Abstract
Desensitization of the cannabinoid CB1 receptor is mediated by the interaction with arrestin. In this study, we report the structural changes of a synthetic diphosphorylated peptide corresponding to residues 419–439 of the CB1 C-terminus upon binding to arrestin-2. This segment is pivotal to the desensitization of CB1. Using high-resolution proton NMR, we observe two helical segments in the bound peptide that are separated by the presence of a glycine residue. The binding we observe is with a diphoshorylated peptide, whereas a previous study reported binding of a highly phosphorylated rhodopsin fragment to visual arrestin. The arrestin bound conformations of the peptides are compared.
Keywords: arrestin, CB1, C-terminus, complex, phosphorylation, NMR
1. INTRODUCTION
An essential feature of G-protein Coupled Receptor (GPCR) function is the regulation of signal transduction by arrestins[1]. G-protein Coupled Receptors(GPCRs) are the largest known family of membrane bound receptors, also referred to as seven-transmembrane receptors (7TM) as they feature seven membrane spanning helices flanked by an extracellular N-terminal region and an intracelluar C-terminal region[2–4]. GPCRs regulate a diverse array of important physiological functions such as phototransduction, olfaction, neurotransmission, vascular tone, cardiac output, digestion, and pain and are widely recognized as the largest class of drug targets for developing pharmaceuticals[5]. Upon activation GPCRs cause heterotrimeric G proteins to mobilize and dissociate, initiating multiple signaling events[6], however, equally important, is the cessation of these signaling events[7,8]. This is accomplished by phosphorylation of the GPCR C-terminus by G-protein coupled receptor kinases, concomitant with binding to cytosolic arrestins. This causes the uncoupling of the GPCR-G-protein signaling complex and a decrease in the intracellular transduction response, a process referred to as desensitization[9–14]. In addition to desensitization, the binding of arrestins also promotes the internalization of GPCRs [15,16]. Once internalized, GPCRs may undergo trafficking towards degradation or recycling pathways. It has more recently been shown that bound arrestin allows the internalized GPCR to interface with a wider range of signaling pathways than previously thought[17,18]. There is biochemical evidence to suggest that the nature of the phosphorylation of the GPCR governs the interaction with arrestin and hence, the eventual fate of the receptor[19–21].
Members of the arrestin family include arrestin-1 (or visual arrestin) and arrestin-4 (or cone arrestin) that specifically modulate the action of rhodopsin, a photosensitive GPCR found in the retina [22,23]. In contrast, arrestin-2 (beta-arrestin-1) and arrestin-3 (beta-arrestin-2) modulate the action of non-retinal GPCRs[1,11,24]. There has been significant work to provide structural information on arrestins[23,25,26], however, direct measurement of the structures formed by arrestin-GPCR complexes is lacking. Such direct biophysical measurements concerning the nature of complexes between arrestins and GPCRs are limited to arrestin-1 and rhodopsin [27–29].
In this study we examine the conformational changes in a segment of the human CB1 cannabinoid receptor upon binding to arrestin-2. CB1 is a G-protein coupled, membrane bound receptor that is currently of great interest as a drug target[30]. It is hypothesized that residues 401–473 comprise the C-terminus of CB1 that extends intracellularly from the cell membrane[31]. The segment of the C-terminus between Gly-418 and Asn-439 is crucial to arrestin mediated desensitization as mutations in this region, particularly at Ser-426 and Ser-430, result in decreased desensitization with no effect on internalization [19,20]. In this study, we employ transferred nuclear overhauser (TrNOE) spectroscopy[32] to show a significant conformational change in a synthetic peptide corresponding to CB1 amino acid residues 419–438 (CB1419-438), upon binding to arrestin-2.
2. MATERIALS and METHODS
2.1 Peptide Synthesis
The diphosphorylated synthetic peptide CB1419-438 (Fig. 1) was obtained from New England Peptides at > 95% purity as shown by HPLC and MALDI-TOF DE mass spectral analysis.
Figure 1.
The amino acid sequences and numbering of CB1419-438 and Rh330-348. The phosphorylated residues are shown in italics and underlined.
2.2 Expression of Arrestin-2
cDNA encoding the human arrestin-2 ARRB1 variant 1 (accession number NM_004041; splice variant containing an additional eight amino acids as compared to variant two) was amplified from a human hypothalamus cDNA library (Quick clone cDNA library, Clontech, Palo Alto, CA) via PCR utilizing ARRB1 sequence specific oligonucleotide primers spanning the start and stop codons of the deduced cDNA. The resultant PCR products were size fractionated by agarose gel electrophoresis. A band of 1257 bp encoding ARRB1 (arrestin-2) was purified by silica-gel membrane adsorption (Qiagen, Chatsworth, CA) and cloned into pCR-Blunt II-TOPO (Invitrogen, Carlsbad, CA) following the procedures outlined in the respective manufacturer user manuals. All constructs were verified for integrity by sequencing (University of Connecticut Biotechnology Center, Storrs, CT). Arrestin-2 was cloned into a commercially available recombinant protein expression system (Invitrogen) utilizing the pTrcHisA vector from Invitrogen for high level expression in E. coli. This vector (pTrcHis.Arr-2) incorporates a hexa-histidine tag allowing for the purification of expressed protein by immobilized metal ion affinity chromatography (IMAC). pTrcHis.Arr-2 was transformed into Escherichia coli strain (DE3) pLysS. To over express 6Xhis-Arrestin-2 in Escherichia coli, 1 L of LB medium was inoculated with 10 ml of overnight cell culture and induced at 30°C with 300 μM isopropyl-1-thio-β-D-galactopyranoside at a cell density of 0.6. After a twelve-hour incubation, cells were harvested by centrifugation at 7500rpm and resuspended in binding buffer (50 mM phosphate, pH 8.0, 300 mM NaCl, 10%(v/v) glycerol, 2 mM 2-mercapto-ethanol, 1 mg/ml lysozyme, 0.1 mg/ml DNAse, 1mM PMSF). The resuspended cells were lysed using a French pressure cell and the lysate was centrifuged at 12000 rpm for 30 min. The supernatant was loaded onto 5ml Talon Resin (Clonetech) washed with 20 volumes of lysis buffer under gravity, washed with a salt buffer (50mM phosphate, pH 8.0, 300mM NaCl, 20mM imidazole) and 6Xhis-Arrestin-2 eluted with an imidazole containing buffer (50mM phosphate, pH 8.0, 300mM NaCl, 150mM imidazole ). A second ion-exchange purification step was performed by loading the elutions onto a 7.5 mm × 7.5 cm Alltech Prosphere P-Wax column with a 50 mM Tris Buffer, pH = 8.0 and eluted with a 30 minute linear gradient to 100% of a buffer comtaining 500mM NaCl, 50 mM Tris pH=8.0. The HPLC fractions containing arrestin-2 were concentrated using a Centricon MWCO=10000 (Mllipore).
2.3 NMR Experiments and Structure Calculations
The purified, concentrated arrestin-2 sample was dialyzed against a buffer containing 80mM phosphate, pH=7.2, 0.02% NaN3 at 4°C. For preparation of the complex sample, 0.95 mg of peptide was dissolved in 70μl dialysis buffer and 30 μl D20 for a final volume of 100 μl. This solution was added to 210 μl of the dialyzed arrestin-2 sample. The final sample volume of, 350 μl containing 0.1 mM arrestin-2, 1.14mM peptide at pH= 7.2 (final protein: peptide ratio of 1:10) was placed in a shigemi tube.
Two-dimensional high-resolution proton spectra were acquired on a Varian Unity 600MHz spectrometer fitted with a cryoprobe. DQF-COSY, TOCSY (mix = 60 & 90 ms) and NOESY (mix = 100, 200, 400 & 600 ms) spectra of the free-peptide, free-arrestin and arrestin2-peptide complexes were recorded with 2k real points, 512 increments and relaxation delay of 1s at two different temperatures of 10°C and 20°C. Water suppression was achieved with the WATERGATE sequence. Spectra were processed using NMRPipe.
The NMR assignment program Sparky was used for assignment and peak integration. Distance restraints used in the structure calculations were derived from TRNOESY experiments performed with mixing times of 100ms at 10°C as the NOE intensities proved to be in the linear range of the NOE buildup curve. Peaks were classified as Strong Medium or Weak based on TRNOE cross peak volumes. Distance restraints of 1.8–2.7Å (1.8–2.9 Å for NOEs involving NH protons), 1.8–3.5Å (1.8–3.7 Å for NOEs involving NH protons), 1.8–5.0 or 6.0 Å were assigned to strong, medium and weak peaks respectively[33,34]. The upper limits for distances involving methyl protons were increased by an additional 0.5 Å[35,36].
Distance restraints were used in a two-stage simulated annealing protocol as implemented in XPLOR-NIH where randomized structures were initially 'heated' to 2000K and then slowly cooled to 100K over 3000steps. The target function minimized during simulated annealing was comprised of quadratic harmonic potential terms for covalent geometry, square-well quadratic potentials for the experimental distance, and a quadratic van der Waals repulsion term for the nonbonded contacts. Only the measured distances were emplyed as restratints in these calculations. No torsion angle restraint, hydrogen-bonding, electrostatic, or 6–12 Lennard–Jones empirical potential energy terms were included in the target function. In the second stage, resulting structures that successfully satisfied the restraints were each used as starting structures in a second round of simulated annealing as described above.
3. RESULTS
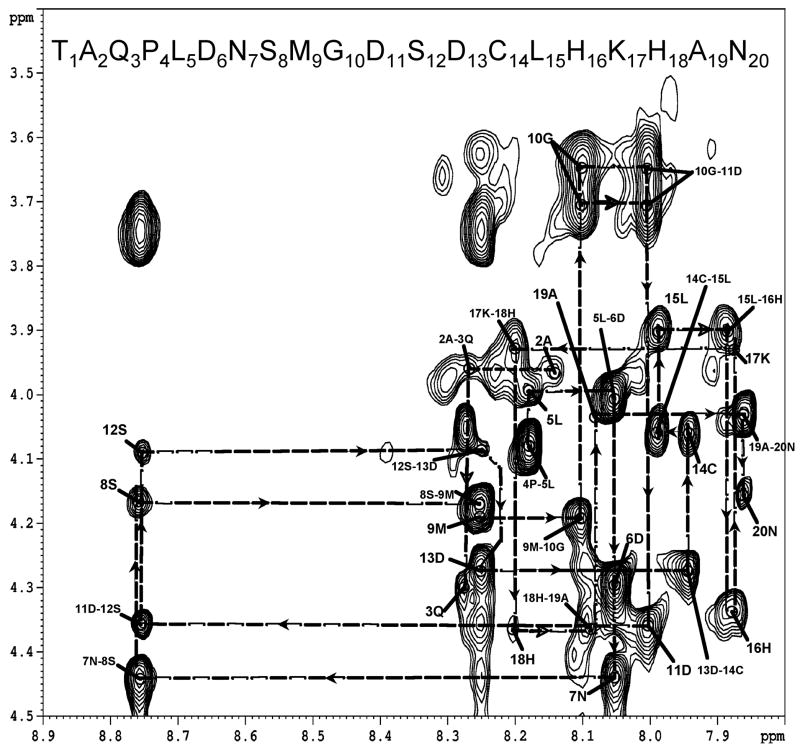
Proton chemical shifts and resonance assignments were established using standard techniques [37] with TOCSY, DQF-COSY and NOESY experiments and are reported in Table 1. The Gly428-spin system could be identified without any difficulty in the DQF-COSY spectrum from the characteristic alpha protons appearing at 3.640 and 3.706 ppm and was used as a starting point for sequential assignments. The phosphorylated residues, Ser426 and Ser430 could be identified through the downfield chemical shifts of the amide protons and the unique β proton chemical shifts while their sequential order was identified in the NOESY fingerprint region (Fig. 2). Similarly, Leu423 and Leu433 spin systems could be identified based on characteristic COSY cross peaks between γ protons (1.248ppm & 1.102ppm) and the two methyl groups (0.490–0.609ppm). Cross peaks between γ and α protons as well as amide protons could also be observed in the TOCSY spectrum and their sequential order identified in the NOESY fingerprint region. The Pro422 spin system was identified based on cross peaks between δ (3.482,3.345ppm) and γ protons (1.897ppm) through to both α and β protons observed in both COSY and TOCSY spectra and the notable absence of couplings from these resonances to the amide region. Asn425 and Asn438 was identified based on the coupling between the two side chain amines with the β-protons at approximately 2.4 ppm (Table 1) observed in TOCSY and NOESY spectra. Met427 and Gln421 were identified by their characteristic γ protons, which appear downfield as compared to β-protons. The N-terminal amide of Thr419 is not observed due to fast exchange with the solvent but the side chains could be assigned based on appearance of strong β and γ proton crosspeaks. His434 and His436 were identified using the β protons present at ~ 2.75ppm and cross peaks between β protons and aromatic protons (~6.8 and 7.9ppm) in the NOESY spectrum. The remaining residues were easily assigned in a similar manner and could clearly be identified through correlations of their sidechains.
Table 1.
The 1H chemical shifts of the free and bound peptide CB1419-438.
| Amino Acid | HN | Alpha | Beta | Gamma | Delta | Epsilon | Methyl | |
|---|---|---|---|---|---|---|---|---|
| THR419 | Free | 3.755 | 3.368 | 0.969 | ||||
| Bound | 3.77 | 3.409 | 0.980 | |||||
| ALA420 | Free | 7.991 | 4.048 | 1.066 | ||||
| Bound | 7.991 | 4.048 | 1.066 | |||||
| GLN421 | Free | 8.271 | 4.299 | 1.782,1.620 | 2.090 | 7.339,6.651 | ||
| Bound | 8.281 | 4.299 | 1.782,1.620 | 2.090 | 7.339,6.651 | |||
| PRO422 | Free | 4.078 | 1.978,1.596 | 1.722 | 3.475,3.335 | |||
| Bound | 4.078 | 1.986,1.601 | 1.729 | 3.482,3.345 | ||||
| LEU423 | Free | 8.177 | 4.007 | 1.333 | 1.248 | 0.610,0.562 | ||
| Bound | 8.184 | 4.003 | 1.333 | 1.248 | 0.609,0.560 | |||
| ASP424 | Free | 8.056 | 4.288 | 2.382,2.274 | ||||
| Bound | 8.056 | 4.288 | 2.382,2.274 | |||||
| ASN425 | Free | 8.051 | 4.44 | 2.515,2.443 | 7.419,6.637 | |||
| Bound | 8.055 | 4.446 | 2.519,2.451 | 7.419,6.638 | ||||
| SER426 | Free | 8.784 | 4.164 | 3.744,3.744 | ||||
| Bound | 8.757 | 4.090 | 3.744,3.744 | |||||
| MET427 | Free | 8.253 | 4.192 | 1.806,1.736 | 2.215 | |||
| Bound | 8.253 | 4.195 | 1.810,1.738 | 2.22 | ||||
| GLY428 | Free | 8.104 | 3.706,3.640 | |||||
| Bound | 8.104 | 3.706,3.640 | ||||||
| ASP429 | Free | 8.014 | 4.360 | 2.418,2.354 | ||||
| Bound | 8.005 | 4.359 | 2.418,2.354 | |||||
| SER430 | Free | 8.764 | 4.095 | 3.744 | ||||
| Bound | 8.757 | 4.090 | 3.744 | |||||
| ASP431 | Free | 8.258 | 4.278 | 2.383,2.383 | ||||
| Bound | 8.254 | 4.274 | 2.388,2.388 | |||||
| CYS432 | Free | 7.933 | 4.07 | 2.641,2.641 | ||||
| Bound | 7.944 | 4.054 | 2.641,2.641 | |||||
| LEU433 | Free | 7.993 | 3.899 | 1.213,1.288 | 1.11 | 0.583,0.494 | ||
| Bound | 7.989 | 3.899 | 1.220,1.3 | 1.102 | 0.584,0.490 | |||
| HIS434 | Free | 7.919 | 4.325 | 2.866.2.793 | 6.795,7.88 | |||
| Bound | 7.919 | 4.325 | 2.866.2.793 | 6.802,7.887 | ||||
| LYS435 | Free | 7.852 | 3.898 | 1.402,1.359 | 0.972 | 2.645,7.851 | ||
| Bound | 7.889 | 3.898 | 1.402,1.359 | 1.011 | 2.646,7.872 | |||
| HIS436 | Free | 8.189 | 4.339 | 2.877,2.788 | 6.769,7.991 | |||
| Bound | 8.207 | 4.357 | 2.903,2.788 | 6.832,7.991 | ||||
| ALA437 | Free | 8.1 | 4.27 | 1.050 | ||||
| Bound | 8.1 | 4.27 | 1.057 | |||||
| ASN438 | Free | 7.862 | 4.152 | 2.442,2.342 | ||||
| Bound | 7.869 | 4.152 | 2.444,2.342 |
Figure 2.
Sequential assignments in the fingerprint region of the NOESY spectrum of the CB1419-438 - arrestin-2 complex. The one letter codes of amino acids and numbering of the peptide from 1–20 is used for the sake of clarity. (See Figure 1 for numbering) Labels such as “5L” indicate the NH-α intra-residue NOEs for each residue while labels such as “5L-6D” indicate the ai-NHi+1 inter-residue NOE’s.
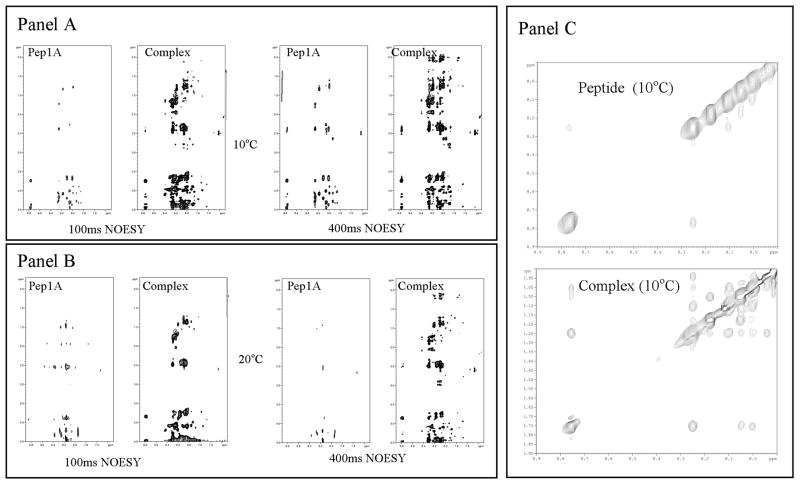
With the addition of arrestin-2 to CB1419-438, broadening and small changes in chemical shift were observed for a several peaks such as the Cys432NH, Asp429NH, and most notably, the phosphorylated residues Ser426NH and Ser430NH (Table 1). This is indicative of a fast exchange between the free and bound peptide state. Figure 3 shows expanded regions encompassing the fingerprint and side chain regions from the NOESY spectra of free Pep1A and the Pep1A-arrestin2 mixtures. The spectra are normalized to aid the comparison. In the NOESY spectrum of free CB1419-438 at 20°C there are several weak NOEs and a significant level of noise in the peptide spectrum. At 10°C there are a few more visible NOEs, most likely due to a decrease in side chain motions, however, there is no indication of secondary structure. The decreased number of NOEs at the higher temperature and the absence of any discernible evidence of structure suggest that the peptide is highly flexible in solution. With the addition of arrestin-2 there is a marked increase in the number and intensity of observed NOE peaks. This observation of peaks, non-existent in free peptide, is a result of the transferred NOE phenomenon and shows that there is a complex formed [38].
Figure 3.
Panel A) NOESY spectra of free CB1419-438 (peptide) and CB1419-438 -arrestin2 mixtures (complex) at 10°C and at two different mixing times. Panel B) NOESY spectra of free CB1419-438 and CB1419-438 -arrestin2 mixtures (complex) at 20°C and at two different mixing times. Panel C) NH-NH region of NOESY spectra of free CB1419-438 (peptide) and CB1419-438 -arrestin2 mixture (complex) at 10°C.
A total of two hundred and sixty six unambiguous NOEs were observed and utilized for structure calculations. Connectivity’s observed, between NH-αH protons from residue 423 to residue 428 (Fig. 2) and NH(i)–NH(i+1) (Fig 3) NOEs are highly suggestive of secondary structure in the bound peptide[38]. In addition to these there are numerous (i, i+1) NOEs between NH-βH, NH-γH, NH-δH, αH–βH, and αH–δH protons. There are numerous (i, i+1) NOEs between NH-NH, NH-NH, NH-αH, NH-βH, NH-γH, NH-δH, and αH–βH , and αH– δH protons. In addition there are numerous (i,i+3) transferred NOE peaks such as Asp424αH-Met427NH, Asn425αH-Gly428NH, Gly428αH-Asp431NH, Asp429αH-Cys432NH, Ser430αH-Leu433NH. Inter-residue NOEs are summarized in figure 4 and argue in favor of a helical structure for the segments of the peptide near the phosphorylated serine sites.
Figure 4.
Summary of inter-resdue NOEs and structural statistics of the ordered regions of the peptide (423–433) generated using PROCHECK. Approximately 90% of these residues occur in favored and allowed regions of the ramachandran plot with less than 5% occurring in disallowed regions. The residues that fall in the allowed or generously allowed regions were the residues that borderer the ordered helices and fell within the flexible region near the glycine.
4. DISCUSSION
Despite the fact that resonances could all be identified from side chain cross peaks, spectral overlap resulted in difficulty in unambiguously assigning certain NOE’s. For example, in the case of expected α N(i, i+1) cross peaks in the fingerprint region, overlap of amide proton chemical shifts of Asp424 and Asn425 result in an ambiguous assignment of the 424Hα–425HN cross peak hence these were omitted from structure calculations. Similarly, overlap of amide chemical shifts of Met427NH and Asp431NH makes it cumbersome to differentiate between an assignment of Met427NH-Asp429NH and Asp429NH-Asp431NH for this crosspeak. Similarly the high degree of overlap between Ser426 and Ser430 amides made assignments of certain potential long range NOEs ambiguous. For example the NOE from Asn425NH-Ser426NH may also be interpreted as Asn425NH-Ser430NH. In these cases the more conservative short-range assignment was used as a restraint in the initial structure calculations.
The conformation of CB1419-438 when bound to arrestin-2 was calculated using the NOEs as distance restraints in a simulated annealing protocol in XPLOR. A set of one hundred structures produced by simulated annealing were refined using no contribution from the van der waals term, followed by checks for agreement with the assigned distance restraints. No violations of distance restraints greater than 0.3–0.5A were allowed. Of the allowed structures, there was a family of thirty-three structures that clearly possessed the lowest energies. When aligned across the entire length of the peptide these structures showed a reasonable RMSD value between them of approximately 2.5. However, closer inspection of this final ensemble of thirty-three structures, showed two highly ordered segments (Fig 5). Alignment of the backbone atoms between the residues Pro423-Met427 gave an RMSD of 0.72 ± 0.2 and a second alignment of backbone atoms between residues Asp429-Leu433 gave an RMSD of 0.44 ± 0.08 (Fig. 4).
Figure 5.
NMR derived structures of CB1419-438 fragment bound to arrestin-2. (A) The low energy structures are aligned from residue 422–427 (B) The same set of structures are aligned from residue 429–433 (C) The ordered regions are shown together to emphasize the flexible GLY428 present in between the two ordered regions.
The results show that the diphosphorylated peptide CB1419-438 undergoes structural changes upon binding to arrestin-2. The lowest energy structure that fits the experimental restraints shows that the N terminal region up to Pro422 is disordered and the rest of the peptide encompasses two helical segments from Leu423-Gly428 and from Asp429-Leu433 (Fig. 5). This is consistent with the role of proline to form an end cap towards the N-terminal side of helices and of glycine to interrupt alpha-helix structures due to its high degree of flexibility and lack of hydrophobic stabilization[39]. The higher RMSD values calculated over the entire length of the peptide is due to the fact that the GLY428 residue acts as a hinge point so that the angle between the two ordered segments can vary as is illustrated in figure 5.
A previous study by Kisselev et al. has determined the conformation of a peptide (Rh330-348)(Fig. 1) mimicking the last nineteen residues of the rhodopsin (Rh) receptor with visual arrestin (or arrestin-1). The similarity of the observed arrestin structures across the family combined with the fact that mutagenic sensitive residues are largely conserved across all arrestins [23] has prompted conclusions from different studies to be applied across the family to in order to gain insights into the in situ mechanism of action. This warrants a comparison of the structures of bound CB1419-438 to arrestin-2 and bound Rh330-348 to arrestin-1. The C-termini of the two GPCRs have no sequence homology. The peptide Rh330-348 mimics a segment of the rhodopsin C-terminus that is immediately adjacent to helix eight, a portion of the C-terminus close to the cell membrane known to form a helix that associates with the membrane surface[40]. The C-terminus of CB1 is approximately twenty five residues longer than that of rhodopsin however CB1419-438 is estimated to also be adjacent to helix eight of CB1[31].
We observe binding of CB1419-438 with a diphosphorylated peptide, whereas, Rh330-348 contained seven phosphorylated sites (Fig. 1). Binding with this low level of phosphorylation is consistent with data indicating that only one or two phosphorylation sites are necessary for an observable interaction with arrestin[41,42]. Furthermore, the structural changes observed in this study bear similarities to the structural changes reported for Rh330-348 upon binding (Fig. 6) [27,28]. In each case, α-helices are induced towards the C-terminal end of each peptide (Fig. 6). In the case of Rh330-348, it was predicted that this structural element would also be observable with a single phosphorylation at Ser343[27]. The current results for CB1419-438 support this prediction. The CB1-Ser430 is analogous to the Rh-Ser343 phosphorylation site in that it is located in approximately the same relative position in the induced helix of the peptide (Fig. 6). This structural similarity between two peptides with no sequence homology that are interacting with two different members of the arrestin family, further argues in favor of the notion that this is a structure imposed by arrestins that is functionally significant rather than a potential artifact caused by an indiscriminant association of the highly phosphorylated Rh330-348 with the many arrestin surface charges.
Figure 6.
Comparison of (A) a minimimum energy conformer of diphosphorylated CB1419-438 fragment bound to arrestin and (B) the seven phosphorylated Rh330-348 fragment bound to visual arrestin. This later structure was downloaded from the protein data bank (www.rcsb.org.1NZS.pdb) and oriented to highlight the similarity of the C-terminal end with CB1419-438. The phosphorylated residues are highlighted by CPK representations.
On the N-terminal side of CB1419-438 there is another short helix, separated from the C-terminal helix by GLY428 (Fig. 5 & 6). This is in contrast to Rh330-348 which shows a loop that brings the N and C-termini of the peptide close to each other (Fig. 6). It is possible that these differences are due to the differences between arrestin-1 and arrestin-2 and/or the sequences of the two peptides. Another interpretation, however, is that the decreased level of phosphorylation in this portion of CB1419-438 compared to Rh330-348 allows the formation of this second helix[43]. The increased phosphorylation in this vicinity of Rh330-348 may prevent a helix from forming in this case, due to the increased association with charges on the surface of the protein.
Previously published data suggests that phosphorylation at both Ser426 and Ser430 is necessary for desensitization of the CB1 receptor [20]. This is consistent with the fact that the arrestin bound CB1419-438 shows structured regions in the vicinity of both the phosphorylated serines. This data taken together suggests that both phosphates are accommodated within the arrestin binding site. Comparison of the conformations of CB1419-438 with Rh330-348 also shows that the induced helices are relatively short, extending over no more than five amino acid residues. This might be suggestive of the size of binding pocket on the arrestin molecule. The flexibility of CB1419-438 between the two helical regions is a particularly striking feature. The fact that two helices are observed in the case of CB1419-438 may point to a functional significance of the flexible hinge point about Gly428. This may affect the affinity of the phosphorylated peptide for the binding site in that it may allow these helices to be better accommodated within the binding site.
In summary, CB1419-438 undergoes a conformational change upon binding to arrestin where helices are induced in the vicinity of the two-phosphorylation sites. The consistencies with the bound structure of Rh330-348 suggest that the arrestin imposes this structure and this imposed structure may be affected by the degree of phosphorylation. Taken together with literature evidence showing that the phosphorylation pattern may affect the functional outcome of the GPCR-arrestin interaction [19–21], our results prompt the speculation that the structural details of the high affinity complex between the C-terminus of GPCRs and arrestin-2 may vary depending on the nature of the phosphorylation, however this remains to be tested.
Acknowledgments
This work was supported in part by the University of Connecticut Research Foundation Large Faculty Grant and by a grant from the National Institute on Drug Abuse (NIDA DA019537)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lohse MJ, Benovic JL, Codina J, Caron MG, Lefkowitz RJ. beta-Arrestin: a protein that regulates beta-adrenergic receptor function. Science. 1990;248:1547–1550. doi: 10.1126/science.2163110. [DOI] [PubMed] [Google Scholar]
- 2.Okada T, Le Trong I, Fox BA, Behnke CA, Stenkamp RE, Palczewski K. X-Ray diffraction analysis of three-dimensional crystals of bovine rhodopsin obtained from mixed micelles. J Struct Biol. 2000;130:73–80. doi: 10.1006/jsbi.1999.4209. [DOI] [PubMed] [Google Scholar]
- 3.Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 4.Yeagle PL, Albert AD. G-protein coupled receptor structure. Biochim Biophys Acta. 2007;1768:808–824. doi: 10.1016/j.bbamem.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Klabunde T, Hessler G. Drug design strategies for targeting G-protein-coupled receptors. Chembiochem. 2002;3:928–944. doi: 10.1002/1439-7633(20021004)3:10<928::AID-CBIC928>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 6.Leifert WR, Aloia AL, Bucco O, McMurchie EJ. GPCR-induced dissociation of G-protein subunits in early stage signal transduction. Mol Membr Biol. 2005;22:507–517. doi: 10.1080/09687860500370604. [DOI] [PubMed] [Google Scholar]
- 7.Premont RT, Gainetdinov RR. Physiological roles of G protein-coupled receptor kinases and arrestins. Annu Rev Physiol. 2007;69:511–534. doi: 10.1146/annurev.physiol.69.022405.154731. [DOI] [PubMed] [Google Scholar]
- 8.Gainetdinov RR, Premont RT, Bohn LM, Lefkowitz RJ, Caron MG. Desensitization of G protein-coupled receptors and neuronal functions. Annu Rev Neurosci. 2004;27:107–144. doi: 10.1146/annurev.neuro.27.070203.144206. [DOI] [PubMed] [Google Scholar]
- 9.Sibley DR, Peters JR, Nambi P, Caron MG, Lefkowitz RJ. Desensitization of turkey erythrocyte adenylate cyclase. Beta-adrenergic receptor phosphorylation is correlated with attenuation of adenylate cyclase activity. J Biol Chem. 1984;259:9742–9749. [PubMed] [Google Scholar]
- 10.Stadel JM, Nambi P, Shorr RG, Sawyer DF, Caron MG, Lefkowitz RJ. Phosphorylation of the beta-adrenergic receptor accompanies catecholamine-induced desensitization of turkey erythrocyte adenylate cyclase. Trans Assoc Am Physicians. 1983;96:137–145. [PubMed] [Google Scholar]
- 11.Attramadal H, et al. Beta-arrestin2, a novel member of the arrestin/beta-arrestin gene family. J Biol Chem. 1992;267:17882–17890. [PubMed] [Google Scholar]
- 12.Lefkowitz RJ, Inglese J, Koch WJ, Pitcher J, Attramadal H, Caron MG. G-protein-coupled receptors: regulatory role of receptor kinases and arrestin proteins. Cold Spring Harb Symp Quant Biol. 1992;57:127–133. doi: 10.1101/sqb.1992.057.01.016. [DOI] [PubMed] [Google Scholar]
- 13.Lohse MJ, Andexinger S, Pitcher J, Trukawinski S, Codina J, Faure JP, Caron MG, Lefkowitz RJ. Receptor-specific desensitization with purified proteins. Kinase dependence and receptor specificity of beta-arrestin and arrestin in the beta 2-adrenergic receptor and rhodopsin systems. J Biol Chem. 1992;267:8558–8564. [PubMed] [Google Scholar]
- 14.Mukherjee S, Palczewski K, Gurevich V, Benovic JL, Banga JP, Hunzicker-Dunn M. A direct role for arrestins in desensitization of the luteinizing hormone/choriogonadotropin receptor in porcine ovarian follicular membranes. Proc Natl Acad Sci U S A. 1999;96:493–498. doi: 10.1073/pnas.96.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferguson SSG, Zhang J, Barak LS, Caron MG. G-protein-coupled receptor kinases and arrestins: Regulators of G-protein-coupled receptor sequestration. Biochem Soc Trans. 1996;24:953–959. doi: 10.1042/bst0240953. [DOI] [PubMed] [Google Scholar]
- 16.Goodman OB, Jr, Krupnick JG, Santini F, Gurevich VV, Penn RB, Gagnon AW, Keen JH, Benovic JL. Beta-arrestin acts as a clathrin adaptor in endocytosis of the beta2-adrenergic receptor. Nature. 1996;383:447–450. doi: 10.1038/383447a0. [DOI] [PubMed] [Google Scholar]
- 17.Lefkowitz RJ. G protein-coupled receptors. III New roles for receptor kinases and beta-arrestins in receptor signaling and desensitization. J Biol Chem. 1998;273:18677–18680. doi: 10.1074/jbc.273.30.18677. [DOI] [PubMed] [Google Scholar]
- 18.Lin FT, Daaka Y, Lefkowitz RJ. beta-arrestins regulate mitogenic signaling and clathrin-mediated endocytosis of the insulin-like growth factor I receptor. J Biol Chem. 1998;273:31640–31643. doi: 10.1074/jbc.273.48.31640. [DOI] [PubMed] [Google Scholar]
- 19.Hsieh C, Brown S, Derleth C, Mackie K. Internalization and Recycling of the CB1 Cannabinoid Receptor. J Neurochem. 1999;73:493–501. doi: 10.1046/j.1471-4159.1999.0730493.x. [DOI] [PubMed] [Google Scholar]
- 20.Jin W, Brown S, Roche JP, Hsieh C, Celver JP, Kovoor A, Chavkin C, Mackie K. Distinct Domains of the CB1 Cannabinoid Receptor Mediate Desensitization and Internalization. J Neurosci. 1999;19:3773–3780. doi: 10.1523/JNEUROSCI.19-10-03773.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohout TA, Nicholas SL, Perry SJ, Reinhart G, Junger S, Struthers RS. Differential desensitization, receptor phosphorylation, beta-arrestin recruitment, and ERK1/2 activation by the two endogenous ligands for the CC chemokine receptor 7. J Biol Chem. 2004;279:23214–23222. doi: 10.1074/jbc.M402125200. [DOI] [PubMed] [Google Scholar]
- 22.Craft CM, Whitmore DH, Wiechmann AF. Cone arrestin identified by targeting expression of a functional family. J Biol Chem. 1994;269:4613–4619. [PubMed] [Google Scholar]
- 23.Hirsch JA, Schubert C, Gurevich VV, Sigler PB. The 2.8 A Crystal Structure of Visual Arrestin: A Model for Arrestin's Regulation. Cell. 1999;97:257–269. doi: 10.1016/s0092-8674(00)80735-7. [DOI] [PubMed] [Google Scholar]
- 24.Benovic JL, Kuhn H, Weyand I, Codina J, Caron MG, Lefkowitz RJ. Functional desensitization of the isolated beta-adrenergic receptor by the beta-adrenergic receptor kinase: potential role of an analog of the retinal protein arrestin (48-kDa protein) Proc Natl Acad Sci U S A. 1987;84:8879–8882. doi: 10.1073/pnas.84.24.8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Granzin J, Wilden U, Choe HW, Labahn J, Krafft B, Buldt G. X-ray crystal structure of arrestin from bovine rod outer segments. Nature. 1998;391:918–921. doi: 10.1038/36147. [DOI] [PubMed] [Google Scholar]
- 26.Han M, Gurevich VV, Vishnivetskiy SA, Sigler PB, Schubert C. Crystal Structure of Beta-Arrestin at 1.9 A: Possible Mechanism of Receptor Binding and Membrane Translocation. Structure. 2001;9:869–880. doi: 10.1016/s0969-2126(01)00644-x. [DOI] [PubMed] [Google Scholar]
- 27.Kisselev OG, Downs MA, McDowell JH, Hargrave PA. Conformational changes in the phosphorylated C-terminal domain of rhodopsin during rhodopsin arrestin interactions. J Biol Chem. 2004;279:51203–51207. doi: 10.1074/jbc.M407341200. [DOI] [PubMed] [Google Scholar]
- 28.Kisselev OG, McDowell JH, Hargrave PA. The arrestin-bound conformation and dynamics of the phosphorylated carboxy-terminal region of rhodopsin. FEBS Lett. 2004;564:307–311. doi: 10.1016/S0014-5793(04)00226-1. [DOI] [PubMed] [Google Scholar]
- 29.Hanson SM, Francis DJ, Vishnivetskiy SA, Kolobova EA, Hubbell WL, Klug CS, Gurevich VV. Differential interaction of spin-labeled arrestin with inactive and active phosphorhodopsin. Proc Natl Acad Sci U S A. 2006;103:4900–4905. doi: 10.1073/pnas.0600733103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pavlopoulos S, Thakur GA, Nikas SP, Makriyannis A. Cannabinoid receptors as therapeutic targets. Curr Pharm Des. 2006;12:1751–1769. doi: 10.2174/138161206776873743. [DOI] [PubMed] [Google Scholar]
- 31.Choi G, Guo J, Makriyannis A. The conformation of the cytoplasmic helix 8 of the CB1 cannabinoid receptor using NMR and circular dichroism. Biochim Biophys Acta. 2005;1668:1–9. doi: 10.1016/j.bbamem.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 32.Clore GM, Gronenborn AM. The two-dimensional transferred nuclear Overhauser effect. Journal of Magnetic Resonance. 1982;48:402–417. [Google Scholar]
- 33.Clore GM, Nilges M, Sukumaran DK, Brunger AT, Karplus M, Gronenborn AM. The three-dimensional structure of alpha1-purothionin in solution: combined use of nuclear magnetic resonance, distance geometry and restrained molecular dynamics. Embo J. 1986;5:2729–2735. doi: 10.1002/j.1460-2075.1986.tb04557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williamson MP, Havel TF, Wuthrich K. Solution conformation of proteinase inhibitor IIA from bull seminal plasma by 1H nuclear magnetic resonance and distance geometry. J Mol Biol. 1985;182:295–315. doi: 10.1016/0022-2836(85)90347-x. [DOI] [PubMed] [Google Scholar]
- 35.Clore GM, Gronenborn AM, Nilges M, Ryan CA. Three-dimensional structure of potato carboxypeptidase inhibitor in solution. A study using nuclear magnetic resonance, distance geometry, and restrained molecular dynamics. Biochemistry. 1987;26:8012–8023. doi: 10.1021/bi00398a069. [DOI] [PubMed] [Google Scholar]
- 36.Wagner G, Braun W, Havel TF, Schaumann T, Go N, Wuthrich K. Protein structures in solution by nuclear magnetic resonance and distance geometry. The polypeptide fold of the basic pancreatic trypsin inhibitor determined using two different algorithms, DISGEO and DISMAN. J Mol Biol. 1987;196:611–639. doi: 10.1016/0022-2836(87)90037-4. [DOI] [PubMed] [Google Scholar]
- 37.Wuthrich K. In: NMR of proteins and Nucleic Acids. Wiley J, editor. New York: 1986. [Google Scholar]
- 38.Chou YT, Gierasch LM. The conformation of a signal peptide bound by Escherichia coli preprotein translocase SecA. J Biol Chem. 2005;280:32753–32760. doi: 10.1074/jbc.M507532200. [DOI] [PubMed] [Google Scholar]
- 39.Blaber M, Zhang XJ, Matthews BW. Structural basis of amino acid alpha helix propensity. Science. 1993;260:1637–1640. doi: 10.1126/science.8503008. [DOI] [PubMed] [Google Scholar]
- 40.Yeagle PL, Alderfer JL, Albert AD. Structure of the Carboxy-Terminal Domain of Bovine Rhodopsin. Nat Struct Biol. 1995;2:832–834. doi: 10.1038/nsb1095-832. [DOI] [PubMed] [Google Scholar]
- 41.Palczewski K, Rispoli G, Detwiler PB. The influence of arrestin (48K protein) and rhodopsin kinase on visual transduction. Neuron. 1992;8:117–126. doi: 10.1016/0896-6273(92)90113-r. [DOI] [PubMed] [Google Scholar]
- 42.Zhang L, Sports CD, Osawa S, Weiss ER. Rhodopsin phosphorylation sites and their role in arrestin binding. J Biol Chem. 1997;272:14762–14768. doi: 10.1074/jbc.272.23.14762. [DOI] [PubMed] [Google Scholar]
- 43.Szilak L, Moitra J, Krylov D, Vinson C. Phosphorylation destabilizes alpha-helices. Nat Struct Biol. 1997;4:112–114. doi: 10.1038/nsb0297-112. [DOI] [PubMed] [Google Scholar]