Abstract
Phosphorylation plays a critical role in controlling the function of cytoskeletal assemblies but no direct method yet exists to measure the phosphorylation state of proteins at the level of individual molecules and assemblies. Herein, we apply scanning transmission electron microscopy in combination with electron energy loss spectroscopy to measure the distributions of mass and phosphorus in neurofilaments (NFs) isolated from the squid giant axon. We find that native squid NFs, in contrast to typical reconstituted intermediate filaments, are a relatively homogeneous population containing only eight coiled-coil dimers per cross section. The measured stoichiometry of ∼1:1 for light/heavy peptides strongly suggests that squid NFs are composed of heterodimers. Furthermore, each heavy chain of the dimers carries at least 100 phosphate groups and is, therefore, near-maximally phosphorylated. These results also demonstrate that scanning transmission electron microscopy combined with electron energy loss spectroscopy at the nanometer scale is capable of characterizing the level and distribution of phosphorylation in individual mass-mapped protein assemblies.
Keywords: electron energy loss spectroscopy/squid axon/intermediate filaments
Neurofilaments (NFs), a neuron-specific subtype of intermediate filament, are an essential structural component of the axonal cytoskeleton (1, 2). The sequence and secondary structure of NF protein subunits in many species are well established (3), but the higher level organization of subunits into filaments, including their degree and pattern of phosphorylation remains unclear, especially in native preparations. Axoplasm from the squid giant axon is a particularly favorable source for preparing native NFs that are assuredly only axonal in origin. This axoplasm can be extruded and separated from the surrounding glia as well as from the cell body, and NFs in a configuration close to their native state can be readily isolated.
Squid NFs contain low (NF-L, also known as NF-60, Mr = 59,000) and high (NF-H, also NF-200, Mr = 131,000) molecular weight proteins (4, 5). As in all intermediate filaments, each NF subunit in the squid contains an α-helical domain favoring formation of parallel coiled-coil dimers (6). Like the nuclear lamins, NF peptides in squid contain six additional heptad repeats in their 348-residue α-helical domains, so that each dimer has a central rod structure with a length 50–52 nm, compared with 44–46 nm for vertebrate cytoplasmic intermediate filament proteins (7). The N-terminal and α-helical domains of the two chains are essentially identical, but NF-H has a C-terminal tail containing many consensus motifs for phosphorylation (5). Phosphorylation is thought to play an essential role in NF function, for example, by regulating NF transport rates and spacing, and thereby determining cytoskeletal stability and axonal caliber (1, 2). However, neither the actual phosphate content nor the subunit organization has ever been determined for squid or any other native NFs. Since NFs from mature squid giant axons are expected to be highly phosphorylated (1, 2), these filaments should be a favorable preparation for directly measuring the level and distribution of phosphorylation in individual native NFs once their subunit organization has been determined.
In this report, we describe the application of electron energy loss spectroscopy (EELS) (8–10) in the scanning transmission electron microscope (STEM) (11–13) to determine the mass and phosphorus distributions along individual NFs from squid giant axons. STEM/EELS has recently gained attention as a near-atomic-resolution structural and nanoanalytical tool for studying materials (14–16), but its application to biological molecules has been largely precluded by the difficulty of extracting information from very weak signals. This study demonstrates that high-efficiency EELS can detect as few as 10 atoms of phosphorus bound to an individual protein assembly. With such sensitivity, EELS can be combined with STEM mass mapping to deduce the level and distribution of subunit phosphorylation, as exemplified by the present results that reveal the arrangement and phosphorylation level of subunits in squid NFs. These findings also indicate that the STEM/EELS approach may be generally applicable to characterizing the phosphorylation level of other protein assemblies.
MATERIALS AND METHODS
Purification and Characterization of Squid Axonal NFs.
NFs from squid giant axon were purified by mildly dissociating extruded axoplasm in potassium aspartate buffer; no significant NF dephosphorylation occurs under these conditions (17). After centrifugation at 125,000 × g (Beckman SW55 Ti rotor) through a sucrose step gradient (30 and 50%) for 2–3 hr at 4°C, a high yield of the NFs were recovered above the 50% sucrose. The purified filaments were composed of two major polypeptides with apparent molecular weights of 60,000 and 200,000, as determined by SDS/PAGE (Coomassie blue staining). To estimate the ratio of NF subunits, quantitative amino acid analysis (Biotechnology Resource Laboratory, Yale University) was carried out after ion exchange chromatography of the hydrolysates of 60-kDa and 200-kDa bands cut from the Coomassie blue-stained SDS gels.
The number and identity of consensus protein phosphorylation sites were estimated by using the ScanProsite tool on the ExPASy World-Wide Web molecular biology server of the University of Geneva and Geneva University Hospital (http://www.expasy.hcuge.ch/). To arrive at a conservative estimate, nonoverlapping sites for only protein kinase C and casein kinase II were added to the XSP plus SEK sites (5, 18); sites within coiled-coil regions were not counted.
Electron Microscopy.
Specimens were prepared by adsorbing NFs suspended in potassium aspartate buffer onto thin (≈3 nm) carbon foils that had been floated onto copper grids and briefly air-dried. Adsorbed NFs were lightly fixed for 1 min by transferring grids onto drops of freshly dissolved 0.1% glutaraldehyde in buffer. Grids were then washed twice on separate drops of 200 mM ammonium acetate and subsequently incubated on a drop of suspended tobacco mosaic virus (TMV) particles, which were adsorbed to the grid to provide mass and phosphorus calibration (19–21). All washes were carried out by stirring the grid floating on the appropriate solution for 20 s. Some specimens were negative-stained in 1% uranyl acetate and imaged in a JEOL 100CX transmission electron microscope; others were blotted to a thin aqueous film and plunge-frozen (Leica KF-80, Deerfield, IL) in liquid ethane at −170°C. These grids were cryotransferred into a VG Microscopes (East Grinstead, England) HB501 field-emission STEM, freeze-dried at −110°C, and recooled to −160°C for imaging. STEM images containing 1024 × 1024 pixels were recorded with single-electron sensitivity at a dose of ∼103 electrons/nm2 as described (21). Images were analyzed on Macintosh desktop computers using the image processing program image available from W. S. Rasband at the National Institutes of Health (http://rsb.info.nih.gov/nih-image/).
EELS.
EELS core-excitation spectra were recorded with a Gatan (Warrendale, PA) model 666 electron spectrometer equipped with a cooled photodiode array detector coupled to a yittrium aluminum garnet (YAG) scintillator that provided a detective quantum efficiency of ∼0.3 at the high electron fluxes used (10, 22). The beam energy was 100 keV (eV = electron volt) and the collection semi-angle referred to the specimen was 10 milliradians. Extraction of the P L2,3 signal (<0.5% of the background) was achieved by using a second-difference acquisition technique in which the mean of two spectra shifted over the detector by ±8 eV was subtracted from a third unshifted spectrum (8). This acquisition mode corrects for the ∼1% (rms) fixed-pattern noise caused by detector channel-to-channel gain variations. Phosphorus in TMV was quantified by using a least-squares procedure to fit a reference spectrum from phosvitin (also recorded in second-difference mode); this reference spectrum had been divided by the signal in the normal phosvitin spectrum (integrated in a 50 eV window above the L2,3 edge) so that the fitting coefficient corresponded to the net phosphorus signal. The spectral background due to valence excitations was modeled as ∝E−3 where E is the energy loss.
RESULTS
Subunit Organization.
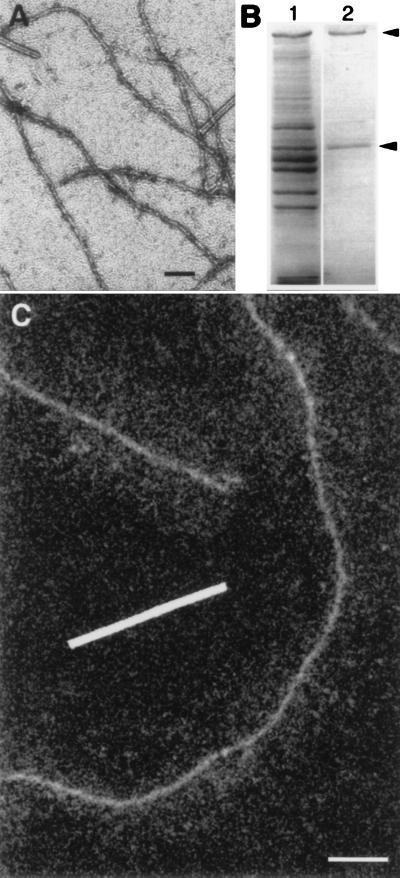
In negatively stained preparations (Fig. 1A), the purified NFs (Fig. 1B) appeared as homogeneous cylinders ∼8 nm in diameter, which is significantly narrower than the 10-nm diameter typical of intermediate filaments (23, 24). Parallel preparations of rapidly frozen NFs were cryotransferred into a 100-keV-field-emission STEM and freeze-dried before collecting digital dark-field images at low dose to avoid radiation damage (Fig. 1C). In addition to the central core of the filaments, these images revealed the NF-H sidearms as a diffuse intensity extending approximately 50 nm on either side of the core.
Figure 1.
Electron micrographs and SDS/PAGE gels of native NFs isolated from squid giant axon. (A) Conventional transmission electron micrograph of negatively stained preparation. NFs are visible as undulating structures 8 ± 1 nm in diameter; as expected, it was not possible to visualize sidearms corresponding to NF-H carboxyl-terminal tails. Rod-like particles of TMV are clearly distinguishable from thinner NFs. Bar = 100 nm. (B) Coomassie blue staining of proteins separated by SDS/PAGE illustrates major proteins present in axoplasm (lane 1) as compared with those in the NF preparation isolated from axoplasm (lane 2). NF-H (NF-200) and NF-L (NF-60) polypeptides are indicated (arrowheads). (C) Digital dark-field STEM micrograph from preparation that was lightly fixed, rapidly frozen, cryotransferred into the STEM, and freeze-dried. The cores of these NFs appear structurally similar to those observed in the negatively stained preparations but sidearms, representing the C-terminal tails of NF-H subunits, are now visible as a diffuse intensity surrounding the central cores. The swath of sidearms was evident, even though the dark-field signal was too noisy to distinguish the individual polypeptide chains of the NF-H tails. Bar = 100 nm.
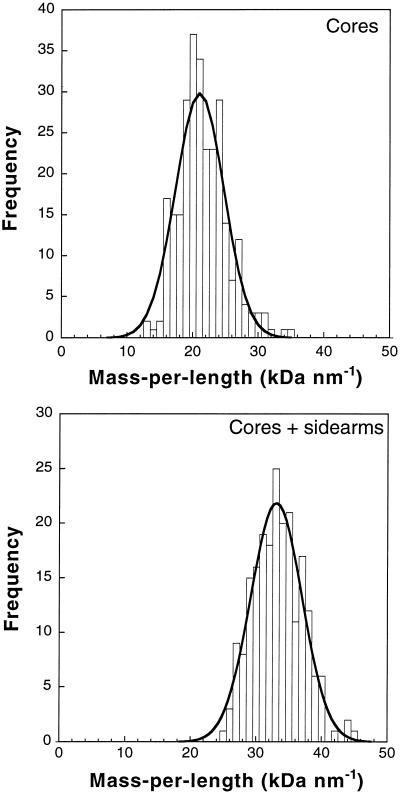
Because the pixel intensities in such STEM images are proportional to the projected mass of the polymer, the filament mass per unit length can be determined (19, 20). In turn, the number of subunits per NF cross section can be deduced from this mass measurement if the subunit molecular weights, NF-L/NF-H protein ratio, and dimer axial repeat distance are also known. From the known length of the central rod domain, 50–52 nm, it can be predicted that the axial repeat distance of the dimers is 46–50 nm because of the short head-to-tail overlap of the dimers (25). Analysis of STEM images yielded a mass per length of 21.0 ± 0.5 kDa/nm (mean ± SEM) for the NF cores (Fig. 2A) and 33.5 ± 0.3 kDa/nm for the cores and sidearms (Fig. 2B). Quantitative amino acid analysis of SDS gel-separated NF subunits (Fig. 1B), revealed an NF-L/NF-H subunit ratio of 1.1 to 1 (±10%), suggesting that the basic building blocks of these NFs, like the keratins (26), are light/heavy (LH) heterodimers. Formation of LH heterodimers may be favored by the different C-terminal sequences of NF-L and NF-H, which are derived from alternatively spliced mRNA. (Although the case for heterodimers is strong, it is still possible that squid NFs are a mixture of homodimers and heterodimers. Nevertheless, this occurrence will not change the conclusions regarding subunit organization and phosphorylation.) STEM mass distribution measurements provided a second independent estimate of the subunit ratio. If it is assumed that the NF-H carboxyl tails are excluded from the region of the NF cores, then the relative mass of cores and sidearms also indicates an NF-L/NF-H ratio of 1.0 to 1 (±20%) in agreement with the biochemical measurements. An axial repeat distance of 48 ± 2 nm and a mass of 201 kDa for one LH dimer (including 9.5 kDa from 100 phosphates, see below) yields 8.0 ± 0.4 dimers per filament cross section. These observations therefore reveal that squid axonal NFs are relatively homogeneous and contain eight coiled-coil dimers per cross section. An eight-dimer structure would explain the unusually small diameter of these filaments; indeed, 7–8 nm is the expected width for a rod composed of eight coiled-coils, each 2 nm in diameter.
Figure 2.
Distribution of squid NF mass-per-length measurements obtained by integrating the background-corrected dark-field pixel intensities. (A) Cores were analyzed by using 276 segments with a width of 10 nm and a length of 50 nm. (B) Whole filaments including sidearms were analyzed using 211 segments with a width of 100 nm and a length of 100 nm. Fits of normal distributions are shown as solid curves. The measured core mass/length, 21.0 ± 4.8 kDa/nm (mean ± SD), directly reflects the number of rod domains per filament cross section, whereas the mass of the whole filament, 33.5 ± 5.5 kDa/nm, reflects the number of rod domains plus NF-H carboxyl-terminal tails, thereby revealing the number of heavy chains.
Subunit Phosphorylation.
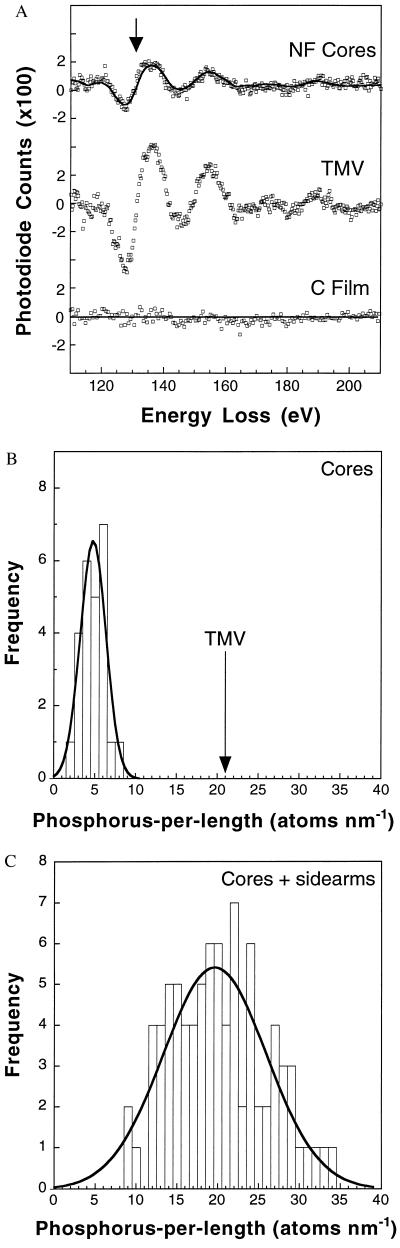
In STEM/EELS, a parallel-detection electron spectrometer records element-specific excitations of inner-shell electrons generated in the specimen volume irradiated by a scanned nanometer-diameter electron probe provided by a cold field-emission source. This technique offers a direct approach to measuring the number of phosphorus atoms per length of filament in squid NFs. This value, combined with the already determined number of LH dimers per filament length, specifies the average phosphorylation level of each NF dimer, which in turn allows an estimate of the phosphorylation level of native NFs. EELS spectra were recorded from segments of freeze-dried NFs and TMVs (Fig. 3A). The measured phosphorus signal recorded in 100 s from 20-nm segments of different TMVs varied by only ±9% (±SD) and was consistent with that predicted from the ionization cross section and probe current. Thus, the known phosphorus content of TMV particles (21 atoms per nm) served as a validated internal standard for quantifying the phosphorus distribution along the NFs (21). By fitting a reference spectrum to the P L2,3 core edge, a value of 4.8 ± 0.5 P atoms per nm (mean ± SEM.) was obtained within a 10-nm width along the NF core (Fig. 3B) and 19.6 ± 0.6 phosphorus atoms per nm within a 100-nm width of the whole filament (Fig. 3C). This indicates that at least three-quarters of the phosphates are situated on the sidearms; this fraction is likely to be even higher because some sidearms are undoubtedly overlaying the cores. From our determination of eight NF-H chains per cross section—and the known value of 48 ± 2 nm for the axial repeat length—we deduce that each dimer contains 117 ± 10 phosphorus atoms, most of which are situated on the sidearms away from the central core.
Figure 3.
(A) Sum of four EELS spectra each recorded in second-difference mode in 100 s with a 1-nA probe current from 10-nm × 10-nm regions of NF cores, TMV, and support film; P L2,3 ionization edge at 132 eV indicated by arrow. The solid curve represents the calculated fit to the NF core spectrum. No phosphorus was detectable in spectra recorded from the adjacent carbon support film. (B) Distribution of phosphorus atoms per nm of NF obtained from 25 EELS measurements on cores of different filaments; fit of normal distribution (solid curve) gives a value of 4.8 ± 2.2 phosphorus atoms/nm (mean ± SD). Corresponding number of phosphorus atoms for the TMV standard is indicated (arrow). (C) Distribution of phosphorus atoms per nm of NF obtained from 84 EELS measurements on cores plus sidearms; analyzed regions were 100 nm × 100 nm for NFs and 20 nm × 20 nm for TMVs. Fit of normal distribution (solid curve) gives 19.6 ± 8.9 phosphorus atoms per nm.
DISCUSSION
The determination of eight coiled-coil dimers per cross section for squid NFs is consistent with a model for intermediate filament assembly based on sequential lateral dimerizations, starting with NF-H/NF-L dimers and progressing through protofilaments (2 dimers), protofibrils (4 dimers), “half-filaments” (8 dimers), and “full filaments” (16 dimers) (25). The present results indicate that native squid NFs are in fact organized as an essentially homogeneous population of “half-filaments.” Intermediate filaments composed of ≈8 dimers per filament cross section have previously been reported for certain human epidermal keratins reconstituted in vitro. Such filaments form a polymorphic population whose mass per length also yielded minor components with 10–13 dimers per cross section (20, 27). In general, intermediate filaments and particularly NFs reconstituted in vitro exhibit a polymorphic character, but typically contain between 14 and 23 dimers per cross section (3, 28). Our results suggest that native intermediate filaments may be more homogeneous than reconstituted intermediate filaments. They also imply that native filaments and reconstituted filaments may be composed of different numbers of dimers, although the 8-dimer arrangement may be a characteristic of the 8-nm lamin-like squid NF.
There is no doubt that squid axonal NFs, like NFs of vertebrate axons, are highly phosphorylated (5), although only the five phosphate groups on the N terminus of NF-L have been directly determined (29). However, the squid NF-H subunit alone contains over 300 potential phosphate acceptor sites (Ser, Thr, and Tyr), of which approximately 100 are consensus sites for known axonal kinases; virtually all of the latter sites reside in the C-terminal tail, including a cluster of 64 KSP, RSP, and SEK motifs (5, 18). Our measured phosphorylation level of 117 ± 10 phosphorus atoms per dimer compares quite well with the maximum level predictable from consensus motifs, as well as with the highest values yet reported from biochemical measurements (30). Thus, we speculate that the heavy chains of all dimers in axonal squid NFs are near-maximally phosphorylated. At the least, squid NF-H is among the most heavily phosphorylated proteins yet reported. Motifs actually phosphorylated are likely to include most of the 32 SEK sites (5), for which in situ phosphorylation has not yet been demonstrated. Taken together with the mass distribution, these results lead to the structural model in Fig. 4.
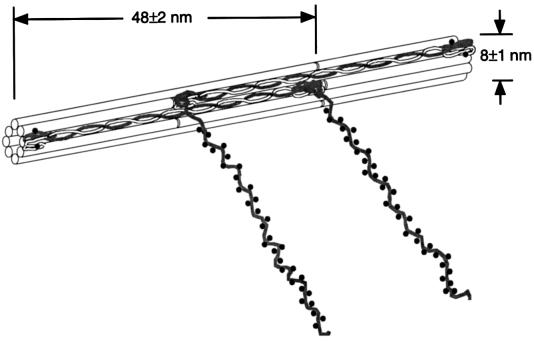
Figure 4.
Schematic model of the organization of squid NFs illustrates parallel coiled-coil polypeptide dimers assembled head-to-tail along the filament with adjacent dimers arranged antiparallel to form an NF with eight dimers per cross section. Each C-terminal chain of the heavy subunit contains approximately 100 phosphates and a few sites are also present on the N-terminal end of both subunits. For simplicity, only two antiparallel heterodimers are shown, staggered by half of the axial repeat distance (doubled-headed arrow), and the NFs are drawn as hollow cylinders. It is equally possible that the dimers are arranged in a nonhollow structure as two protofibrils, each with four dimers per cross section (36). Light chains, white; heavy chains, dark; phosphorylated residues represented by dots. Model is to scale, but for clarity only a third of the actual phosphorylated residues are represented.
In a more general context, the present results demonstrate that STEM/EELS is capable of measuring the phosphorylation state of individual protein assemblies, while mass-mapping the molecular weight distributions in the same molecules. The complementary technique of energy-filtered transmission electron microscopy (EFTEM) has been more widely applied to mapping elements in biological specimens, and has proven especially useful for mapping higher concentrations of phosphorus in nucleic acids polymers (31–33). However, EFTEM cannot at present be used to quantitate low levels of phosphorus bound to proteins because the full electron energy-loss spectrum at each pixel is not available. To illustrate this point, the NFs plus support film in our preparations contain a total of ∼5 × 103 carbon atoms per nm length of the filament core. Thus, measurement of NF phosphorus at the required sensitivity of ∼0.5 phosphorus atom per nm necessitates detection of ∼1 phosphorus atom per 104 matrix atoms. Such low levels of phosphorus—corresponding to an L2,3-edge signal that is only ∼0.1% of the background (21, 34)—can only be quantitated by STEM/EELS. Nanoanalysis at this trace level does require very high electron doses (>109 electrons per nm2), which entails considerable specimen damage. However, we have previously shown that phosphorus is retained in biological specimens at cryogenic temperatures even though beam damage degrades the spatial information to ∼10 nm, i.e., approximately the NF diameter (21).
Our STEM/EELS analysis attained a sensitivity of ∼10 phosphorus atoms in a single measurement from a 10-nm segment of NF, and a further improvement in sensitivity (at least 2-fold) can be expected in the future with the use of more efficient charge-couple device detectors (35). It is anticipated that STEM/EELS will find further applications for characterizing phosphorylation and quaternary structure of other large protein assemblies, including cytoskeletal filaments and enzymatic complexes.
Acknowledgments
We are indebted to Drs. H. C. Pant, H. Gainer, and A. C. Steven for stimulating discussions and to Prof. U. Aebi for his very helpful suggestions and comments.
ABBREVIATIONS
- NF
neurofilament
- NF-H
high molecular weight NF protein
- NF-L
low molecular weight NF protein
- STEM
scanning transmission electron microscope
- EELS
electron energy loss spectroscopy
- TMV
tobacco mosaic virus
References
- 1.Fliegner K H, Liem R K. Int Rev Cytol. 1991;131:109–167. doi: 10.1016/s0074-7696(08)62018-5. [DOI] [PubMed] [Google Scholar]
- 2.Pant H C, Veeranna Biochem Cell Biol. 1995;73:575–592. doi: 10.1139/o95-063. [DOI] [PubMed] [Google Scholar]
- 3.Heins S, Wong P C, Müller S, Goldie K, Cleveland D W, Aebi A. J Cell Biol. 1993;123:1517–1533. doi: 10.1083/jcb.123.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szaro B G, Pant H C, Way J, Battey J. J Biol Chem. 1991;266:15035–15041. [PubMed] [Google Scholar]
- 5.Way J, Hellmich M R, Jaffe H, Szaro B, Pant H C, Gainer H, Battey J. Proc Natl Acad Sci USA. 1992;89:6963–6967. doi: 10.1073/pnas.89.15.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aebi U, Haner M, Troncoso J, Eichner R, Engel A. Protoplasma. 1988;145:73–81. [Google Scholar]
- 7.Aebi U, Cohn J, Buhle L, Gerace L. Nature (London) 1986;323:560–564. doi: 10.1038/323560a0. [DOI] [PubMed] [Google Scholar]
- 8.Shuman H, Kruit P. Rev Sci Instrum. 1985;56:231–239. [Google Scholar]
- 9.Shuman H. Ultramicroscopy. 1981;6:163–168. doi: 10.1016/0304-3991(81)90056-5. [DOI] [PubMed] [Google Scholar]
- 10.Krivanek O L, Ahn C C, Keeney R B. Ultramicroscopy. 1987;22:103–116. [Google Scholar]
- 11.Crewe A V, Wall J, Langmore J. Science. 1970;168:1338–1340. doi: 10.1126/science.168.3937.1338. [DOI] [PubMed] [Google Scholar]
- 12.Crewe A V, Isaacson M, Johnson D. Nature (London) 1971;231:262–263. doi: 10.1038/231262a0. [DOI] [PubMed] [Google Scholar]
- 13.Isaacson M, Johnson D. Ultramicroscopy. 1975;1:33–52. doi: 10.1016/s0304-3991(75)80006-4. [DOI] [PubMed] [Google Scholar]
- 14.Krivanek O L, Mory C, Tencé M, Colliex C. Microsc Microanal Microstruct. 1991;2:257–267. [Google Scholar]
- 15.Browning N D, Chisholm M F, Pennycook S J. Nature (London) 1993;366:143–146. [Google Scholar]
- 16.Muller D A, Tzou Y, Raj R, Silcox J. Nature (London) 1993;366:725–727. [Google Scholar]
- 17.Pant H C, Gallant P E, Gainer H. J Biol Chem. 1986;261:2968–2977. [PubMed] [Google Scholar]
- 18.Elhanany E, Jaffe H, Link W T, Sheeley D M, Gainer H, Pant H C. J Neurochem. 1994;63:2324–2335. doi: 10.1046/j.1471-4159.1994.63062324.x. [DOI] [PubMed] [Google Scholar]
- 19.Wall J S, Hainfeld J F. Annu Rev Biophys Biophys Chem. 1986;15:355–376. doi: 10.1146/annurev.bb.15.060186.002035. [DOI] [PubMed] [Google Scholar]
- 20.Engel A, Eichner R, Aebi U. J Ultrastruct Res. 1985;90:323–335. doi: 10.1016/s0022-5320(85)80010-1. [DOI] [PubMed] [Google Scholar]
- 21.Leapman R D, Andrews S B. J Microsc. 1992;165:225–238. doi: 10.1111/j.1365-2818.1992.tb01482.x. [DOI] [PubMed] [Google Scholar]
- 22.Egerton R F, Yang Y Y, Cheng S C. Ultramicroscopy. 1993;48:239–250. [Google Scholar]
- 23.Hisanaga S, Hirokawa N. J Mol Biol. 1988;202:297–305. doi: 10.1016/0022-2836(88)90459-7. [DOI] [PubMed] [Google Scholar]
- 24.Nakagawa T, Chen J, Zhang Z, Kanai Y, Hirokawa N. J Cell Biol. 1995;129:411–429. doi: 10.1083/jcb.129.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heins S, Aebi U. Curr Opin Cell Biol. 1994;6:25–33. doi: 10.1016/0955-0674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 26.Steinert P M, Marekov L N, Fraser R D B, Parry D A D. J Mol Biol. 1993;230:436–452. doi: 10.1006/jmbi.1993.1161. [DOI] [PubMed] [Google Scholar]
- 27.Steven A C, Hainfeld J F, Trus B L, Wall J S, Steinert P M. J Cell Biol. 1983;97:1939–1944. doi: 10.1083/jcb.97.6.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herrmann H, Häner M, Brettel M, Müller S A, Goldie K N, Fedtke B, Lustig A, Franke W W, Aebi U. J Mol Biol. 1996;264:933–953. doi: 10.1006/jmbi.1996.0688. [DOI] [PubMed] [Google Scholar]
- 29.Steinert P M, Wantz M L, Idler W W. Biochemistry. 1982;21:172–183. doi: 10.1021/bi00530a030. [DOI] [PubMed] [Google Scholar]
- 30.Jones S M, Williams R C. J Biol Chem. 1982;157:9902–9905. [PubMed] [Google Scholar]
- 31.Bazett-Jones D P, Ottensmeyer F P. Science. 1981;211:169–170. doi: 10.1126/science.7444457. [DOI] [PubMed] [Google Scholar]
- 32.Ottensmeyer F P. J Ultrastruct Res. 1984;88:121–134. doi: 10.1016/s0022-5320(84)80004-0. [DOI] [PubMed] [Google Scholar]
- 33.Bazett-Jones D P, Leblanc B, Herfort M, Moss T. Science. 1994;264:1134–1137. doi: 10.1126/science.8178172. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y Y, Ho R, Shao Z, Somlyo A P. Ultramicroscopy. 1992;41:11–31. doi: 10.1016/0304-3991(92)90091-w. [DOI] [PubMed] [Google Scholar]
- 35.Tang Z, Ho R, Xu Z, Shao Z, Somlyo A P. J Microsc. 1994;175:100–107. doi: 10.1111/j.1365-2818.1994.tb03473.x. [DOI] [PubMed] [Google Scholar]
- 36.Steven A C. In: Cellular and Molecular Biology of Intermediate Filaments. Goldman R D, Steinert P M, editors. New York: Plenum; 1990. pp. 233–263. [Google Scholar]