of Young and Davis 1983, using mAb 6C4 ascites fluid at 1:200 dilution. Detection of immunopositive plaques was by Vectastain ABC (Vector Labs Inc.).
RNA-mediated Inhibition
Oligos forward T7 (TAATACGACTCACTATAGGGggtggcgacgactcctgg), forward (ggtggcgacgactcctgg), reverse T7 (TAATACGACTCACTATAGGGttgacaccagaccaactgg), and reverse (ttgacaccagaccaactgg) were used to generate PCR products corresponding to the cDNA inserts obtained from the expression screening. This region included the COOH-terminal 324 amino acids of HCP-1 and 3′UTR. Each T7 oligo contains a T7 polymerase promoter (shown in capital letters) and sequence complementary to the flanking polylinker region of λgt11. Oligos HCP2-1 T7 (TAATACGACTCACTATAGGGgacctgaatgcgaagcttg), HCP2-1 (gacctgaatgcgaagcttg), HCP2-2 T7 (TAATACGACTCACTATAGGGgctggttgcagtttgagcgg), and HCP2-2 (gctggttgcagtttgagcgg) were used to PCR amplify a region of the HCP-2 mRNA corresponding to the COOH-terminal 384 amino acids of HCP-2, from total RNA, after first strand cDNA synthesis as described (Sambrook et al. 1989). 1 μg of PCR product was used to synthesize double-stranded RNA (dsRNA) using T7 polymerase as described (Grodberg and Dunn 1988). dsRNA was resuspended in water at a concentration of 5 mg/ml. dsRNA derived from the COOH-terminal region of HCP-1 or from the corresponding region of HCP-2 was injected into the syncytial gonad of wild-type hermaphrodites as described (Guo and Kemphues 1995; Fire et al. 1998).
Results
mAb 6C4 Recognizes the Poleward Face of Metaphase Chromosomes
We generated a mouse mAb, 6C4, that stains mitotic chromosomes in C. elegans. Holocentric chromosomes orient perpendicular to the spindle axis during metaphase and anaphase, with their respective kinetochores facing the spindle (Hughs-Schrader and Ris 1941; Albertson and Thomson 1982). Immunofluorescence with mAb 6C4 showed reactivity on the poleward face of metaphase chromosomes (Fig. 1). This staining pattern suggests that mAb 6C4 recognizes a protein important for holocentric chromosome structure and function.
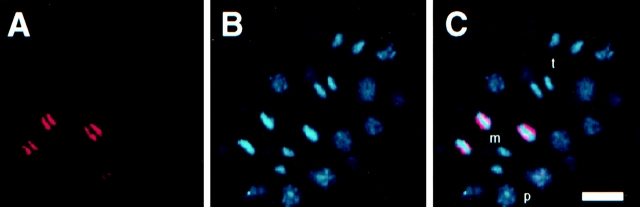
Figure 1.
mAb 6C4 stains the poleward face of C. elegans metaphase chromosomes. A 28-cell C. elegans embryo was fixed and stained according to methods described in Materials and Methods with mAb 6C4 (red in A and C) and DAPI (blue in B and C). Nuclei in metaphase (m), prophase (p), and late anaphase/telophase (t) are indicated. Bar, 10 μm.
mAb 6C4 Staining Is Temporally and Spatially Regulated on C. elegans Chromosomes
To determine the mAb 6C4 staining pattern on individual chromosomes, we used deconvolution microscopy to generate high resolution images of the nuclei of two-cell embryos stained with mAb 6C4 and DAPI (Hiraoka et al. 1991; Carrington et al. 1995). The relative time of each nucleus in the cell cycle was inferred by comparing the staining patterns of the AB and P1 blastomeres; the AB blastomere division occurs before that of the P1 blastomere (Deppe et al. 1978). The morphology of DAPI staining and presence of a nuclear membrane were used as additional landmarks of cell cycle progression. In these experiments, interphase nuclei had no detectable mAb 6C4 reactivity. In addition, nuclei that are in the earliest stages of chromosome condensation do not stain with mAb 6C4 (data not shown). Staining with mAb 6C4 was first observed in prophase nuclei that contain condensed chromosomes. At this stage, mAb 6C4 staining was observed as dots distributed throughout the nucleus; many of these dots colocalize with the chromosomes (Fig. 2, A–C). These chromosome-associated dots are widely dispersed along the entire length of each chromosome. Later in prophase, mAb 6C4–stained structures located on opposite sides of each chromosome (Fig. 2, D–F). In metaphase, when the chromosomes are aligned at the equatorial plane, chromosomes were oriented with each mAb 6C4–stained structure facing one centrosome (Fig. 1 and Fig. 2, G–I). At anaphase, sister chromatids separated with the mAb 6C4–stained side on the poleward face of each sister chromatid (Fig. 2, J–L). No mAb 6C4 staining was detectable by late anaphase. These results show that there is dynamic localization of the mAb 6C4 antigen to chromosomes throughout mitosis.
Figure 2.
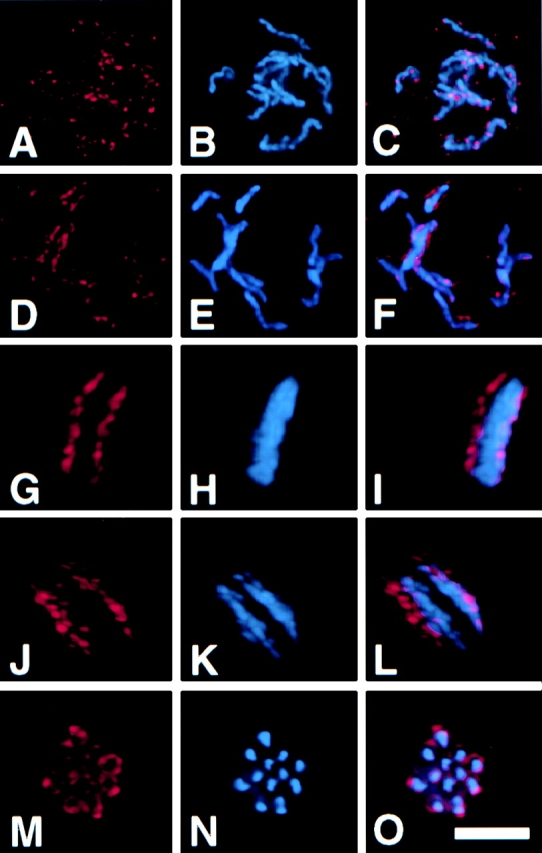
Dynamic localization of mAb 6C4 antigen throughout the cell cycle. Two-cell C. elegans embryos were fixed and stained with mAb 6C4 (A, D, G, J, and M), DAPI (B, E, H, K, and N). C, F, I, L, and O show the merged mAb 6C4 (red) and DAPI (blue) staining. Coverslips were imaged on Deltavision microscope and 0.2-μm optical sections were obtained as described in Materials and Methods. The images were deconvolved and analyzed as a projection of the entire stack. Nuclei at different positions in the cell cycle are shown: A–C, early prophase; D–F, late prophase; G–I, metaphase; J–L, anaphase; and M–O, metaphase/anaphase I of meiosis. Bar, 5 μm.
mAb 6C4 Staining during Meiosis
During meiosis, C. elegans chromosomes lack a trilaminar kinetochore structure, and attachment to the meiotic spindle is mediated through the chromosome ends (Albertson and Thomson 1993). We stained adult hermaphrodite gonads with mAb 6C4. No mAb 6C4 nuclear staining was detected in the mitotic region of the gonad of the hermaphrodite nor in oocytes arrested at diakinesis of prophase I (data not shown). After fertilization, embryos exit prophase I arrest and undergo two consecutive rounds of division to form two polar bodies. Staining with mAb 6C4 was detected on meiotic chromosomes after fertilization and before first polar body formation (Fig. 2, M–O). The mAb 6C4 staining pattern was observed as a halo surrounding the meiotic chromosomes.
The mAb 6C4 Antigen Is HCP-1
To identify the antigen recognized by mAb 6C4, we screened a λgt11 C. elegans cDNA expression library with mAb 6C4. Three overlapping cDNAs were isolated that correspond to a predicted gene, ZK1055.1. We refer to this gene as hcp-1. The hcp-1 gene can encode a 1,475–amino acid protein predicted to be composed primarily of coiled-coil domains (Fig. 3 A). HCP-1 has a direct repeat of 132 amino acids that is 45% similar to a direct repeat of 179 amino acids present in CENP-F (Fig. 3 B) (Liao et al. 1995; Zhu et al. 1995). A search of C. elegans sequence databases with the HCP-1 amino acid sequence revealed a second C. elegans coiled-coil protein, T06E4.2, that is 54% similar to HCP-1. We refer to T06E4.1 as HCP-2. The highest levels of similarity between HCP-1 and HCP-2 is seen at the two termini. HCP-2 lacks the tandem repeats observed in HCP-1 and CENP-F. A low level (<20%) of similarity is observed between HCP-1 and HCP-2 with several other coiled-coil proteins in the databases, probably due to conservation of the coiled-coil structure.
Figure 3.
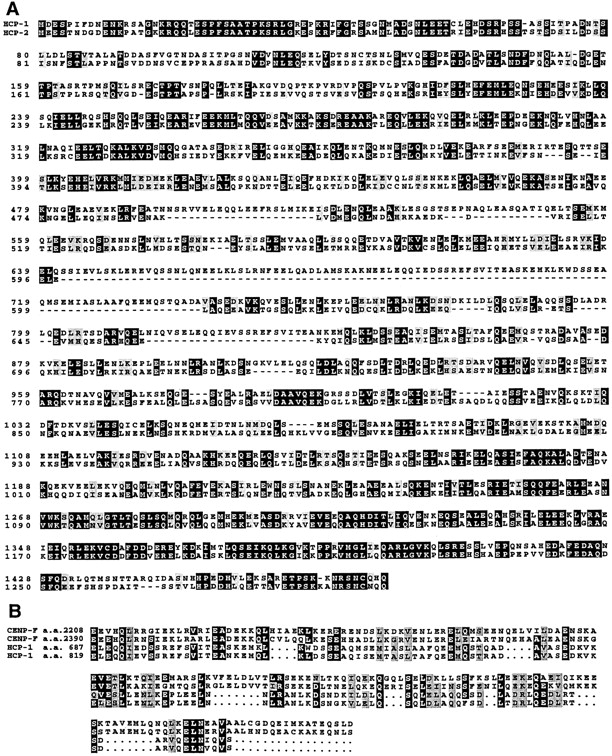
Sequence alignment of HCP-1 and HCP-2 deduced amino acid sequences, and alignment of HCP-1 and CENP-F repeats. (A) The predicted sequence of HCP-1 was used to search the C. elegans genome database, and one similar protein, HCP-2, was identified. HCP-1 and HCP-2 share 42% identity and 54% similarity. (B) Alignment of the HCP-1 repeated motifs with CENP-F repeated sequences. The HCP-1 repeated sequences are 89% identical to one another and 29% identical (45% similar) to the CENP-F repeats. Sequence alignments were constructed using CLUSTAL W (Thompson et al. 1994). Sequence identities are shaded in black and similarities are shaded gray. These sequence data are available from GenBank/EMBL/DDBJ under accession nos. AF068721 for cosmid ZK1055 (HCP-1) and Z70756 for cosmid T06E4 (HCP-2).
To test whether hcp-1 encodes the antigen recognized by mAb 6C4, we characterized mAb 6C4 immunofluorescence in embryos in which HCP-1 expression has been reduced by RNA interference (RNAi) (Guo and Kemphues 1995; Fire et al. 1998). RNAi has proved useful for studies of both maternally and zygotically expressed proteins. The hcp-1 (RNAi) embryos were fixed and stained with mAb 6C4. In these experiments, 77% of the hcp-1 (RNAi) embryos failed to stain with mAb 6C4. In contrast, only 0–2% of embryos from control injections with water or hcp-2 (RNAi) embryos failed to stain with mAb 6C4 (Fig. 4 and Table ). In embryos with no mAb 6C4 staining, all mitotic cells failed to stain, i.e., the staining was never observed to be mosaic. The failure of hcp-1 (RNAi) embryos to stain with mAb 6C4 was specific for this antibody, because the embryos stained positively with mAb K76, a P granule antibody (Fig. 4) (Strome and Wood 1983). These results indicate that HCP-1 is the major antigen for mAb 6C4.
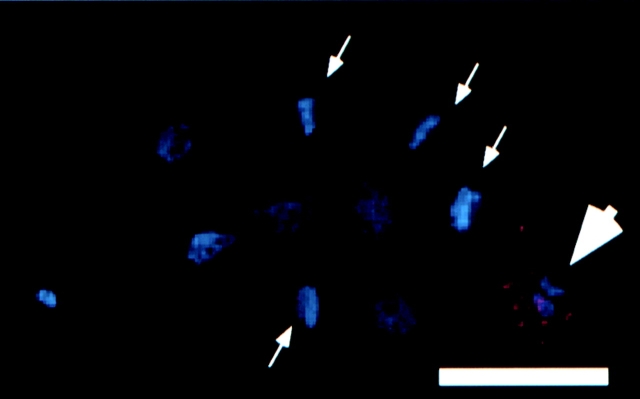
Figure 4.
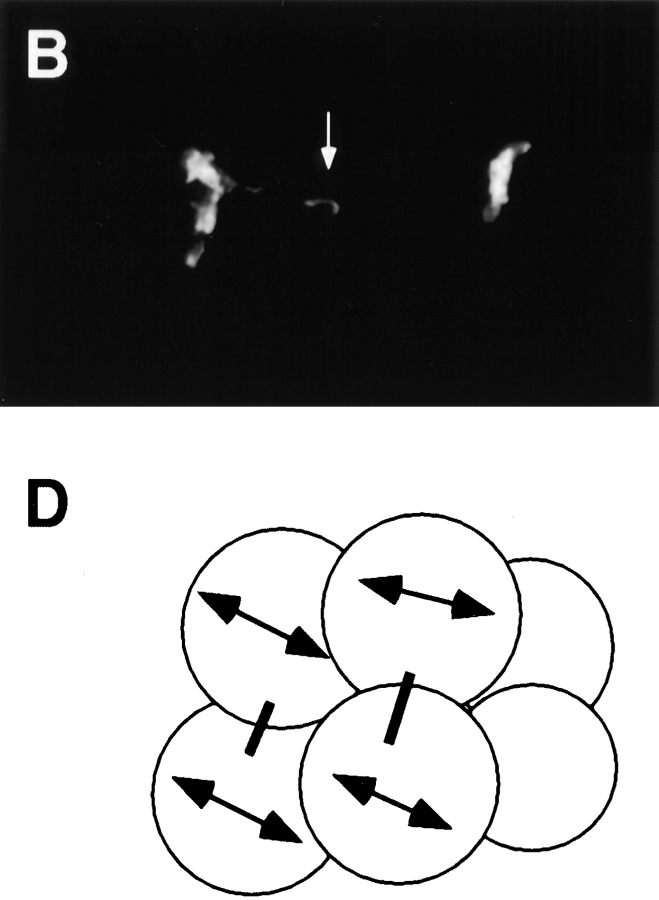
RNAi with hcp-1 abolishes staining with mAb 6C4. hcp-1 (RNAi) embryos were fixed and stained with mAb 6C4, mAb K76, and DAPI (blue). Both primary antibodies were visualized by rhodamine-conjugated (red) goat anti–mouse secondary antibody. K76 recognizes P-granules in the germline and was used as a positive control for staining. An ∼10-cell C. elegans hcp-1 (RNAi) embryo is shown with several cells in metaphase (arrows) that fail to stain with mAb 6C4. The germ line blastomere is selectively stained by mAb K76 (arrowhead). Bar, 10 μm.
Table 1.
hcp-1 and hcp-2 RNAi Embryo Phenotypes
| dsRNA | Brood size ± SE | Percentage of embryonic lethality ± SE | Percentage of mAb 6C4–stained embryos |
|---|---|---|---|
| None | 71 ± 13 | 0.8 ± 0.5 (10) | 100 (96) |
| hcp-1 | 74 ± 7 | 0.5 ± 0.3 (19) | 23 (103) |
| hcp-2 | 84 ± 6 | 2.1 ± 0.5 (26) | 98 (51) |
| hcp-1/hcp-2 | 100 ± 11 | 99.2 ± 0.2 (11) | 20 (31) |
Wild-type hermaphrodites were injected with the RNA indicated in the column labeled dsRNA. After 12 h, injected worms were moved to new plates and their brood sizes and percentage of embryonic lethality were determined over the next 36 h. Embryos laid during the first 12 h were not included in the analysis. The number of injected hermaphrodites is shown in parentheses in the percentage of embryonic lethality column. P values for embryonic lethality relative to no dsRNA injected are P = 0.5 for hcp-1, P = 0.05 for hcp-2, and P < 0.001 for hcp-1/hcp-2. In a separate experiment, all embryos from injected hermaphrodites were stained with mAb 6C4 and a control antibody, K76. Only embryos that stained with K76 were scored for staining with mAb 6C4. The number of embryos scored is indicated in parentheses. SE, standard error.
Colocalization on C. elegans Metaphase Chromosomes of mAb 6C4 with MPM-2 and a Centromeric Histone H3–like Protein
The staining pattern observed by mAb 6C4 suggested that HCP-1 might be located at or near the kinetochore on C. elegans chromosomes. Currently, no proteins have been localized to the kinetochore in C. elegans. We tested several anticentromeric antibodies for cross-reactivity in C. elegans, and only MPM-2 was able to cross-react (Davis et al. 1983). MPM-2 recognizes a conserved phosphoepitope present during mitosis on many mitotic proteins. Using MPM-2, others found that the most intensely stained structure on metaphase chromosomes in a variety of organisms is the kinetochore (Vandre et al. 1984; Renzi et al. 1997). In C. elegans, MPM-2 identifies a structure adjacent to the condensed chromosomes at metaphase that is also recognized by mAb 6C4 (Fig. 5, A–C). This structure is likely to be the kinetochore, since it is present only on the poleward face of metaphase chromosomes and MPM-2 has been observed to identify the kinetochore in a variety of organisms (Vandre et al. 1984; Renzi et al. 1997). To further determine whether HCP-1 is localized to the kinetochore, we used an antibody raised against a centromeric histone H3–variant, HCP-3, in a double labeling experiment with mAb 6C4. HCP-3 was identified as a histone H3–variant in C. elegans, that like the centromeric H3-like proteins, chromosome segregation (CSE) 4 in yeast, and CENP-A in mammals, is localized to the centromere and affects the fidelity of chromosome segregation (Sullivan et al. 1994; Stoler et al. 1995; Buchwitz et al. 1999). Anti–HCP-3 antibody staining is present as continuous lines of reactivity all along each chromosome (Fig. 5 E). This is consistent with the centromere being present along the long axis of each holocentric chromosome (Buchwitz et al. 1999). Staining of each chromosome with mAb 6C4 is also present along each chromosome and adjacent to the centromere, as identified by anti–HCP-3 antibody (Fig. 5, D–F). Furthermore, this staining is restricted to opposite surfaces of the holocentric chromosome and is analogous to centromere/kinetochore staining of the primary constriction of mammalian chromosomes (Fig. 5, G–H). The localization of HCP-1 to the region of the chromosome immediately adjacent to the centromere, and its colocalization with an antibody, MPM-2, which recognizes the kinetochore in many organisms, indicates that HCP-1 is likely to be a structural component of the C. elegans kinetochore.
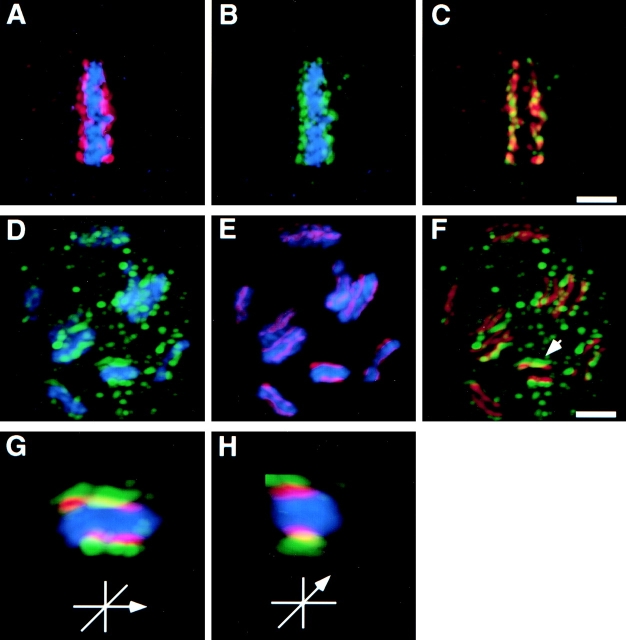
Figure 5.
HCP-1 is localized to the centromere/kinetochore. Embryos were fixed in cold methanol and stained with mAb 6C4 and either MPM-2 or anti–HCP-3 antibody as described in Materials and Methods. Chromosomes were visualized by staining with DAPI. Images were collected using the Deltavision microscopy and analyzed as stacks. (A) A one-cell embryo in metaphase is shown stained with MPM-2 antibody (red) and DAPI (blue). The MPM-2 antibody primarily stained the poleward face of metaphase chromosomes. (B) The same embryo stained with mAb 6C4 (green) and DAPI (blue). (C) The two channels for MPM-2 (red) and mAb 6C4 (green) staining were merged using Photoshop (Adobe Systems Inc.) to identify the colocalization (yellow). D and E show a late prophase nucleus stained with mAb 6C4 (green), anti–HCP-3 (red), and DAPI (blue). F shows the two channels for anti–HCP-3 (red) and mAb 6C4 (green) staining merged (yellow). G depicts a single prophase chromosome (arrow in F) enlarged fourfold and rotated 90° into the plane of the paper (H) to show the specific localization to only opposite surfaces of the chromosome. Bar, 2 μm.
HCP-1 and HCP-2 Function during Chromosome Segregation
Embryos derived from hermaphrodites injected with hcp-1 dsRNA are viable and exhibit no visible phenotype other than loss of mAb 6C4 staining. We considered the possibility that HCP-1 might be functionally redundant with HCP-2. Wild-type staining with mAb 6C4 was observed with hcp-2 (RNAi) embryos which were also viable (Table ). However, coinjection of both hcp-1 and hcp-2 dsRNAs resulted in 99% lethality of progeny embryos (Table ). The synthetic lethality of hcp-1 (RNAi) and hcp-2 (RNAi), and the localization of HCP-1 to the kinetochore region of mitotic chromosomes, together suggest that both HCP-1 and HCP-2 function in mitotic chromosome segregation.
To determine whether the synthetic lethality correlates with a defect in chromosome segregation, we stained hcp-1 (RNAi)/hcp-2 (RNAi) embryos with an antitubulin antibody and DAPI. In these experiments, we observed that 80% (n = 60) of all anaphases showed signs of defective chromosome segregation, including lagging chromosomes and anaphase bridges (Fig. 6B and Fig. C). This defect was observed as early as the first mitotic cleavage (Fig. 6 B). In these anaphase nuclei, the spindle as observed by antitubulin staining appeared wild-type, in that a normal bipolar orientation is present (Fig. 6 A). As observed by DAPI, chromosome condensation appeared to also not be grossly altered, as chromosomal structures are visible (Fig. 6 C). In many instances, some chromosomes were not located between the centrosomes; for example, Fig. 6 C shows an early embryo in which the four ABxx blastomeres are in prophase. Several chromosomes are located outside of the spindles and along the previous spindle axis, indicating that they failed to segregate at the previous division (see Fig. 6 D for orientation). The hcp-1 (RNAi)/hcp-2 (RNAi) embryos arrested with ∼50–100 cells. At this stage, the DNA content of individual nuclei was variable, suggesting that some nuclei have more and others have less than the diploid set of chromosomes (Fig. 7). In contrast, DNA content in wild-type embryos was uniform among nuclei. These results further support the idea that HCP-1 and HCP-2 function together in chromosome segregation.
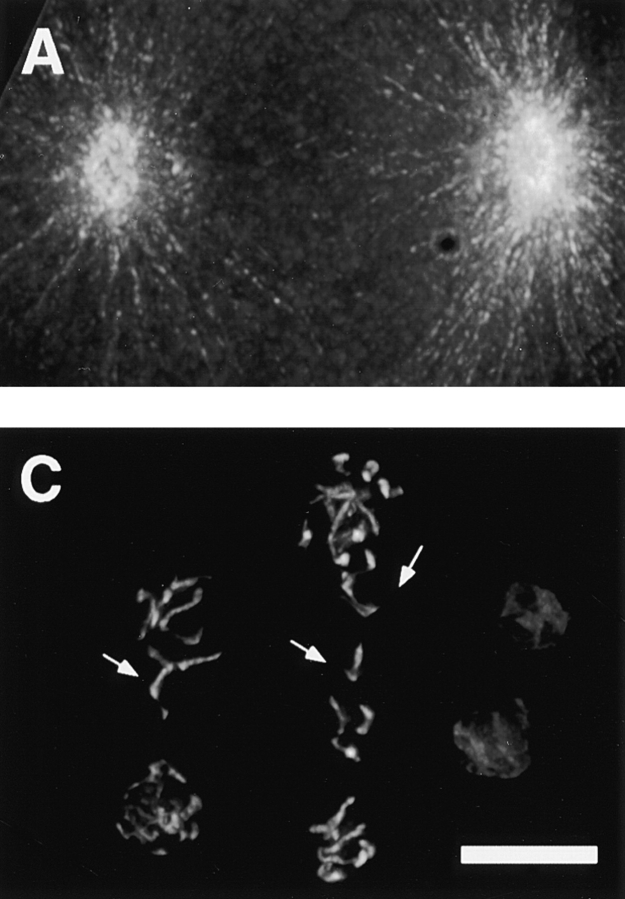
Figure 6.
Chromosome segregation is affected in hcp-1/hcp-2 (RNAi) embryos. Embryos were obtained from injected hermaphrodites 12 h after injection and processed for immunofluorescence as described in Materials and Methods. Antitubulin antibody (Amersham Pharmacia Biotech) was used at 1:100 dilution and was visualized with fluorochrome conjugated goat anti–mouse secondary antibody (A). DNA staining was by DAPI (B and C). (A) hcp-1/hcp-2 (RNAi) one-cell embryo also in anaphase stained with antitubulin antibody or DAPI; (B) same embryo as in A stained with DAPI to visualize the chromosomes. (C) A six-cell hcp-1/hcp-2 (RNAi) embryo stained with DAPI. (D) Diagram showing division axis of the six-cell embryo shown in C. Anterior is to the left. Double arrow lines (D) indicate current spindle axis of ABxx blastomeres and diagonal bars indicate the previous spindle axis of ABx blastomeres. Arrows in B and C indicate lagging chromosomes. Bar, 10 μm.
Figure 7.
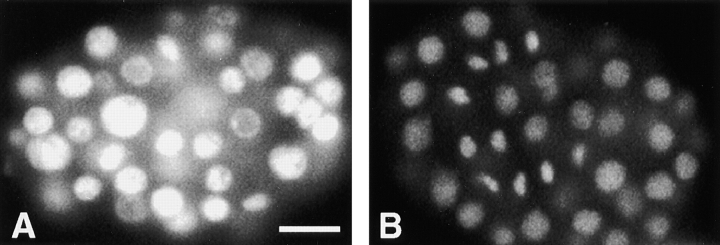
Terminal arrest phenotype of hcp-1/hcp-2 (RNAi) embryos. (A) An arrested embryo from an hcp-1/hcp-2 dsRNA injected hermaphrodite was stained with DAPI to visualize DNA content. (B) A wild-type embryo at approximately the same stage as the arrested embryo is included for comparison. Photographs were taken with identical exposure times for each embryo. Bar, 10 μm.
Discussion
mAb 6C4 as a Marker for the C. elegans Kinetochore
We have identified mAb 6C4, which has a staining pattern on mitotic chromosomes in C. elegans that is analogous to that of anticentromeric antibodies in mammals. Anticentromeric antibodies stain two structures which flank mitotic chromosomes in mammalian cells (Zinkowski et al. 1991). Similarly, mAb 6C4 recognizes two structures which flank mitotic chromosomes in C. elegans. These two structures are oriented towards the centrosomes at metaphase and anaphase in both mammals and C. elegans. This relationship of the C. elegans and mammalian staining patterns is consistent with the structural characterization of their chromosomes by EM. In mammals, the centromere, as defined by the presence of a kinetochore, is localized to the primary constriction (Reider 1982). Conversely, in C. elegans the kinetochore is present on opposing sides and extends nearly the length of the chromosome (Albertson and Thomson 1982). These similarities suggested that mAb 6C4 may be used to identify the C. elegans kinetochore.
To test whether mAb 6C4 stains at or near the kinetochore, we performed a colocalization experiment using an antibody (MPM-2) previously shown to stain the kinetochore in many different organisms, and an antibody directed against a centromeric histone H3–variant, HCP-3, (Vandre et al. 1984; Renzi et al. 1997; Buchwitz et al. 1999). MPM-2 recognizes a highly conserved phosphoepitope present on kinetochore-associated proteins during mitosis (Taagepera et al. 1997). MPM-2 antibody did cross-react in C. elegans, further indicating the conserved nature of its epitope. MPM-2 staining was observed to be strongest in the region adjacent to the mitotic chromosomes in a pattern overlapping with that observed for mAb 6C4. This overlap was specific for the region adjacent to the mitotic chromosomes, although both antibodies showed staining not associated with the chromosomes. Elimination of the mAb 6C4 antigen by RNAi did not affect the staining with MPM-2, indicating that the colocalization is not due to the presence of the MPM-2 epitope on HCP-1 (data not shown). Thus, the colocalization of MPM-2 and mAb 6C4 is likely the result of their epitopes being present at the same cytological structure, the kinetochore.
Furthermore, mAb 6C4 staining is present adjacent, but not coincident with the staining of chromatin by anti–HCP-3 antibody. Both staining with anti–HCP-3 and mAb 6C4 is analogous to that observed on monocentric chromosomes by anticentromeric antibodies that recognize different regions of the trilaminar kinetochore (Pluta et al. 1995). Taken together, these two colocalization experiments indicate that the staining pattern observed with mAb 6C4 along mitotic chromosomes in C. elegans represents the kinetochore of the holocentric chromosomes.
HCP-1 and HCP-2 Are Involved in Kinetochore Function
We have used mAb 6C4 to identify two proteins, HCP-1 and HCP-2, which together are involved in kinetochore function. When we reduced the expression of both HCP-1 and HCP-2 by RNA-mediated inhibition, we observed that embryos arrested with ∼50–100 nuclei containing variable amounts of DNA. By observing embryos at earlier stages, we observed that chromosomes were often found to be unattached to the mitotic spindle. Not all chromosomes failed to attach to the spindle. This may reflect low levels of expression of HCP-1 or HCP-2. The inability of chromosomes to attach to the mitotic spindle does not appear to be the result of a defect in chromosome condensation. When viewed by DAPI staining, chromosomes are observed and appear normal when compared with wild-type. This is consistent with the localization of HCP-1 to the chromosomes occurring after chromosome condensation has begun, suggesting HCP-1 is not generally required for chromosome condensation.
The hcp-1 (RNAi)/hcp-2 (RNAi) defects are similar to those observed when human centromere components are inhibited, which include lagging chromosomes in anaphase and inefficient spindle attachment to chromosomes (Bernat et al. 1990). These defects are also similar to defects observed when expression of the C. elegans homologue of the Zeste White 10 protein (CeZW10) is reduced by RNAi (Starr et al. 1997). Homologues of ZW10 have been localized to the kinetochore in both Drosophila and humans (Williams et al. 1992, Williams et al. 1996; Starr et al. 1997). Mutations in Drosophila zw10 result in aberrant chromosome segregation, which is observed during anaphase as lagging chromatids or chromosomes remaining in the vicinity of the metaphase plate during anaphase. Reduction of CeZW10 expression in C. elegans embryos results in a similar phenotype in which chromosomes are often observed to be improperly attached to both spindles and results in anaphase bridges during anaphase. Anaphase bridges and even lagging chromatids or chromosomes are observed with hcp-1 (RNAi)/hcp-2 (RNAi) embryos, suggesting that like ZW10, HCP-1 and HCP-2 are required for proper kinetochore function during mitosis.
mAb 6C4 Defines Intermediates in Mitotic Chromosome Structure
The dynamic mAb 6C4 staining pattern on mitotic chromosomes suggests that there are intermediate stages of holocentric kinetochore assembly. As cells progress from early to late prophase, the mAb 6C4 staining pattern changes from discontinuous points dispersed along chromosomes to structures on opposite sides of each chromosome. The temporal relationship of these images indicates that the dispersed dots may be used to assemble the larger structures seen at late prophase. The visualization of a disperse mAb 6C4 staining pattern is consistent with a model in which there are discrete regions of C. elegans chromosomes that act as the primary points of spindle microtubule capture. This model is similar to the repeat subunit model proposed for the mammalian centromere, which proposes that the centromere is composed of short segments distributed along the DNA that bind spindle microtubules (Zinkowski et al. 1991). These segments are brought into parallel register as a result of chromatin condensation to form the observed kinetochore structure. The C. elegans chromosomes may also contain such repeats discontinuously distributed along the entire chromosome. Our results support the model suggested by Albertson and Thomson 1982 that the holocentric chromosome may be thought of as an extended centromere.
Acknowledgments
The authors would like to thank B. Earnshaw, J. Stark, D. Frank, J. Stear, R. Hill, B. Paige, and J. Priess for critical reading of the manuscript, and A. Quintanilla for assistance with the Deltavision microscope and deconvolution software.
L.L. Moore was supported by a National Institutes of Health (NIH) training grant (T32CA09657). M. Morrison was supported by training grant (2T32HD07183-16) from the NIH. This work was supported by an NIH grant (GM48435-01A2) to M.B. Roth.
Footnotes
1.used in this paper: CENP, centromeric protein; DAPI, 4′,6-diamidino-3-phenylindole dihydrochloride; dsRNA, double-stranded RNA; HCP, holocentric protein; RNAi, RNA interference
References
- Albertson D.G. Formation of the first cleavage spindle in nematode embryos. Dev. Biol. 1984;101:61–72. doi: 10.1016/0012-1606(84)90117-9. [DOI] [PubMed] [Google Scholar]
- Albertson D.G., Thomson J.N. The kinetochores of Caenorhabditis elegans . Chromosoma. 1982;86:409–428. doi: 10.1007/BF00292267. [DOI] [PubMed] [Google Scholar]
- Albertson D.G., Thomson J.N. Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans . Chromosome Res. 1993;1:15–26. doi: 10.1007/BF00710603. [DOI] [PubMed] [Google Scholar]
- Allshire R.C. Centromeres, checkpoints and chromatid cohesion. Curr. Opin. Genet. Dev. 1997;7:264–273. doi: 10.1016/s0959-437x(97)80137-2. [DOI] [PubMed] [Google Scholar]
- Bernat R.L., Borisy G.G., Rothfield N.F., Earnshaw W.C. Injection of anticentromeric antibodies in interphase disrupts events required for chromosome movement at mitosis. J. Cell Biol. 1990;111:1519–1533. doi: 10.1083/jcb.111.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernat R.L., Delannoy M.R., Rothfield N.F., Earnshaw W.C. Disruption of centromere assembly during interphase inhibits kinetochore morphogenesis and function in mitosis. Cell. 1991;66:1229–1238. doi: 10.1016/0092-8674(91)90045-z. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans . Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., Pepper D., Berns M.W., Tan E., Brinkley B.R. Kinetochore structure, duplication, and distribution in mammalian cellsanalysis by human autoantibodies from scleroderma patients. J. Cell Biol. 1981;91:95–102. doi: 10.1083/jcb.91.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwitz B.J., Ahmad K., Moore L.L., Roth M.B., Henikoff S. A histone H3-like protein in C. elegans . Nature. 1999;401:547. doi: 10.1038/44062. [DOI] [PubMed] [Google Scholar]
- Buck R.C. Mitosis and meiosis in Rhodnius prolixusthe fine structure of the spindle and diffuse kinetochore. J. Ultrastruct. Res. 1967;18:489–501. doi: 10.1016/s0022-5320(67)80199-0. [DOI] [PubMed] [Google Scholar]
- Carrington W.A., Lynch R.M., Moore E.D.W., Isenberg G., Fogarty K.E., Fay F.S. Superresolution three-dimensional images of fluorescence in cells with minimal light exposure. Science. 1995;268:1483–1487. doi: 10.1126/science.7770772. [DOI] [PubMed] [Google Scholar]
- Comings D.E., Okada T.A. Fine structure of the kinetochore in the Indian muntjac. Exp. Cell Res. 1971;67:97–110. doi: 10.1016/0014-4827(71)90625-2. [DOI] [PubMed] [Google Scholar]
- Comings D.E., Okada T.A. Holocentric chromosomes in Oncopeltuskinetochore plates are present in mitosis but absent in meiosis. Chromosoma. 1972;37:177–192. doi: 10.1007/BF00284937. [DOI] [PubMed] [Google Scholar]
- Davis F.M., Tsao T.Y., Fowler S.K., Rao P.N. Monoclonal antibodies to mitotic cells. Proc. Natl. Acad. Sci. USA. 1983;80:2926–2930. doi: 10.1073/pnas.80.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deppe U., Schierenberg E., Cole T., Krieg C., Schmitt D., Yoder B., von Ehrenstein G. Cell lineages of the embryo of the nematode Caenorhabditis elegans . Proc. Natl. Acad. Sci. USA. 1978;75:376–380. doi: 10.1073/pnas.75.1.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W.C., Mackay A.M. Role of nonhistone proteins in the chromosomal events of mitosis. FASEB J. 1994;8:947–956. doi: 10.1096/fasebj.8.12.8088460. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans . Nature (Lond.). 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Goday C., Ciofi-Luzzatto A., Pimpinelli S. Centromere ultrastructure in germ-line chromosomes of Parascaris . Chromosoma. 1985;91:121–125. doi: 10.1007/BF00294055. [DOI] [PubMed] [Google Scholar]
- Goday C., González-García J.M., Estban M.R., Giovinazzo G., Pimpinelli S. Kinetochores and chromatin diminution in early embryos of Parascarus univalins . J. Cell Biol. 1992;118:23–32. doi: 10.1083/jcb.118.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grodberg J., Dunn J.J. ompT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J. Bacteriol. 1988;170:1245–1253. doi: 10.1128/jb.170.3.1245-1253.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Kemphues K.J. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- Harlow E., Lane D. AntibodiesA Laboratory Manual 1988. Cold Spring Harbor Laboratory Press; Plainview, NY: pp. 726 [Google Scholar]
- Herman R.K., Albertson D.G., Brenner S. Chromosome rearrangements in Caenorhabditis elegans . Genetics. 1976;83:91–105. doi: 10.1093/genetics/83.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman R.K., Madl J.E., Kari C.K. Duplications in Caenorhabditis elegans . Genetics. 1979;92:419–435. doi: 10.1093/genetics/92.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y., Swedlow J.R., Paddy M.R., Agard D.A., Sedat J.W. Three dimensional multiple-wavelength fluorescence microscopy for the structural analysis of biological phenomena. Semin. Cell Biol. 1991;2:153–165. [PubMed] [Google Scholar]
- Hughs-Schrader S., Ris H. The diffuse spindle attachment of coccids verified by the mitotic behavior of induced chromosome fragments. J. Exp. Zool. 1941;87:429–456. [Google Scholar]
- Hughs-Schrader S., Schrader F. The kinetochore of the Hemiptera . Chromosoma. 1961;12:327–350. doi: 10.1007/BF00328928. [DOI] [PubMed] [Google Scholar]
- Jokelaninen P.T. The ultrastructure and spatial organization of the metaphase kinetochore in mitotic rat cells. J. Ultrastruct. Res. 1967;19:19–44. doi: 10.1016/s0022-5320(67)80058-3. [DOI] [PubMed] [Google Scholar]
- Liao H., Winkfein R.J., Mack G., Rattner J.B., Yen T.J. CENP-F is a protein of the nuclear matrix that assembles onto kinetochores at late G2 and is rapidly degraded after mitosis. J. Cell Biol. 1995;130:507–518. doi: 10.1083/jcb.130.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather K., Stone L.H.A. The effect of X-radiation upon somatic chromosomes. J. Genet. 1933;28:1–24. [Google Scholar]
- Moroi Y., Peebles C., Fitzler M.J., Steigerwald J., Tan E.M. Autoantibody to centromere (kinetochore) in scleroderma sera. Proc. Natl. Acad. Sci. USA. 1980;77:1627–1631. doi: 10.1073/pnas.77.3.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta A.F., Mackay A.M., Ainsztein A.M., Goldberg I.G., Earnshaw W.C. The centromerehub of chromosome activities. Science. 1995;270:1591–1594. doi: 10.1126/science.270.5242.1591. [DOI] [PubMed] [Google Scholar]
- Reider C.L. The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int. Rev. Cytol. 1982;79:1–58. doi: 10.1016/s0074-7696(08)61672-1. [DOI] [PubMed] [Google Scholar]
- Renzi L., Gersch M.S., Campbell M.S., Wu L., Osmani S.A., Gorbsky G.J. MPM-2 antibody-reactive phophorylations can be created in detergent-extracted cells by kinetochore-bound and soluble kinases. J. Cell Sci. 1997;110:2013–2025. doi: 10.1242/jcs.110.17.2013. [DOI] [PubMed] [Google Scholar]
- Ris H. A cytological and experimental analysis of the meiotic behavior of the univalent X chromosome in the bearberry aphid Tamalia (Phyllaphis) coweni (Ck.11) J. Exp. Zool. 1942;90:267–330. [Google Scholar]
- Ris H., Witt P.L. Structure of the mammalian kinetochore. Chromosoma. 1981;82:153–170. doi: 10.1007/BF00286101. [DOI] [PubMed] [Google Scholar]
- Roos U.-P. Light and electron microscopy of rat kangaroo cells in mitosis. II. Kinetochore structure and function. Chromosoma. 1973;41:195–220. doi: 10.1007/BF00319696. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. Molecular CloningA Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1989. [Google Scholar]
- Schrader F. Notes on the mitotic behavior of long chromosomes. Cytology. 1935;6:422–430. [Google Scholar]
- Starr D.A., Williams B.C., Li Z., Etemad-Moghadam B., Dawe R.K., Goldberg M.L. Conservation of the centromere/kinetochore protein ZW10. J. Cell Biol. 1997;138:1289–1301. doi: 10.1083/jcb.138.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S., Keith K.C., Curnick K.E., Fitzgerald-Hayes M. A mutation in Cse4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- Strome S., Wood W.B. Generation of asymmetry and segregation of germ-line granules in early Caenorhabditis elegans embryos. Cell. 1983;35:5–25. doi: 10.1016/0092-8674(83)90203-9. [DOI] [PubMed] [Google Scholar]
- Sullivan K.F., Hechenberger M., Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J. Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taagepera S., Campbell M.S., Gorbsky G.J. Cell-cycle-regulated localization of tyrosine and threonine phosphoepitopes at the kinetochores of mitotic chromosomes. Exp. Cell Res. 1997;221:249–260. doi: 10.1006/excr.1995.1373. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL Wimproving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandre D.D., Davis F.M., Rao P.N., Borisy G.G. Phosphoproteins are components of mitotic microtubule organizing centers. Proc. Natl. Acad. Sci. USA. 1984;81:4439–4443. doi: 10.1073/pnas.81.14.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B.C., Karr T.L., Montgomery J.M., Goldberg M.L. The Drosophila l(l)zw10 gene product, required for accurate mitotic chromosome segregation, is redistributed at anaphase onset. J. Cell Biol. 1992;118:759–773. doi: 10.1083/jcb.118.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B.C., Gatti M., Goldberg M.L. Bipolar spindle attachments affect redistributions of ZW10, a Drosophila centromere/kinetochore component required for accurate chromosome segregation. J. Cell Biol. 1996;134:1127–1140. doi: 10.1083/jcb.134.5.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R.A., Davis R.W. Efficient isolation of genes by using antibody probes. Proc. Natl. Acad. Sci. USA. 1983;80:1194–1198. doi: 10.1073/pnas.80.5.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Mancini M.A., Chang K.-H., Liu C.-Y., Chen C.-F., Shan B., Jones D., Yang-Feng T.L., Lee W.-H. Characterization of a novel 350-kilodalton nuclear phosphoprotein that is specifically involved in mitotic-phase progression. Mol. Cell. Biol. 1995;15:5017–5029. doi: 10.1128/mcb.15.9.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkowski R.P., Meyne J., Brinkley B.R. The centromere-kinetochore complexa repeat subunit model. J. Cell Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]