Abstract
Peptide deformylase (PDF, E.C. 3.5.1.88) catalyzes the removal of N-terminal formyl groups from nascent ribosome-synthesized polypeptides. PDF contains a catalytically essential divalent metal ion, which is tetrahedrally coordinated by three protein ligands (His, His, and Cys) and a water molecule. Previous studies revealed that the metal cofactor is a Fe2+ ion in Escherichia coli and many other bacterial PDFs. In this work, we found that PDFs from two iron-deficient bacteria, Borrelia burgdorferi and Lactobacillus plantarum, are stable and highly active under aerobic conditions. The native B. burgdorferi PDF (BbPDF) was purified 1200-fold and metal analysis revealed that it contains ~1.1 Zn2+ ion/polypeptide but no iron. Our studies suggest that PDF utilizes different metal ions in different organisms. These data have important implications in designing PDF inhibitors and should help address some of the unresolved issues regarding PDF structure and catalytic function.
Keywords: Peptide deformylase, hydrolase, iron, zinc, metal specificity, catalysis
Peptide deformylase (PDF) catalyzes the removal of N-terminal formyl groups from nascent ribosome-synthesized polypeptides [1]. This co-translational event takes place on essentially all bacterial proteins and is essential for bacterial survival [2–4]. Since N-formylation is not involved in eukaryotic cytoplasmic protein synthesis, PDF has been heralded as one of the most promising targets for the development of novel antibacterial agents [5, 6]. More recent studies indicate that active PDF is present in some eukaryotic organelles including human mitochondrion [7–12]. The human PDF gene contains a mutation at a highly conserved active-site residue and the purified recombinant human PDF is weakly active in vitro [10–12]. While the function of eukaryotic PDF is currently under debate, a large number of PDF inhibitors have been synthesized and two of them have been advanced to phase I clinical trials [13].
Early biochemical characterization of PDF was hampered by its extreme instability [14–16]. Later studies revealed that PDF is a metalloenzyme and contains a ferrous ion (Fe2+) in the active site of E. coli PDF (EcPDF) [17, 18]. The instability is caused by oxidation of the Fe2+ ion into Fe3+, which is catalytically inactive [19]. Since PDFs isolated from many other bacterial species shared the instability of EcPDF, it is generally accepted that bacterial PDFs utilize Fe2+ as the metal cofactor. Interestingly, substitution of Ni2+ or Co2+ for the Fe2+ ion in EcPDF gives highly stable PDF variants that retain essentially full catalytic activity, whereas the Zn2+-substituted PDF form is highly stable but catalytically compromised (>100-fold less active than the native enzyme) [18, 20–22]. More than 30 high-resolution structures of various metal-bound PDF forms (Zn2+, Fe2+, Co2+, and Ni2+), either free or bound with inhibitors and reaction products, have been determined [20, 23–33]. In the free enzyme, the metal ion is always tetrahedrally coordinated with the side chains of a cysteine (Cys-90 in EcPDF) and two histidines (His-132 and His-136 of an HEXXH motif), plus a water molecule/hydroxide. The proposed mechanism involves a nucleophilic attack of the metal-bound hydroxide onto the formyl carbonyl group to form a tetrahedral intermediate [21, 34]. A highly conserved glutamate residue (Glu-133 of the HEXXH motif) acts as a general acid to donate a proton to the leaving amide ion during the subsequent decomposition of the tetrahedral intermediate.
The above observations raise the question of what metal ion, if any, would serve as PDF metal cofactor in bacteria that grow under Fe-limited conditions or contain limited intracellular concentrations of Fe. For example, Borrelia burgdorferi, the spirochete that causes Lyme disease, has bypassed a critical, innate host defense (Fe sequestration) by eliminating the need of iron. It has been reported that the intracellular concentration of iron in B. burgdorferi was less than 10 atoms per cell, a level well below physiological relevance [35]. Likewise, the lactic acid bacterium Lactobacillus plantarum is also deficient in iron [36]. Recently, mitochondrial PDF1A of Arabidopsis thaliana (AtPDF1A) and Leptospira interrogans PDF (LiPDF) have been proposed to use Zn2+ as the catalytic metal ion, based on observations that their recombinant Zn2+-containing forms are catalytically proficient [37, 38]. However, these studies did not examine whether Zn2+ is the metal cofactor in their native forms. In this work, we describe the biochemical and kinetic characterization of PDFs from B. burgdorferi (BbPDF) and L. plantarum (LpPDF). We show that native BbPDF contains a Zn2+ ion as the catalytic metal cofactor and, unlike PDF from other bacteria, it is both extremely stable and catalytically active.
Materials and methods
Materials
Formate dehydrogenase, thiamine, glucose, and Aeromonas aminopeptidase (AAP) were purchased from Sigma (St. Louis, MO.). All other chemicals including isopropyl-β-D-thiogalactopyranoside (IPTG), phenylmethanesulfonyl fluoride, kanamycin, biotin, tris(2-carboxyethyl)phosphine (TCEP) were purchased from Aldrich (Milwaukee, WI). High purity casamio acids were from Difco (Detroit, MI). Talon resin was purchased from Clontech (Mountain View, CA). B. burgdorferi strain B31 and L. plantarum (ATCC BAA-793) genomic DNAs were obtained from American Type Culture Collection (Manassas, Virginia).
Buffers
Buffer A: 25 mM Tris, pH 8.0, 5 mM NaCl; Buffer B: 25 mM MES, pH 6.5, 5 mM NaCl; Buffer C: 50 mM HEPES, pH 7.0, 100 mM NaCl; Buffer D: 50 mM HEPES, pH 7.0, 150 NaCl; Buffer E: 20 mM HEPES, pH 7.0, 10 mM NaCl.
Cloning, expression, and purification of recombinant BbPDF and LpPDF
BbPDF contains three N-terminal methionine residues at positions 1, 8, and 10 [39]. Previous gene annotation by others predicted Met-1 as the translational start site (http://pedant.gsf.de/cgi-bin/wwwfly.pl?Set=Borrelia_burgdorferi_B31&Page=index). Open-reading frame BB0065 encoding amino acids 1-172 of BbPDF (designated as “BbPDF1”) was amplified by polymerase chain reaction (PCR) with B. burgdorferi genomic DNA as template and using primers 5′-GGAGTTACATATGAAAGGGGGATGGGTTTTTATGG-3′ and 5′-TCCGCCTCGAGTTTTGCCTTAAGCCCCCTTTCTC-3′. The PCR product was digested with NdeI and XhoI and cloned into prokaryotic expression vector pET-22b to give plasmid pET22b-BbPDF1-HT. This cloning procedure resulted in the addition of a six-histidine tag (HT) to the C-terminus of BbPDF. Construction of N-terminally truncated BbPDF variants, BbPDF2 (amino acids 8-172) and BbPDF3 (amino acids 10-172), was similarly performed with the same 3′ PCR primer but using 5′-GCCGTGGTGCATATGGAAATGGTATTTTATCCT-3′ and 5′-GCCGTGGTGCATATGGTATTTTATCCTAATGATTT-3′ as the 5′ primers, respectively. The resulting plasmids were designated as pET22b-BbPDF2-HT and pET22b-BbPDF3-HT. To express BbPDF2 without a C-terminal histidine tag, a 3′ primer 5′-TCCGCCTCGAGTCATTTTGCCTTAAGCCCCCTTTCTC-3′ was used in conjunction with the above 5′ primer to amplify the BbPDF coding sequence by PCR and the resulting DNA fragment was cleaved with NdeI and XhoI and cloned into pET-22b to produce plasmid pET-BbPDF2. The gene coding LpPDF (LP2155) was similarly amplified by PCR using primers 5′-GGAGTTACCATATGATTAAAATGCGCGACATTATCCGC-3′ and 5′-ACCGACTCGAGAGAAATTAGAACTAAATCATCATCAGC-3′ and cloned into pET-22b (pET-LpPDF-HT). All DNA constructs were confirmed by dideoxy sequencing and transformed into E. coli BL21(DE3) Rosetta cells (Novagen).
To prepare BbPDF or LpPDF enriched in a specific metal (Zn, Co, and Fe), recombinant E. coli cells were grown in minimal media to an OD600 nm of 0.6 and induced for 3 h at 28 °C by the addition of 60–100 μM IPTG (final concentration) and 100 μM of the desired metal (ZnCl2, CoCl2, or FeCl3). Cells were harvested by centrifugation, lysed and the crude protein sample was fractionated by metal affinity chromatography on a cobalt affinity column (Talon resin, Novagen) as previously described [21]. The fractions with PDF activity from the Talon column were pooled and precipitated by ammonium sulfate (80% saturation). The precipitate was dissolved in a minimal volume of buffer C and loaded on an FPLC equipped with a Superdex G-75 10/300 column (Pharmacia) that had been equilibrated in buffer C. The protein was eluted with buffer C at a flow rate at 0.5 mL/min. Fractions containing active PDF were pooled, rapidly frozen, and stored at −80 °C. For the purification of Fe-BbPDF and Fe-LpPDF, 500 μM TCEP was included in all buffers and the gel-filtration step was omitted. Metal contents were analyzed by inductively coupled plasma emission spectrometry (ICP-ES) at the Chemical Analysis Laboratory at the University of Georgia, Athens, GA. Protein concentration was determined by Bradford assay using bovine serum albumin as a standard and corrected by a factor of 0.77 on the basis of absorbance at 280 nm of pure Zn and Co-BbPDF proteins (actual concentration = 0.77 × concentration as measured by Bradford).
Zn-BbPDF2 (which has no C-terminal histidine tag) was similarly overexpressed in E. coli Rosetta cells. The crude lysate was loaded on a Q-Sepharose column (2.5 cm × 10 cm, Pharmacia). The column was washed with two column volumes of buffer A and eluted with buffer A plus a linear gradient of 10 mM-500 mM NaCl. Fractions containing deformylase activity were pooled and precipitated with ammonium sulfate (80% saturation). The pellet was dissolved in a minimal volume of buffer E (20 mM HEPES, pH 7.0, 10 mM NaCl) and loaded on a Superdex G-75 column (3.6 cm × 60 cm, Pharmacia) and eluted with the same buffer at a flow rate of 0.5 mL/min. The PDF fractions were pooled and loaded on an FPLC equipped with a mono Q column (Pharmacia) that had been pre-equilibrated in buffer E. The column was washed with 5 volumes of buffer E and elution was carried out with a linear gradient of 10–500 mM NaCl in buffer E. PDF fractions were pooled, concentrated, and stored at −80 °C.
Purification of native BbPDF
B. burgdorferi strain B31A was grown in modified Barbour-Stoenner-Kelly (BSK-II) medium at 34 °C under atmospheric oxygen as previously described [40]. When the cell density reached approximately 1 × 108 cells/ml, bacteria were harvested by centrifugation (5,000 g, 15 min), washed 2x with HEPES buffer (20 mM HEPES; 50 mM NaCl, pH 7.6) and cell pellets were stored at −80 °C. Cell density was determined using dark-field microscopy. Before purification, all glassware was cleaned with 2% HNO3. All buffers were prepared with Millipore purified water with a dissipation resistance at 18.3 MΩ. B. burgdorferi cells from a 10-L culture were lysed in 25 mL of buffer A containing 1% triton X-100, 0.5% protamine sulfate, and a protease inhibitor cocktail consisting of 40 μg/mL trypsin inhibitor, 180 μg/mL phenylmethylsulfonyl fluoride, 10 μM pepstatin A, and 5 μM bestatin. After centrifugation to remove any cell debris, the crude lysate was fractionated with ammonium sulfate precipitation (30–45% saturation). The protein pellet was dissolved in buffer A and loaded on an FPLC Mono Q HR 5/5 column (Pharmacia) pre-equilibrated in Buffer A. The column was washed with 5-column volumes of buffer A and then eluted with buffer A plus a linear gradient of 5–1000 mM NaCl. The PDF active fractions were pooled (3 mL), diluted into 9 mL of buffer B, and loaded on an FPLC Mono S GL 5/50 column (Pharmacia) that had been equilibrated in buffer B. After being washed with 5-column volumes of buffer B, the column was eluted with buffer B plus a linear gradient of 5–1000 mM NaCl. The PDF-containing fractions were pooled, concentrated, and loaded on an FPLC Superdex G-75 10/300 gel-filtration column (Pharmacia) and eluted in buffer C. The purified BbPDF was quickly frozen and stored at −80 °C until use.
PDF assays
Two assay methods were employed. When f-ML-pNA was used as substrate, the PDF reaction was coupled to that of Aeromonas aminopeptidase (AAP) [41]. A typical assay reaction (total volume of 600-μL) was carried out in buffer D containing 500 μM TCEP, 0–125 μM f-ML-pNA, and 0.5 units AAP. The reaction was initiated by the addition of 0.030 μg of PDF. The reaction was monitored continuously at 405 nm on a Perkin-Elmer UV/Vis Lambda 25 spectrophotometer. The initial rates were calculated from the first 60 s of the reaction progress curves. A formate dehydrogenase coupled assay was used with f-MAS as substrate and the assay reactions were performed as previously described [42, 43]. PDF inhibition assays were similarly performed. Typically, Zn- or Co-BbPDF1-HT (10 nM) was preincubated with an inhibitor (0–150 nM) in buffer D for 2 h on ice. The solution was then warmed to room temperature and f-ML-pNA was added to a final concentration of 100 μM. The reaction was allowed to proceed for 15 min before being terminated by heating to 95 °C for 15 min (the inactivation process is usually complete within the first 30 s) and cooled to room temperature. After the addition of AAP (1 unit) and incubation for 15 min, the absorbance increase at 405 nm was measured on a UV-Vis spectrophotometer. The inhibition constant (KI) was calculated by using the equation:
PDF stability
At time zero, 20 μL of a purified Zn-, Fe, or Co-BbPDF1-HT stock solution (typically 0.3–1.0 mg/mL) was diluted into 1.0 mL of buffer D containing 0, 100, or 300 μM H2O2. The resulting solution was incubated at room temperature with exposure to air. At varying time points, 10-μL aliquots were withdrawn and added into a reaction mixture containing buffer D, 125 μM f-ML-pNA, and 0.5 U of AAP (total volume of 500 μL). The PDF reaction was monitored continuously at 405 nm on a UV-Vis spectrophotometer. The initial rates (relative to that at time 0) were plotted as a function of time. For native BbPDF, 25 μL of partially purified B. burgdorferi cell lysate (after Mono S column) was treated with 0 or 100 μM H2O2 for varying lengths of time and used to initiate the PDF reaction as described above. To assess the stability of PDFs in crude cell lysates, E. coli BL21(DE3) Rosetta cells harboring plasmid pET-BbPDF1-HT or pET22b-EcPDF [43] were grown in 100 mL of media (LB medium or minimal medium plus 100 μM ZnCl2 and/or FeCl3) to an OD600 of 0.6 and induced at 28 °C with 100 μM IPTG. After 3 h, the cells were spun down at 5000 rpm (GS-3 rotor, Sorvall), resuspended in 2.5 mL of lysis buffer (20 mM Tris, pH 8.0, 250 mM NaCl, and 1 mM TCEP), and lysed by sonication. After centrifugation at 16,000 rpm (SS-34 rotor, Sorvall) for 15 min, the clear supernatant was used in activity assays as described above.
Metal analysis of native BbPDF
The metal content in native BbPDF was analyzed by atomic absorption spectrometry. Prior to use, all glassware, tubes, and tips were washed with 6 g/L EDTA and 2% HNO3. A Barnstead “EASYpure LF” filtration system was used to purify water with a resistance at dispensation of 18.3MΩ. Ammonium dihydrogen phosphate (ADHP, GFS 99.999% pure) was used as the matrix modifier. In a typical analysis, the sample (1 mL) contains 1 g/L ADHP, 10 g/L HNO3, and the diluted protein samples. Twenty μL aliquots were loaded with an AS40 auto sampler. Atomic absorption was carried out on a Perkin Elmer Z5000 graphite furnace atomic absorption spectrometer equipped with an HGA500 graphite furnace controller. The furnace was programmed to stay at 150 °C for 60 s to dry samples, at 1000 °C for 60 s to clear inorganic ash, at 1800 °C for 8 s for atomization, and at 2700 °C for 5 s to clean graphite tube [44–46]. Purge was performed with argon gas (4.8 grade) at a flow rate of 300 mL/min. Absorption at 213.9 nm was measured for Zn, 248.3 nm for Fe, and 242.5 nm for Ni and Co. Background correction was performed by a Zeeman (transverse) method. Each analysis was carried out in triplicate and the average value was calculated. Zinc concentrations from protein samples were calculated by comparison against a known zinc standard curve.
Results
Overexpression and purification of recombinant BbPDF and LpPDF
The genes encoding BbPDF1 (BB0065, amino acids 1-172) and LpPDF (LP2155, amino acids 1-183) were amplified from their respective genomic DNAs by PCR and overexpressed in E. coli with a C-terminal six-histidine tag (BbPDF1-HT and LpPDF-HT, respectively). Both enzymes were prepared in various metal-substituted forms (Fe, Co, Zn) by growing the recombinant cells in minimal media supplemented with the desired metal ion. The majority of the expressed BbPDF1-HT protein was found in the insoluble fraction, regardless of the identity of the metal cofactor. Our attempts to purify BbPDF1-HT by metal affinity chromatography always resulted in two species of ~60 and ~22 kDa, respectively, on SDS-PAGE gels (Figure 1a). The 22-kDa is presumably BbPDF1-HT, which has a calculated molecular weight of 20,986 Da. The 60-kDa band was excised from the gel, digested by trypsin, and analyzed by LC-MS/MS. The 60-kDa protein was identified as E. coli chaperonin GroEL. Further purification by gel filtration chromatography produced two protein fractions. The first fraction contained a GroEL-BbPDF complex and the relative levels of the two proteins were consistent with one BbPDF molecule bound to each functional GroEL tetradecamer (Figure 1b). The second fraction contained apparently homogeneous BbPDF1-HT protein (Figure 1c, lanes 1 and 2). Both Zn2+-and Co2+-substituted BbPDF1-HT were purified in this manner yielding ~1 mg of pure protein from 1 L of culture. The Fe2+-substituted BbPDF1-HT could not be subjected to the gel filtration chromatography due to its extreme instability (vide infra). Therefore, the most active fractions from the metal affinity column, which contained a mixture of BbPDF and GroEL-BbPDF complex (Figure 1c, lane 3), were used directly in biochemical and kinetic studies. The protein samples were analyzed for the presence of 20 common divalent metal ions and elements (e.g., Ba, Ca, Cd, Co, Cu, Fe, Mn, Ni, Pb, and Zn). Both Zn-BbPDF1-HT and Co-BbPDF1-HT contained 1.0 atom of the desired metal ion/polypeptide (Table 1). The cobalt protein also contained a trace amount of Zn (0.02 atom/polypeptide) but no detectable amounts of the other metals tested. The Fe-BbPDF1-HT sample had a lower metal content of 0.59 Fe atom/polypeptide plus a small amount of Zn (0.08 atom/polypeptide). The lower metal content is likely due to some loss of Fe2+ during purification and the fact that the Fe protein sample was less pure (Figure 1c, lane 3).
Figure 1.
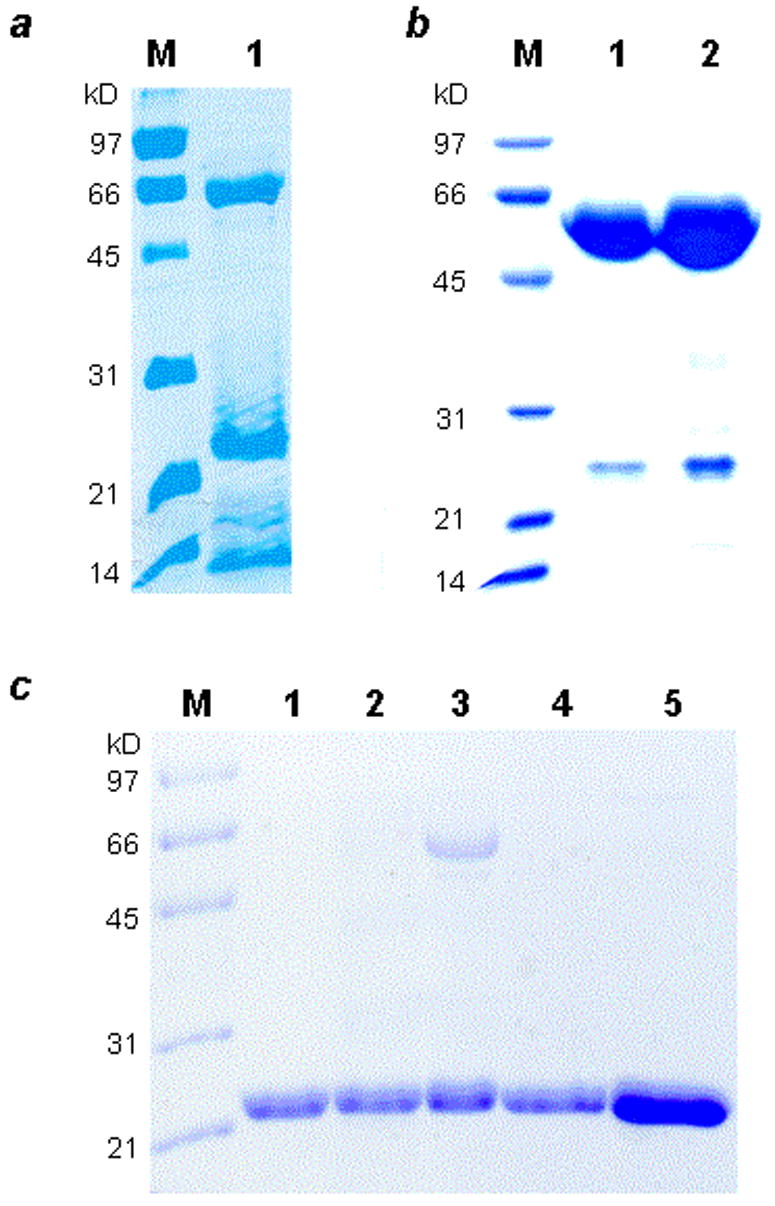
SDS-PAGE gels showing the purified BbPDF variants. Panel a: M, molecular weight markers; lane 1, Zn-BbPDF1-HT after Talon column. Panel b: Purified GroEL-BbPDF1-HT complex (after gel-filtration chromatography). M, molecular weight markers; lane 1, 5 μg of the complex; lane 2, 10 μg of the complex. Panel c: Purified recombinant BbPDF variants. M, molecular weight markers; lane 1, Zn-BbPDF1-HT; lane 2, Co-BbPDF1-HT; lane 3, Fe-BbPDF1-HT; lane 4, Zn-BbPDF2-HT; lane 5, Zn-BbPDF3-HT.
Table 1.
Catalytic Activity of BbPDF and LpPDF Variantsa
| enzyme | metal content | kcat (s−1) | KM (μM) | kcat/KM (M−1s−1) | ||
|---|---|---|---|---|---|---|
| Zn | Co | Fe | ||||
| Zn-BbPDF1-HT | 1.0 | 0 | 0 | 5.4 ± 0.1
20 ± 1b |
2.5 ± 0.2
510 ± 70b |
2.2 × 106
3.9 × 104b |
| Co-BbPDF1-HT | 0.02 | 1.0 | 0 | 10 ± 1
26 ± 1b |
3.9 ± 0.7
580 ± 60b |
2.6 × 106
4.5 × 104b |
| Fe-BbPDF1-HT | 0.08 | 0 | 0.59 | 3.8 ± 0.1 | 1.9 ± 0.2 | 2.0 × 106 |
| Zn-BbPDF2-HT | ND | ND | ND | 5.3 ± 0.5 | 2.8 ± 1.0 | 2.0 × 106 |
| Zn-BbPDF3-HT | ND | ND | ND | 0.062 ± 0.011 | 6.3 ± 1.2 | 1.0 × 104 |
| Zn-BbPDF2 | ND | ND | ND | 5.6 ± 0.9 | 2.3 ± 0.6 | 2.5 × 106 |
| Native BbPDF | 1.1 | 0 | 0 | 4.5 ± 1.2 | 3.3 ± 0.4 | 1.4 × 106 |
| Zn-LpPDF-HT | ND | ND | ND | 0.9 ± 0.1 | 17 ± 0 | 5.4 × 104 |
| Co-LpPDF-HT | ND | ND | ND | 5.8 ± 0.3 | 22 ± 1 | 2.6 × 105 |
| Fe-LpPDF-HT | ND | ND | ND | 4.3 ± 0.1 | 15 ± 1 | 3.0 × 105 |
Unless otherwise noted, the activities reported were determined with f-ML-pNA as substrate;
activity toward f-MAS; ND, not determined.
In light of the folding problems associated with the BbPDF1, we considered the possibility of alternative translational initiation sites. The putative BbPDF open reading frame contains three methionine residues at position 1, 8, and 10 [39]. We therefore constructed two “truncation mutants” that lack the N-terminal 7 (BbPDF2-HT) or 9 amino acid residues (BbPDF3-HT). Both truncation mutants were expressed in the soluble form with high yields. Moreover, we did not observe any association between the truncated proteins and GroEL. These proteins were prepared in Zn-substituted form and purified to apparent homogeneity (Figure 1c, lanes 4 and 5). To ascertain that the C-terminal six-histidine tag does not affect BbPDF properties, we also expressed and purified Zn-BbPDF2, which lacks the C-terminal histidine tag. Likewise, Fe, Co, and Zn-substituted LpPDF-HT variants were readily produced as a soluble protein and purified to near homogeneity.
Catalytic properties of BbPDF and LpPDF
The catalytic activities of BbPDF and LpPDF variants were assessed with f-ML-pNA and f-MAS as substrates [41–43] and the results are summarized in Table 1. First, Zn-, Co-, and Fe-substituted BbPDF1-HT were all highly active and had similar kinetic constants toward f-ML-pNA (kcat/KM = 2.0–2.6 × 106 M−1s−1). These activities are comparable to that of Fe-EcPDF (kcat/KM of 3.5 × 106 M−1s−1) [17, 21]. Similar activities were also observed for Zn- and Co-substituted enzymes when f-MAS was used as substrate. Second, removal of the N-terminal 7 amino acids from BbPDF1 resulted in a soluble, stable protein with no reduction in enzymatic activity (compare Zn-BbPDF1-HT and Zn-BbPDF2-HT). However, further truncation of 2 amino acids from the N-terminus produced a much less active variant (kcat/KM = 1.0 × 104 M−1s−1 toward f-ML-pNA for Zn-BbPDF3-HT). We propose that the translational starting site for BbPDF had been incorrectly annotated in the database (http://pedant.gsf.de/cgi-bin/wwwfly.pl?Set=Borrelia_burgdorferi_B31&Page=index) and that BbPDF2 (amino acids 8-172) represents the authentic BbPDF. Note that Zn-BbPDF2-HT (which contains a C-terminal histidine tag) and Zn-BbPDF2 (no histidine tag) have the same catalytic activity, indicating that the C-terminal histidine tag does not affect the catalytic properties of BbPDF. Fe2+- and Co2+-substituted LpPDF variants had similar activities, although the Zn-LpPDF had a 5-fold lower activity (Table 1).
Stability of metal-substituted BbPDF forms
We compared the stability of recombinant BbPDF1-HT containing different metal ions (Zn2+, Fe2+, and Co2+) and native BbPDF (partially purified B. burgdorferi lysate) under ambient conditions (room temperature and with exposure to atmospheric oxygen) or in the presence of H2O2. Under ambient conditions, the catalytic activity of recombinant Fe-BbPDF1-HT rapidly decreased with a half-life (t½) of ~18 min and was nearly completely lost after 120 min, presumably due to oxidation of the Fe2+ ion into Fe3+ (Figure 2a). In contrast, the native BbPDF and recombinant Zn- and Co-BbPDF1-HT forms were highly stable. When treated with 100 μM H2O2, the recombinant Fe-BbPDF enzyme was completely inactivated within 8 min while the native BbPDF, the recombinant Co- and Zn-BbPDF1-HT retained full catalytic activity after 16 min (Figure 2b). These results indicate that native BbPDF contains a metal ion other than Fe2+.
Figure 2.
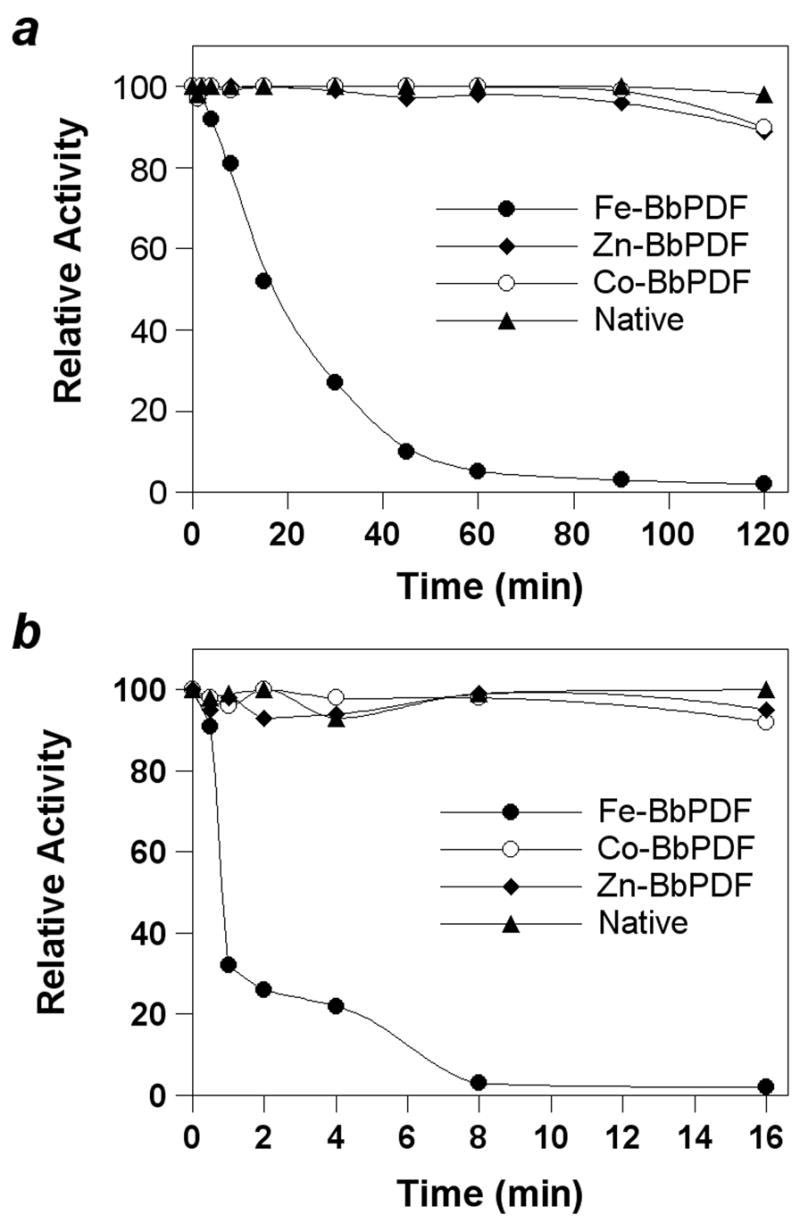
Stability profile of native BbPDF (partially purified B. burgdorferi cell lysate) and recombinant Zn-, Co, and Fe-substituted BbPDF1-HT under ambient condition (panel a) and upon treatment with 100 μM H2O2 (panel b).
Purification and metal analysis of native BbPDF
To determine the physiological metal cofactor of BbPDF, we purified the native enzyme ~1200-fold from B. burgdorferi cells. The crude cell lysate was fractionated by ammonium sulfate precipitation, followed by ion exchange (Mono Q and Mono S) and gel-filtration chromatography (Table 2). Approximately 100 μg of BbPDF was obtained from a 10-L B. burgdorferi culture. The purified protein showed predominantly a single band of ~22 kDa on SDS-PAGE gels (Figure 3, lane 5) and had an apparent kcat value of 4.5 s−1, a KM value of 3.3 μM, and a kcat/KM value of 1.4 × 106 M−1s−1 toward f-ML-pNA (Table 1). This activity is slightly lower than that of the recombinant Zn-BbPDF2 (in kcat value), likely reflecting the still heterogeneous nature of the enriched protein sample (estimated 80% purity on the basis of specific activity). As expected, the purified native BbPDF was highly stable under both ambient conditions and upon treatment with H2O2 (data not shown). The purified protein sample was analyzed by atomic absorption spectroscopy for the presence of Zn, Fe, Ni, and Co. Significant amounts of Zn (corresponding to ~1.1 Zn atom/polypeptide) but no Fe, Ni, or Co were found in the sample. Attempts of N-terminal sequencing of the native BbPDF were not successful so the proposed translational start site could not be confirmed experimentally.
Table 2.
Purification of native BbPDF from Borrelia burgdorferi
| Purification Step | Total protein (mg) | Specific Activitya (nmol/min per mg protein) | Purification (fold) | Recovery (%) |
|---|---|---|---|---|
| Crude lysate | 1555 | 2.7 | 1 | 100 |
| Ammonium Sulfate | 1200 | 61 | 23 | 80 |
| Mono Q | 412 | 560 | 207 | 56 |
| Mono S | 60 | 1290 | 477 | 29 |
| Gel Filtration | 0.1 | 3235 | 1198 | 11 |
The substrate used to assess activity was f-Met-Leu-p-NA.
Figure 3.
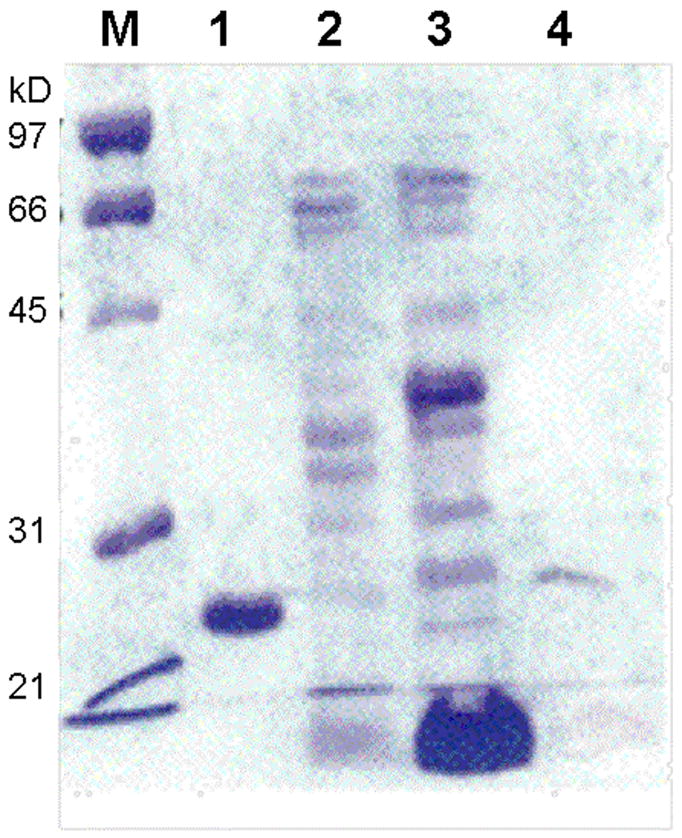
SDS-PAGE gel showing the purification of native BbPDF. Lane M, molecular weight markers; lane 1, purified recombinant Co-BbPDF1-HT; lane 2, pooled active fractions after Mono Q column; lane 3, active fraction after Mono S column; and lane 4, active fraction after G-75 column.
Metal preference of BbPDF
The use of Zn as the metal cofactor by native BbPDF may be a result of iron deficiency in B. burgdorferi [35]. Alternatively, the active-site structure of BbPDF may bind Zn2+ preferentially for thermodynamic or kinetic reasons (e.g., enzyme-catalyzed metal insertion during protein folding). To test these possibilities, E. coli cells harboring the BbPDF-overproducing plasmid (pET-BbPDF1-HT) were grown in LB media or minimal media supplemented with ZnCl2 and/or FeCl3 and the PDF activities in the crude cell lysates were determined after varying times of incubation in the presence of H2O2. Regardless of the growth media (LB, minimal + Zn, and minimal + Zn + Fe), the overproduced BbPDF had virtually identical stability profiles (Figure 4). In the absence of H2O2, the crude lysates retained ~90% of the original PDF activity after incubation at room temperature for 12 h. Treatment with 300 μM H2O2 for 12 h resulted in ~30% decrease in enzyme activity. As a control, a crude EcPDF preparation lost 40% of its catalytic activity after incubation for 4 h under ambient conditions (no H2O2) and all of the catalytic activity immediately after the addition of 300 μM H2O2 (Figure 4). It has previously been shown that overproduction of EcPDF in rich media gives a mixture of Fe-and Zn-EcPDF [17]. It was proposed that EcPDF binds Fe2+ preferentially and Zn-EcPDF is formed when the overproduced polypeptides deplete the Fe supply in rich media. The observed instability of EcPDF activity is consistent with this notion. The insensitivity of the crude BbPDF samples to H2O2 indicates that BbPDF contained mostly Zn2+ and little Fe2+ (if any) in the active site under the conditions tested. Therefore, when heterologously expressed in E. coli, BbPDF bound preferentially to Zn2+ during protein expression and folding. The slight loss of activity after 12 h is likely due to enzyme inactivation via other mechanisms (e.g., proteolysis).
Figure 4.
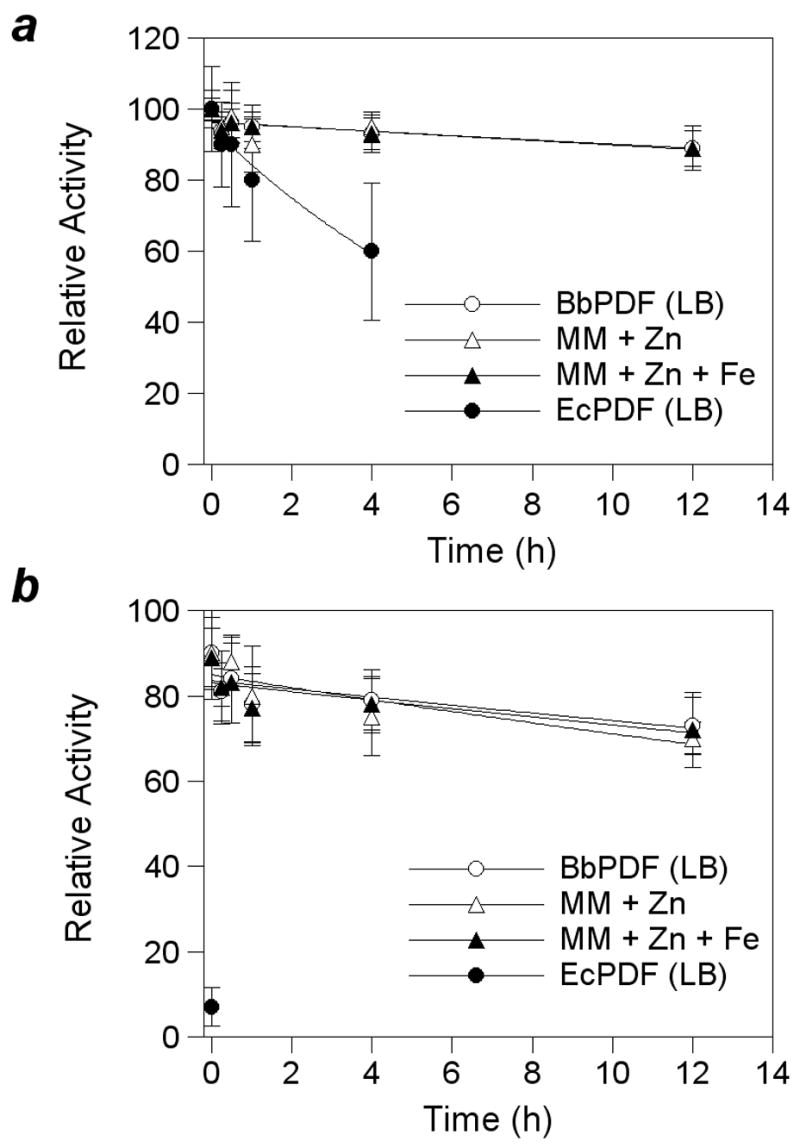
Stability profiles of recombinant EcPDF and BbPDF1-HT produced in LB medium (LB), minimal medium plus 100 μM ZnCl2 (MM + Zn), or minimal medium plus 100 μM ZnCl2 and FeCl3 (MM + Zn + Fe). Panel a, under ambient condition; panel b, upon treatment with 300 μM H2O2.
Absorption spectra and inhibition of BbPDF
The absorption spectrum of Co-BbPDF1-HT was recorded at pH 7.4 and compared with that of Co-EcPDF. The two proteins have remarkably similar spectra, both showing two d-d transition bands at 565 and 650 nm and a ligand-to-metal charge transfer (LMCT) band at 325 nm (Figure 5). The intensity of the LMCT band is consistent with the ligation of a single cysteine side chain to the metal ion. The apparently stronger absorption at 325 nm of BbPDF is due to overlap with absorption peaks by its aromatic side chains (BbPDF has a Trp and 9 Tyr as opposed to just 2 Tyr in EcPDF). The extinction coefficient of ~500 M−1 cm−1 (at 565 nm) suggests that the metal ion in BbPDF is tetrahedrally coordinated [47]. On the basis of the spectral data as well as the sequence conservation, we conclude that the metal ion in BbPDF has the same ligand environment as other structurally characterized PDFs [20, 23–33].
Figure 5.
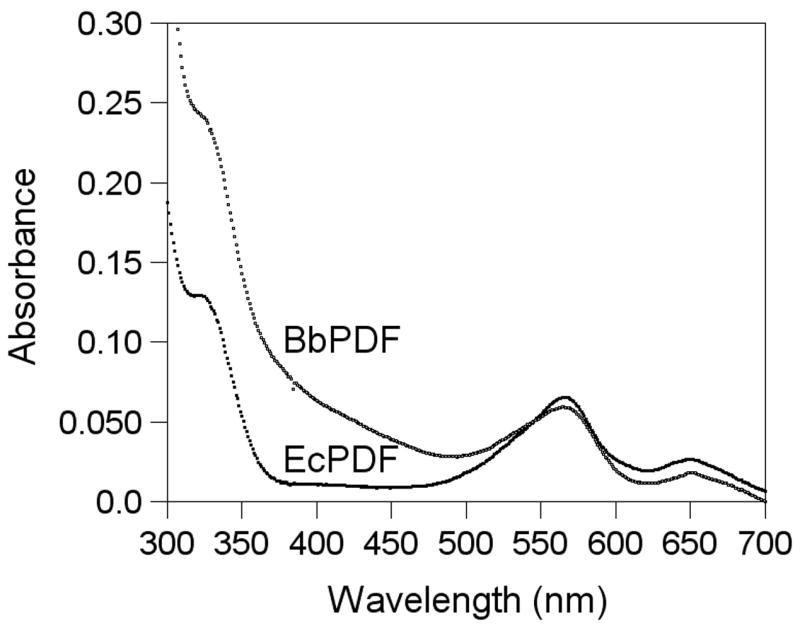
Absorption spectra of Co-EcPDF and Co-BbPDF (100 μM in buffer C).
Two previously reported PDF inhibitors (Table 3, compounds 1 and 2) [48, 49] were tested against Zn- and Co-BbPDF1-HT to determine whether their potencies are affected by the metal identity. Compound 1 is a competitive inhibitor against EcPDF (KI = 7–11 nM), while compound 2 is a tight, slow-binding inhibitor (KI = 109 nM and KI* = 0.33 nM against EcPDF). Both compounds are broad-spectrum antibacterial agents and bind bidentately to the catalytic metal ion of PDF through their N-formylhydroxylamine moiety. Compounds 1 and 2 inhibit both Co- and Zn-BbPDF1-HT with similar KI values of 17–39 nM (Table 3). Thus, as previously observed with other bacterial PDFs, the nature of the metal cofactor has minimal effect on the inhibitor potency. Unlike its slow-binding behavior with EcPDF, compound 2 did not exhibit any time-dependent inhibition toward BbPDF1-HT.
Table 3.
Inhibition Constants (KI, nM) of Zn- and Co-BbPDF1-HT
DISCUSSION
Several reasons prompted us to conduct this work. First, in contrast to the majority of metallopeptidases, which are most frequently zinc enzymes [50], EcPDF utilizes Fe2+ as the catalytic metal. It was not clear why Fe has been selected to “replace” Zn in EcPDF. It was tempting to suggest that Fe2+ might be required for high catalytic activity, since Zn-EcPDF is catalytically compromised. Second, the large difference in catalytic efficiency between Fe-EcPDF and Zn-EcPDF is itself an interesting, unresolved mechanistic puzzle. Finally, PDF is an important target for antibacterial drug design and the metal identity might be critical for designing high-affinity inhibitors. It was recently reported that metal-chelating inhibitors inhibited different metal-substituted forms of methionine aminopeptidase with dramatically different potencies [51, 52].
Determination of the physiologically relevant metal ion of a metalloprotein can be a challenging task, especially when the metal ion is loosely bound. A case in point is methionine aminopeptidase, whose metal identity is still under debate after decades of research [51]. We were able to demonstrate that EcPDF is an iron enzyme by comparing the activity and stability profiles of the native enzyme (in fresh cell lysates) with those of purified recombinant proteins enriched in various metal ions [17]. The properties of recombinant Fe-EcPDF (e.g., extreme instability) matched all of the properties of native EcPDF, whereas Zn-, Co-, Cu, or Ni-EcPDF did not. In this work, we applied the same strategy to BbPDF. Unlike EcPDF, native BbPDF (and recombinant Zn-BbPDF) is very stable in the presence of O2 and/or H2O2. This property allowed us to purify the native BbPDF to near homogeneity. The purified native BbPDF contained approximately one Zn2+ per polypeptide but no detectable levels of iron. The presence of high levels of PDF activity in the fresh cell lysate and its resistance to O2 and H2O2 rule out the possibility that Zn2+ had replaced Fe2+ during the purification process. It was previously shown that EcPDF binds its metal cofactor tightly and it is not possible to replace the metal cofactor by simple dialysis [21]. Furthermore, our studies show that BbPDF selectively binds zinc when zinc and iron are both abundant in the medium, whereas EcPDF binds preferentially Fe2+ when overexpressed in rich medium and recruits Zn2+ into its active site only when iron is depleted from the medium [17]. Thus, we conclude that Zn2+ is the physiological metal cofactor in BbPDF. To our knowledge, this is the first time that a native PDF has been purified to any significant extent and conclusively shown to be a zinc enzyme. Previously, Meinnel and coworkers showed that the recombinant Zn-PDF1A from A. thaliana mitochondrion could function effectively with a Zn2+ ion in the active site [37]. Gong et al. also reported that recombinant L. interrogans Zn-PDF was catalytically quite active [38]. It was suggested that the physiological metal cofactor in AtPDF1A and LiPDF might be zinc; however, neither study addressed this issue experimentally.
Our results demonstrate that PDF uses different metal cofactors in different organisms. Besides EcPDF, native PDFs from Haemophilus influenzae [17], Bacillus stearothermophilus [14], and A. thaliana (PDF1B) [37] have been reported to be extremely labile under aerobic conditions. Recombinant Zn-PDFs from Staphylococcus aureus [32], B. stearothermophilus [22], and Thermus thermophilus [22] have catalytic activities that are deemed to be too low to sustain bacterial growth. Recombinant PDFs from S. aureus, Streptococcus pneumoniae, and Thermotoga maritima underwent spontaneous oxidation of their metal-binding cysteine (corresponding to Cys-90 in EcPDF) into cysteic acid, when the proteins were overproduced in LB medium [33]. We propose that PDFs from these organisms are likely Fe2+ enzymes. On the basis of iron deficiency in L. plantarum [36] and the high catalytic activity of Zn-LpPDF, we believe that native LpPDF is also a Zn2+ protein. A. thaliana PDF1A and L. interrogans PDF may also be zinc proteins [37, 38]. The use of different metal ions by a protein in different organisms has other precedence. For example, human MetAP2 is believed to be a Mn2+ enzyme [51], whereas yeast and bacterial MetAPs have been reported to contain Zn2+, Co2+, or Fe2+ as cofactor [53–55]. Human and yeast Glyoxalase I have been shown to be a Zn2+ enzyme, but the enzyme from E. coli has maximal activity when Ni2+ is bound to the active site [56]. Recent work by Fierke and coworkers [57, 58] suggested that many previously reported “zinc” hydrolytic enzymes might actually use iron as the cofactor in vivo.
Why do organisms use different metal ions in their PDFs? For B. burgdorferi and L. plantarum, the answer may simply be that these organisms do not have significant intracellular iron concentrations. Another reason may be the fact that Zn2+ generally binds to a tetrahedral binding site (especially ones that do not undergo substantial structural changes upon metal binding) more tightly than other divalent transition metals with partially filled 3d orbitals [59]. This may explain why BbPDF preferentially bound Zn2+ when it was expressed in E. coli, even when both zinc and iron were readily available in the medium. What is puzzling is the utilization of Fe2+ by E. coli PDF, given that EcPDF binds Zn2+ much more tightly than Fe2+ [17] and both metal ions are available inside the cell. We hypothesize that Fe2+ insertion during PDF folding in E. coli and other bacteria is catalyzed by a specific protein(s) (an insertase) and that BbPDF is a poor substrate of the E. coli Fe2+ insertase. This hypothesis is consistent with the recent finding that the intracellular concentrations of free divalent metal ions are extremely low [60]. Although the identity of the putative PDF iron insertase is yet unknown, assembly of heme and iron-sulfur proteins has been shown to require an iron chaperone (e.g., frataxin) and/or iron insertase (e.g., ferrochelatase) [61, 62]. It has been suggested that Fe2+ may be selected because of its availability and its ability to impart high catalytic activity (as opposed to Zn-PDF). Our finding that Zn-BbPDF is highly active argues against this notion, as it should have been possible for E. coli and other bacteria to evolve a more active Zn-PDF. We propose that Fe-PDF may offer other advantages, for example, it may serve as a regulatory mechanism. It is conceivable that under oxidative stress, Fe-PDF would be quickly inactivated. Because of its essential function during protein synthesis and maturation, inactivation of PDF would stop cells from further growth, until the oxidative stress is relieved.
Our study also sheds some new light on the activity difference between Fe-EcPDF and Zn-EcPDF. Previous investigators attributed this difference to the ability of Fe2+ (also Co2+ and Ni2+) to form pentacoordinated transition states [25, 26]. In light of the high catalytic activities of Zn-BbPDF, Zn-LpPDF, and Zn-AtPDF1A, we conclude that the activity difference cannot be due to the intrinsic properties of Zn2+ vs Fe2+ (or Co2+, Ni2+) ions. Rather, we believe that the combination of the metal ion and its ligand environment (especially the outer shell ligands) is responsible for their different catalytic activities. It is likely that binding of Zn2+ and Fe2+ (or Co2+, Ni2+) to EcPDF results in slightly different metal-ligand bond lengths [25] and/or ligand geometry, which in turn produce active sites of slightly different dimensions and/or shapes. Consequently, Zn-EcPDF and Fe-EcPDF may bind to the same substrate differently such that the formyl group is less optimally aligned against the Zn-OH for nucleophilic attack (in terms of distance and/or orientation). Although such subtle structural differences have eluded detection by X-ray crystallography, they clearly exist, since Zn-EcPDF and Fe-EcPDF were completely separated into two different fractions on a phenyl-Sepharose column [17]. Finally, our study suggests that metal usage by metalloenzymes such as PDF is more complicated than we have previously anticipated and may be quite different in different organisms in order to adapt to their natural environments.
Acknowledgments
We thank Dr. Gordon Renkes at the Analytical Spectroscopy Laboratory (The Ohio State University) for assistance in the atomic absorption spectrometry experiments.
Abbreviations
- AAP
Aeromonas aminopeptidase
peptide deformylase
- BbPDF
B. burgdorferi PDF
- EcPDF
E. coli PDF
- LpPDF
L. plantarum PDF
- f-ML-pNA
N-formylmethionylleucyl p-nitroanilide
- HT
six-histidine tag
- TCEP
tris(carboxyethyl)phosphine
Footnotes
This work was supported by the National Institutes of Health (AI40575 and AI62901 to DP).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Meinnel T, Mechulam Y, Blanquet S. Biochimie. 1993;75:1061–1075. doi: 10.1016/0300-9084(93)90005-d. [DOI] [PubMed] [Google Scholar]
- 2.Mazel D, Pochet S, Marliere P. EMBO J. 1994;13:914–923. doi: 10.1002/j.1460-2075.1994.tb06335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meinnel T, Blanquet S. J Bacteriol. 1994;176:7387–7390. doi: 10.1128/jb.176.23.7387-7390.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Margolis PS, Hackbarth CJ, Young DC, Wang W, Chen D, Yuan Z, White R, Trias J. Antimicrob Agents Chemother. 2000;44:1825–1831. doi: 10.1128/aac.44.7.1825-1831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan Z, White RJ. Biochem Pharmacol. 2006;71:1042–1047. doi: 10.1016/j.bcp.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 6.Leeds JA, Dean CR. Curr Opin Pharmacol. 2006;6:445–452. doi: 10.1016/j.coph.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Giglione C, Serero A, Pierre M, Boisson B, Meinnel T. EMBO J. 2000;19:5916–5929. doi: 10.1093/emboj/19.21.5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bracchi-Ricard V, Nguyen KT, Zhou Y, Rajagopalan PT, Chakrabarti D, Pei D. Arch Biochem Biophys. 2001;396:162–170. doi: 10.1006/abbi.2001.2631. [DOI] [PubMed] [Google Scholar]
- 9.Dirk LMA, Williams MA, Houtz RL. Plant Physiol. 2001;127:97–107. doi: 10.1104/pp.127.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen KT, Hu X, Colton C, Chakrabarti R, Zhu MX, Pei D. Biochemistry. 2003;42:9952–9958. doi: 10.1021/bi0346446. [DOI] [PubMed] [Google Scholar]
- 11.Lee MD, Antczak C, Li Y, Sirotnak FM, Bornmann WG, Scheinberg DA. Biochem Biophys Res Commun. 2003;312:309–315. doi: 10.1016/j.bbrc.2003.10.123. [DOI] [PubMed] [Google Scholar]
- 12.Serero A, Giglione C, Sardini A, Martinez-Sanz J, Meinnel T. J Biol Chem. 2003;278:52953–52963. doi: 10.1074/jbc.M309770200. [DOI] [PubMed] [Google Scholar]
- 13.Ramanathan-Girish S, McColm J, Clements JM, Tauoin P, Barrowcliffe S, Hevizi J, et al. Antimicrob Agents Chemother. 2004;48:4835–4842. doi: 10.1128/AAC.48.12.4835-4842.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams JM. J Mol Biol. 1968;33:571–89. doi: 10.1016/0022-2836(68)90307-0. [DOI] [PubMed] [Google Scholar]
- 15.Livingston DM, Leder P. Biochemistry. 1969;8:435–443. doi: 10.1021/bi00829a059. [DOI] [PubMed] [Google Scholar]
- 16.Takeda M, Webster RE. Proc Natl Acad Sci USA. 1968;60:1487–1494. doi: 10.1073/pnas.60.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajagopalan PTR, Yu XC, Pei D. J Am Chem Soc. 1997;119:12418–12419. [Google Scholar]
- 18.Groche D, Becker A, Schlichting I, Kabsch W, Schultz S, Wagner AFV. Biochem Biophys Res Comm. 1998;246:342–346. doi: 10.1006/bbrc.1998.8616. [DOI] [PubMed] [Google Scholar]
- 19.Rajagopalan PTR, Pei D. J Biol Chem. 1998;273:22305–22310. doi: 10.1074/jbc.273.35.22305. [DOI] [PubMed] [Google Scholar]
- 20.Meinnel T, Blanquet S, Dardel F. J Mol Biol. 1996;262:375–386. doi: 10.1006/jmbi.1996.0521. [DOI] [PubMed] [Google Scholar]
- 21.Rajagopalan PTR, Grimme S, Pei D. Biochemistry. 2000;39:779–790. doi: 10.1021/bi9919899. [DOI] [PubMed] [Google Scholar]
- 22.Ragusa S, Blanquet S, Meinnel T. J Mol Biol. 1998;280:515–523. doi: 10.1006/jmbi.1998.1883. [DOI] [PubMed] [Google Scholar]
- 23.Chan MK, Gong W, Rajagopalan PTR, Hao B, Tsai CM, Pei D. Biochemistry. 1997;36:13904–13909. doi: 10.1021/bi9711543. [DOI] [PubMed] [Google Scholar]
- 24.Hao B, Gong W, Rajagopalan PTR, Zhou Y, Pei D, Chan MK. Biochemistry. 1999;38:4712–4719. doi: 10.1021/bi982594c. [DOI] [PubMed] [Google Scholar]
- 25.Becker A, Schlichting I, Kabsch W, Groche D, Schultz S, Wagner AF. Nat Struct Biol. 1998;5:1053–1058. doi: 10.1038/4162. [DOI] [PubMed] [Google Scholar]
- 26.Jain R, Hao B, Liu RP, Chan MK. J Am Chem Soc. 2005;127:4558–4559. doi: 10.1021/ja0503074. [DOI] [PubMed] [Google Scholar]
- 27.Dardel F, Ragusa S, Lazennec C, Blanquet S, Meinnel T. J Mol Biol. 1998;280:501–513. doi: 10.1006/jmbi.1998.1882. [DOI] [PubMed] [Google Scholar]
- 28.Becker A, Schlichting I, Kabsch W, Schultz S, Wagner AFV. J Biol Chem. 1998;273:11413–11416. doi: 10.1074/jbc.273.19.11413. [DOI] [PubMed] [Google Scholar]
- 29.Zhou C, Song X, Li Y, Gong W. J Mol Biol. 2004;339:207–215. doi: 10.1016/j.jmb.2004.03.045. [DOI] [PubMed] [Google Scholar]
- 30.Guilloteau JP, Mathieu M, Giglione C, Blanc V, Dupuy A, Chevrier M, Gil P, Famechon A, Meinnel T, Mikol V. J Mol Biol. 2002;320:951–962. doi: 10.1016/s0022-2836(02)00549-1. [DOI] [PubMed] [Google Scholar]
- 31.Fieilaine S, Juillan-Binard C, Serero A, Dardel F, Giglione C, Meinnel T, Ferrer JL. J Biol Chem. 2005;280:42315–42324. doi: 10.1074/jbc.M507155200. [DOI] [PubMed] [Google Scholar]
- 32.Baldwin ET, Harris MS, Yem AW, Wolfe CL, Vosters AF, Curry KA, Murray RW, Bock JH, Marshall VP, Cialdella JI, Merchant MH, Choi G, Deibel MR., Jr J Biol Chem. 2002;277:31163–31171. doi: 10.1074/jbc.M202750200. [DOI] [PubMed] [Google Scholar]
- 33.Kreusch A, Spraggon G, Lee CS, Klock H, McMullan D, Ng K, Shin T, Vincent J, Warner I, Ericson C, Lesley SA. J Mol Biol. 2003;330:309–321. doi: 10.1016/s0022-2836(03)00596-5. [DOI] [PubMed] [Google Scholar]
- 34.Deng H, Callende R, Zhu J, Nguyen KT, Pei D. Biochemistry. 2002;41:10563–10569. doi: 10.1021/bi026137e. [DOI] [PubMed] [Google Scholar]
- 35.Posey JE, Gherardini FC. Science. 2000;288:1651–1653. doi: 10.1126/science.288.5471.1651. [DOI] [PubMed] [Google Scholar]
- 36.Archibald FS, Duong MN. J Bacteriol. 1984;158:1–8. doi: 10.1128/jb.158.1.1-8.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serero A, Giglione C, Meinnel T. J Mol Biol. 2001;314:685–708. doi: 10.1006/jmbi.2001.5175. [DOI] [PubMed] [Google Scholar]
- 38.Li Y, Chen Z, Gong W. Biochem Biophys Res Commun. 2002;295:884–889. doi: 10.1016/s0006-291x(02)00777-5. [DOI] [PubMed] [Google Scholar]
- 39.Fraser CM, et al. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 40.Bledsoe HA, Carroll JA, Whelchel TR, Farmer MA, Dorward DW, Gherardini FC. J Bacteriol. 1994;176:7447–7455. doi: 10.1128/jb.176.24.7447-7455.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei Y, Pei D. Anal Biochem. 1997;250:29–34. doi: 10.1006/abio.1997.2194. [DOI] [PubMed] [Google Scholar]
- 42.Lazennec C, Meinnel T. Anal Biochem. 1997;244:180–182. doi: 10.1006/abio.1996.9910. [DOI] [PubMed] [Google Scholar]
- 43.Rajagopalan PTR, Datta A, Pei D. Biochemistry. 1997;36:13910–13918. doi: 10.1021/bi971155v. [DOI] [PubMed] [Google Scholar]
- 44.Kumamaru T, Riordan JF, Vallee BL. Anal Biochem. 1982;126:214–221. doi: 10.1016/0003-2697(82)90132-4. [DOI] [PubMed] [Google Scholar]
- 45.Vinas P, Lopez-Garcia I, Lanzon M, Hernandez-Cordoba M. J Agric Food Chem. 1997;45:3952–3956. [Google Scholar]
- 46.Campillo N, Vinas P, Lopez-Garcia I, Hernandez-Cordoba M. Talanta. 1998;46:615–622. doi: 10.1016/s0039-9140(97)00306-8. [DOI] [PubMed] [Google Scholar]
- 47.Bertini I. In: Coordination Chemistry of Metalloenzymes, I. Bertini I, et al., editors. D. Reidel Publishing Company; Dordrecht, The Netherlands: pp. 1–18. [Google Scholar]
- 48.Clements JM, Beckett RP, Brown A, Catlin G, Lobell M, Palan S, Thomas W, Whittaker M, Wood S, Salama S, Baker PJ, Rodgers HF, Barynin V, Rice DW, Hunter MG. Antimicrob Agents Chemother. 2001;45:563–570. doi: 10.1128/AAC.45.2.563-570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu X, Nguyen KT, Jiang VC, Lofland D, Moser HE, Pei D. J Med Chem. 2004;47:4941–4949. doi: 10.1021/jm049592c. [DOI] [PubMed] [Google Scholar]
- 50.Vallee BL, Auld DS. Biochemistry. 1990;29:5647–5659. doi: 10.1021/bi00476a001. [DOI] [PubMed] [Google Scholar]
- 51.Wang J, Sheppard GS, Lou P, Kawai M, Park C, Egan DA, Schneider A, Bouska J, Lesniewski R, Henkin ]J. Biochemistry. 2003;42:5035–5042. doi: 10.1021/bi020670c. [DOI] [PubMed] [Google Scholar]
- 52.Li J-Y, Chen L-J, Cui Y-M, Luo Q-L, Li J, Nan F-J, Ye ]Q-Z. Biochem Biophys Res Commun. 2003;307:172–179. doi: 10.1016/s0006-291x(03)01144-6. [DOI] [PubMed] [Google Scholar]
- 53.Arfin ST, Kendall RL, Hall L, Weaver LH, Stewart AT, Matthews BW, Bradshaw RA. Proc Natl Acad Sci USA. 1995;92:7714–7718. doi: 10.1073/pnas.92.17.7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walker KW, Bradshaw RA. Protein Sci. 1998;7:2684–2687. doi: 10.1002/pro.5560071224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.D’souza VM, Holz RC. Biochemistry. 1999;38:11079–11085. doi: 10.1021/bi990872h. [DOI] [PubMed] [Google Scholar]
- 56.Clugston SL, Barnadr JFJ, Kinach R, Miedema D, Ruman R, Daub E, Honek JF. Biochemistry. 1998;37:8754–8763. doi: 10.1021/bi972791w. [DOI] [PubMed] [Google Scholar]
- 57.Gantt SL, Gattis SG, Fierke CA. Biochemistry. 2006;45:6170–6178. doi: 10.1021/bi060212u. [DOI] [PubMed] [Google Scholar]
- 58.Henrick M, Fierke CA. Biochemistry. 2006;45:14573–14581. doi: 10.1021/bi061625y. [DOI] [PubMed] [Google Scholar]
- 59.Lachenmann MJ, Ladbury JE, Dong J, Huang K, Carey P, Weiss MA. Biochemistry. 2004;43:13910–13925. doi: 10.1021/bi0491999. [DOI] [PubMed] [Google Scholar]
- 60.Finney LA, O’Halloran TV. Science. 2003;300:931–936. doi: 10.1126/science.1085049. [DOI] [PubMed] [Google Scholar]
- 61.Bulteau AL, O’Neill HA, Kennedy MC, Ikeda-Saito M, Isaya G, Szweda LI. Science. 2004;305:242–245. doi: 10.1126/science.1098991. [DOI] [PubMed] [Google Scholar]
- 62.Hansson MD, Karlberg T, Rahardja MA, Al-Karadaghi S, Hansson M. Biochemistry. 2007;46:87–94. doi: 10.1021/bi061760a. [DOI] [PubMed] [Google Scholar]