Abstract
We examined the effects of spinal cord injury (SCI) on alterations in gene expression and respective protein products in human skeletal muscle 2 days and 5 days post-SCI. Biopsies were taken from skeletal muscle of 9 men and 1 woman (n = 10) (43.9 ± 6.7 years) 2 days and 5 days post-SCI and from 5 healthy young men who served as controls (20.4 ± 0.5 years). Global changes in gene expression were analysed using Affymetrix GeneChips on a subsample of subjects (n = 3). Candidate genes were then pursued via qRT-PCR. Western blotting (WB) was used to quantify protein products of candidate genes. Immunohistochemistry (IHC) was used to localize proteins. Groups of transcripts showing the greatest percentage of altered expression, the most robust fold-changes, and indicative of involvement of an entire pathway using the GeneChip included genes involved in the ubiquitin proteasome pathway (UPP), metallothionein function, and protease inhibition. qRT-PCR analysis confirmed increases in gene expression for UPP components (UBE3C, Atrogin-1, MURF1, and PSMD11), the metallothioneins (MT1A, MT1F, MT1H), and the protease inhibitor, SLPI (P < 0.05) at 2 days and 5 days post-SCI. Protein levels of the proteasome subunit (PSMD11) and the metallothioneins were increased 5 days post-SCI. Protein levels of UBE3C, Atrogin-1, MURF1 and SLPI were unchanged (P > 0.05). IHC showed increased staining for PSMD11 and the metallothioneins 5 days post-SCI, along the peripheral region of the cells. IHC also showed altered staining for Atrogin-1 at 5 days post-SCI along the membrane region. Thus, there was a profound increase in gene expression of UPP components, the metallothioneins, and the protease inhibitor, SLPI, within 5 days of SCI. Increased protein levels for PSMD11 and the metallothioneins 5 days post-SCI, specifically along the cell periphery, indicate that proteins in this region may be early targets for degradation post-SCI.
Skeletal muscle loss following spinal cord injury (SCI) is rapid, with losses in muscle cross-sectional area reaching approximately 25% within the first six weeks of injury (Dudley et al. 1999). This severe muscle atrophy is attributed to a decline in muscle protein synthesis and an increase in muscle protein breakdown (Goldspink, 1976; Booth & Gollnick, 1983; Hornberger et al. 2001; Batt et al. 2006). Denervation models in animals have shown that muscle protein loss occurs within the first 24 h after nerve transection (Dupont-Versteegden et al. 1998; Raffaello et al. 2006). However, how protein loss is controlled in skeletal muscle, including triggers and signalling mechanisms at the molecular level that regulate the early stages of the atrophy process, is largely unknown, especially in humans. Analysis of alterations in gene expression and respective protein products that occur within the first few days of SCI in humans is important for developing targeted interventions to attenuate the onset of muscle atrophy and prevent the profound losses in muscle mass.
Previous work in animal models has shown that denervation produces an upregulation of genes controlling the ubiquitin proteasome pathway, a multistep process that functions in skeletal muscle to tag and degrade proteins for destruction (Tang et al. 2000; Batt et al. 2006; Raffaello et al. 2006). A pioneering study using the high-throughput approach to gene expression profiling in response to denervation, hindlimb suspension and immobilization in a rat model established that all three atrophy stimuli resulted in an increase in gene expression of two key ligases involved in the ubiquitin proteasome pathway, muscle atrophy F-box (MAFbx/Atrogin-1) and muscle specific RING finger (MURF1) (Bodine et al. 2001). This work highlighted the importance of these components of the ubiquitin proteasome pathway in the atrophy process, leading to a number of subsequent investigations that focused on the role of this pathway in skeletal muscle atrophy (Jagoe et al. 2002; Jones et al. 2004; Lecker et al. 2004; Whitman et al. 2005; Urso et al. 2006). However, little work has been done to explore these or other molecular events involved in the early stages of the muscle atrophy programme in humans, specifically in the first days following SCI.
In this study, we sought to extend previous work in animal models of denervation to a human SCI model, to identify genes and proteins involved in the early stages of muscle atrophy. Only a small number of studies have used human models to explore the molecular events associated with muscle atrophy, thus it is not clear whether results from animal models can be extended to humans (Chen et al. 2003). As a consequence, the development of effective countermeasures to attenuate the atrophy processes is delayed. Therefore, we followed the expression of several genes and proteins thought to be involved in the muscle atrophy programme at two and five days post-SCI in humans. We hypothesized that after two days post-SCI, we would document an increase in gene expression of essential components of the ubiquitin proteasome pathway, and that this increase would be greater at five days post-SCI, resulting in an increase in protein products at this time point. Although our focus was on genes involved in the ubiquitin proteasome pathway, we used the microarray analysis as a screening tool to identify novel pathways affected in humans in the first days following SCI. The results of this work provided us with a snapshot of changes in gene expression and relative protein products in the early stages of SCI in humans.
Methods
Subjects
Nine male and one female SCI patients (mean ±s.e.m.) (43.9 ± 6.7 years, 174.4 ± 2.1 cm, 83.0 ± 10.1 kg) were recruited from Baystate Medical Center (Springfield, MA, USA). Six patients were quadriplegics and four patients were paraplegics. All patients gave oral consent to their ward, who signed an informed consent document that was approved by the University of Massachusetts and the Baystate Medical Center Institutional Review Boards. All patients had a clinical workup to determine if they met inclusion criteria. Inclusion criteria were as follows: patients above the age of 18 years, absence of severe brain injury (Glasgow Coma Scale > 13), absence of muscle-crush injury or compartment syndrome, absence of all of the following conditions: hypoxic injury, systemic sepsis, systemic inflammatory or autoimmune disease, and malignancy. Muscle biopsies were obtained from the vastus lateralis muscles of the SCI patients at two and five days post-SCI. Muscle biopsies were also taken from the biceps muscles of the paraplegic patients at two days post-SCI (n = 4). Biopsies were taken from contralateral limbs so that the healing process from the biopsy taken two days post-SCI would not skew the results for the biopsy taken five days post-SCI. Control muscle biopsies were obtained from the vastus lateralis of five (n = 5) healthy volunteers (20.5 ± 0.4 years, 173.2 ± 2.2 cm, 77.7 ± 1.5 kg).
Muscle biopsy
Percutaneous muscle biopsies were obtained using a Bergstrom 5 mm biopsy needle (Depuy, Warsaw, IN, USA). Skin was first lightly anaesthetized with 4 ml of 2% lidocaine hydrochloride solution, a small (5–6 mm) incision was made in the skin and fascia, the biopsy needle was inserted ∼3–4 cm below the skin, and ∼200 mg of tissue was removed. Tissue was divided into at least two 50 mg aliquots, snap frozen in liquid nitrogen, and stored at −80°C until analysis. Because we had limited muscle tissue from several SCI patients, some analyses were only performed on a subset of muscle biopsy specimens from patients, and in some cases statistical analysis was not possible (e.g. protein analysis of biceps samples).
Gene expression profiling
For a subsample of three subjects, microarray analysis was performed using Affymetrix GeneChip® technology (Human Genome U133 Plus 2.0 Array; Affymetrix, Santa Clara, CA) with standard operating procedures and quality control as recently described (Chen et al. 2000, 2003; Urso et al. 2006).
Absolute analysis of Affymetrix ‘raw’ data was conducted using Affymetrix MAS5.0 and DNA-Chip Analyser software (dCHIP) (Li & Hung Wong, 2001; Schadt et al. 2001; Hoffman et al. 2004). Values of intensity differences as well as ratios of each probe pair were used to determine (statistically) whether a gene was called present or absent. Data analysis required >10% of profiles to show a present call to be carried into the next statistical analysis. GeneSpring 7.0 (Silicon Genetics, Redwood City CA, USA) was then used to filter potential candidate genes for statistical significance using Welch's t test, and only genes with P values ≤0.01 were retained for further analysis. Combined, these criteria increased the reliability of our microarray analysis allowing us to reduce false positives (Schadt et al. 2001; Hoffman et al. 2004).
Candidate genes were selected only if they fulfilled four criteria: (1) They met the Affymetrix criteria described above for each subject (two and five days-post SCI versus healthy controls); (2) they showed a mean fold-change >1.5 or <−1.5; (3) they were chronically up- or downregulated at both two and five days post-SCI; and (4) a Welch's t test detected a significant (P≤ 0.01) increase or decrease in average fold-change for each subject in the control versus two and five days-post SCI comparison. Genes that met all four criteria were then ranked according to P value and fold-change, respectively. Groups of transcripts showing the greatest percentage of altered expression, the most robust fold-changes, and indicative of involvement of an entire pathway using the GeneChip at two and five days post-SCI were selected for further analysis. This approach allowed us to narrow our focus to the most robust and probably most physiologically relevant changes in response to SCI. Additionally, since our sample size was small, incorporating these stringent inclusion criteria increased the reliability of our results. The genes that were chosen were those involved in the ubiquitin proteasome pathway (Proteasome Subunit 11 (PSMD11), Ubiquitin E3 Ligase C (UBE3C), Muscle-specific Ring Finger (MURF1) and Atrogin-1 (MAFbx/FBXO32)); metallothionein function (metallothionein 1A (MT1A), metallothionein 1F (MT1F) and metallothionein 1H (MT1H)); and protease inhibition (Secretory Leukocyte Protease Inhibitor (SLPI)). Statistical analysis was performed on log transformed data to reduce inequalities of variance(Chen et al. 2000; Hoffman et al. 2004; Schadt et al. 2001).
Quantitative real-time polymerase chain reaction (qRT-PCR)
We had sufficient sample to isolate RNA from nine patients at two days post-SCI and seven patients five days post-SCI. Total RNA was isolated and purified according to manufacturer's instructions using TRIZOL® reagent (Invitrogen) and measured on a Nanodrop ND-1000 spectrophotometer (Nanodrop, Wilmington DE, USA). Equal amounts of total RNA were synthesized into cDNA using a First-Strand cDNA Synthesis kit (Fermentas, Hanover, MD, USA). ABgene Absolute qPCR SYBR Green Master Mix (ABgene, Surrey, UK) with ROX dye was used for all PCR protocols. Forward and Reverse RT-PCR primers (Integrated DNA Technologies, Coralville, IA, USA) were designed using Affymetrix® and NCBI gene sequences with the Primer Express program v 2.0 (Applied Biosystems, CA, USA) for genes of interest (Table 1).
Table 1.
Primer sequences for qRT-PCR
| Gene symbol | Forward primer | Reverse primer |
|---|---|---|
| Atrogin-1/MAFbx/FBXO32 | 5′-TCA CAG CTC ACA TCC CTG AG-3′ | 5′-AGA CTT GCC GAC TCT TTG GA-3′ |
| MURF1 | 5′-TGA GCC AGA AGT TTG ACA CG-3′ | 5′-TGA TGA GTT GCT TGG CAG TC-3′ |
| UBE3C | 5′-TGA CCA GTG GTG GGA GTG TA-3′ | 5′-ATC AGC TGT CCG ATC CAT TC-3′ |
| PSMD11 | 5′-ACA CCC CAG AAG ATG TCC AG-3′ | 5′-TGC CAG TGA TCT GTT CTT GC-3′ |
| MT1A | 5′-GCA AAT GCA AAG AGT GCA AA-3′ | 5′-ATG GGT CAG GGT TGT ATG GA-3′ |
| MT1F | 5′-GCA AGT GCA AAG AGT GCA AA-3′ | 5′-AGA GCT GTT CCC ACA TCA GG-3′ |
| MT1H | 5′-GCT CCT GCA AGT GCA AAA AG-3′ | 5′-CAG CAG CTG CAC TTC TCT GA-3′ |
| SLPI | 5′-GTG ACT TGA AGT GTT GCA TGG GCA-3′ | 5′-AAC TGG CAC TTC TTG AAA GCC TGC-3′ |
| GAPDH | 5′-CAT TGC CCT CAA CGA CCA CTT TGT-3′ | 5′-TCT CTC TCT TCC TCT TGT GCT CTT GC-3′ |
qRT-PCR reactions were run in triplicate, in 96-well plates with cDNA samples from five healthy controls, and SCI patients at two (n = 9) and five (n = 7) days post-SCI for each gene of interest, such that one gene of interest and one reference standard were analysed per plate according to methods previously described (Urso et al. 2006). At the end of each reaction, a melting curve analysis was performed to identify possible primer dimers. Appropriate product size was determined via 2% agarose gel electrophoresis with ethidium bromide staining. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Integrated DNA Technologies) was used as a reference standard to detect the expression level of genes of interest. GAPDH is a constitutively expressed housekeeping gene that has been validated in previous studies in humans (Mahoney et al. 2004; Urso et al. 2006) and was not found to be differentially expressed in our preliminary analysis of gene expression using the microarray.
Western blotting
Muscle samples (∼50 mg) used for Western blot analysis were homogenized in buffer (100 mm Tris, pH 7.4/250 mm sucrose/protease inhibitor mixture (1 mm sodium pyrophosphate/1 mm sodium orthovanadate/10 μg leupeptin ml−1/10 μg aprotinin ml−1/1 μm microcystin-LR/1 mm phenylmethylsufonyl fluoride/10 mm sodium fluoride)) and analysed for total protein concentration (Lowry method) using commercially available reagents (Sigma, St Louis, MO, USA). Equal amounts of protein sample (30 μg, lane) were subjected to SDS-PAGE (sodium dodecyl sulphate-polyacrylamide gel electrophoresis) using 7–15% gradient gels (Bio-Rad Laboratories, Hercules, CA, USA). Precision Plus Kaleidoscope Protein Standards (Bio-Rad Laboratories) were used as molecular weight markers on each gel. Proteins were then electrophoretically transferred to Polyvinylidene Difluoride membranes (GE Healthcare, Piscataway, NJ, USA), which were then incubated for 60min at room temperature in a solution of TBS containing 5% non-fat dry milk (Carnation, Nestle, Solon, OH, USA) and 0.1% Tween-20 (Bio-Rad Laboratories).
Membranes were incubated overnight at 4°C in primary antibodies against PSMD11 (1: 1000, rabbit polyclonal, EMD Biosciences, San Diego, CA, USA), UBE3C (1: 1000, mouse polyclonal, AbNova, Taipei City, Taiwan), MURF1 (1: 1000, goat polyclonal, Abcam, Cambridge, MA, USA), Atrogin-1 (1: 1000, rabbit polyclonal), MT-I + II (1: 500, mouse monoclonal, against metallothionein isoforms I + II, M. Penkowa, University of Copenhagen, Denmark), SLPI (1: 500 goat polyclonal, R&D Systems, Minneapolis, MN), GAPDH (1: 2500, rabbit polyclonal, Abcam), in a solution of TBS/0.1% Tween-20 containing 5–7% bovine serum albumin (BSA) (Sigma). Primary antibodies to Atrogin-1 were generated by using a synthetic 23-amino acid peptide having a sequence derived from the predicted carboxyl-terminal region of the protein. Rabbits were immunized with peptide coupled to keyhole limpet haemocyanin (Sevetson & Lawrence, 1993). Primary antibodies were affinity-purified before use by means of a column prepared with peptide coupled to Sulfolink resin (Pierce Biotechnology, Rockford, IL, USA). The specificity of the Atrogin-1 antibody was established by performing immunoblotting experiments with preimmune serum, which resulted in no signal. In addition, competition experiments were performed whereby the affinity-purified antibody was incubated with the immunizing peptide for 2 h at room temperature. After hybridizing the blots with this peptide–antibody complex, the signal observed with the antiAtrogin-1 antibody was absent.
Membranes were then incubated in horseradish peroxidase-conjugated secondary antibodies (Bio-Rad Laboratories) for 1 h at room temperature and washed five times in 0.1% Tween-20 in TBS before antibody binding was detected by using enhanced chemiluminescence (ECL) kits (GE Healthcare, Piscataway, NJ, USA). The intensity of bands was quantified using a Fotodyne Image analysis System (Fotodyne Inc., Hartland, WI, USA) and TotalLab software (Nonlinear Dynamics, Durham, NC, USA).
Immunohistochemistry (IHC)
Serial cross-sectional slices (10 μm) of muscle biopsy samples immersed in OCT embedding medium (Fisher Scientific, Pittsburgh, PA, USA) were generated on a MICROM HM 505E Cryostat at −25°C (Richard Allan Scientific, Kalamazoo, MI, USA) and placed on glass slides coated with Vectabound reagent (Vector Laboratories, Burlingame, CA, USA). All samples from healthy controls and patients two days post-SCI and five days post-SCI were processed at the same time. Sections were washed in phosphate-buffered serum (PBS), pH 7.4 (Sigma), blocked with goat normal serum (Jackson ImmunoResearch Laboratories Inc., West Grove, PA, USA) or rabbit normal serum (Zymed Laboratories, San Francisco, CA, USA) and stained overnight at 4°C with primary antibodies against Dystrophin (1: 250, mouse monoclonal, Abcam) and those used for Western blotting (see above), diluted 1: 250–1: 500 in 5% BSA in PBS. Sections were washed in PBS and were incubated for 30 min at 37°C with secondary fluorochrome-labelled antibodies against primary antibodies (Alexa-green, Alexa-red, goat anti-mouse, goat anti-rabbit, rabbit anti-goat) (Molecular Probes, Eugene, OR, USA). Fluorochrome-labelled secondary antibodies, without primary antibodies, were used as controls. After washing with PBS, ∼10 μl of 3,3′-diaminobenzidine (Sigma) and a coverslip were added to each section.
Samples were visualized and photographed using a Nikon Eclipse E600 Phase Contrast Microscope (Nikon Corporation, Tokyo, Japan) equipped with a Spot insight QE camera and EclipseNET software, version 1.16. Scion IMAGE for Windows (Scion Corporation, Frederick, MD, USA) was used for image analysis. Exposure time and image intensity were maintained between conditions.
Results
Subjects
No statistical differences existed in height or weight measurements between the healthy controls and the SCI patients. SCI patients were significantly older than the healthy controls (P < 0.03). Thus, due to the considerable age range among the SCI patients, prior to grouping our data we evaluated the results of the microarray and qRT-PCR on an individual basis and determined that outliers were not restricted to a particular age group.
Microarray analysis
As a preliminary analysis, we obtained a global snapshot of which genes were consistently up- or downregulated two and five days post-SCI, from a subset of three patients. Of the 12 000 genes that met inclusion criteria of a fold-change ≥ 1.5 or ≤−1.5, and a P value ≤ 0.01, approximately 50 genes were found to be consistently up- or downregulated. Of these 50 genes, only 25 had known biological functions as determined through the Affymetrix NetAffyx Gene Ontology (GO) database. Figure 1 presents a schematic of gene categories that were differentially expressed according to criteria outlined in the methods. Groups of transcripts showing the greatest percentage of altered expression, the most robust fold-changes, and indicative of involvement of an entire pathway using the GeneChip at two and five days post-SCI included genes involved in the ubiquitin proteasome pathway, including the protease inhibitor, SLPI and metallothionein function. Our next step was to perform additional work at the mRNA (qRT-PCR) and protein (Western blotting and IHC) level on a subset of genes that have previously been identified and associated with muscle atrophy, and for which primary antibodies to the gene product were available.
Figure 1.
Global changes in gene expression at two and five days post-SCI: microarray analysis Candidate genes grouped according to biological function based on GO criteria (https://www.affymetrix.com/analysis/netaffx/go_analysis_netaffx4.affx). Genes were considered candidate genes if they met the following criteria: (chronically up- or downregulated at two and five days post-SCI, P < 0.01, fold-change > 1.5).
qRT-PCR
Ubiquitin proteasome pathway
Upregulation of the ubiquitin proteasome pathway augments proteolysis and muscle protein breakdown (Taillandier et al. 2004). Thus, the latest experiments have largely focused on the expression of two ubiquitin genes that are specific to skeletal muscle, muscle RING finger 1 (MURF1) and muscle atrophy F-box (MAFbx/Atrogin-1) (Mitch et al. 1999; Bodine et al. 2001; Gomes et al. 2001). In the present experiment, qRT-PCR analysis revealed that Atrogin-1 gene expression was increased two and five days post-SCI (P < 0.05, Table 2). MURF1 is not included on the Human U133 GeneChip, so we used targeted gene expression analysis (custom primers (Table 1)) and show that MURF1 gene expression was increased two and five days post-SCI (P < 0.05, Table 2). UBE3C, an E3 ligase which renders specificity to the ubiquitin proteasome system by tagging and degrading proteins for destruction, was also found to be upregulated via qRT-PCR analysis (P < 0.05, Table 2). However, gene expression of UBE3C was also found to be upregulated in qRT-PCR analysis of biceps samples taken two days post-SCI, suggesting that this alteration was not directly related to the SCI/denervation. The fourth component of the ubiquitin proteasome pathway chosen for further analysis from the microarray results was PSMD11. PSMD11 is a component of the 26S proteasome, which is a catalytic proteinase complex responsible for the cleavage of peptides in an ATP/ubiquitin-dependent process. qRT-PCR showed an increase in gene expression at two and five days post-SCI (P < 0.05, Table 2).
Table 2.
qRT-PCR analysis of gene expression two and five days post-SCI
| Gene name | Gene symbol/Affymetrix ID | Fold-change qRT-PCR day 2 post-SCI vastus | Fold-change qRT-PCR day 2 post-SCI biceps (paraplegics) | Fold-change qRT-PCR day 2 post-SCI vastus |
|---|---|---|---|---|
| Ubiquitin proteasome pathway | ||||
| Atrogin-1 | MAFbx/FBXO32 | 11.4 ± 2.0 | 3.6 ± 2.3 n.s | 12.9 ± 3.9 |
| 241763_at | ||||
| Muscle-specific ring finger | MURF1 | 6.0 ± 1.4 | 3.8 ± 1.3 n.s | 5.6 ± 0.6 |
| (not on U133 GeneChip) | ||||
| Ubiquitin E3 ligase C | UBE3C | 2.4 ± 0.2 | 2.1 ± 0.7 | 2.5 ± 0.4 |
| 201817_at | ||||
| Proteasome subunit 11 | PSMD11 | 2.7 ± 0.3 | 1.8 ± 1.7 n.s | 3.2 ± 0.7 |
| 208776_at | ||||
| Metallothioneins | ||||
| Metallothionein 1A | MT1A | 3.9 ± 1.7 | 2.1 ± 0.8 n.s | 5.5 ± 0.7 |
| 216336_at | ||||
| Metallothionein 1F | MT1F | 20.7 ± 1.4 | 10.9 ± 20.1 n.s | 9.9 ± 9.4 |
| 217165_at | ||||
| Metallothionein 1H | MT1H | 6.4 ± 2.7 | 5.6 ± 2.9 | 9.3 ± 2.1 |
| 206461_at | ||||
| Protease inhibitor | ||||
| Secretory leukocyte | SLPI | 21.8 ± 11.7 | 11.5 ± 25.6 n.s | 35.5 ± 30.3 |
| protease inhibitor | 201323_at | |||
Values are means ±s.e.m. All values are significant (P < 0.01), unless otherwise noted (n.s. = non-significant).
Metallothioneins
Previous work in muscle atrophy models has shown increased expression of the metallothioneins (Kondo et al. 1992; Lecker et al. 2004; Urso et al. 2006). Several genes involved in metallothionein function (MT2A, MT1A, MT1E, MT1F, MT1G, MT1H, MT1M, MT1R, and MT1X) were upregulated in the microarray dataset from the subsample of SCI patients. We chose to confirm the expression of three metallothionein isoforms using qRT-PCR. qRT-PCR analysis showed increases in gene expression for metallothioneins 1A, 1F and 1H two and five days post-SCI in the vastus lateralis (P < 0.05, Table 2). However, MT1H gene expression was also found to be significantly upregulated in biceps muscles of SCI patients, suggesting that this increase is not entirely due to denervation.
Protease inhibitor
Secretory leucocyte protease inhibitors (SLPI) inhibit proteolysis in skeletal muscle by binding to proteases and rendering them inactive. SLPI attenuates proteolysis and macrophage infiltration when muscle is being degraded. qRT-PCR analysis revealed a 21.8-fold and 35.5-fold increase in gene expression at two and five days post-SCI, respectively (P < 0.05, Table 2).
qRT-PCR on muscle biopsies from the biceps two days post-SCI, on all genes except UBE3C and MT1H, revealed that gene expression was always higher for the vastus lateralis compared with the biceps, indicating that the SCI itself induced a greater response in the paralysed muscle than did other factors affecting the entire body, including medications, nutritional status and bedrest.
Western blotting
Ubiquitin proteasome pathway
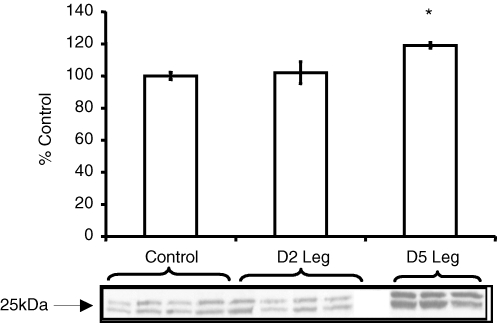
We performed Western blotting on protein products of candidate genes selected from the preliminary microarray analysis involved in the ubiquitin proteasome pathway. Interestingly, three of the four components involved in the ubiquitin proteasome pathway, Atrogin-1, MURF1, and UBE3C, are members of a group of enzymes known as E3 ligases. These proteins function in the pathway to tag proteins for destruction prior to inserting them into the proteolytic core, the proteasome. The fourth protein, PSMD11 encodes proteins that comprise the proteolytic core. Despite increases in gene expression for these components of the ubiquitin proteasome pathway, Western blotting showed that only one protein product from our candidate gene list involved in the ubiquitin proteasome pathway increased following SCI. PSMD11 protein levels increased by 20% five days post-SCI as compared to healthy controls (P < 0.001) and two days post-SCI (P < 0.05, Fig. 2). Protein levels for Atrogin-1, MURF1, and UBE3C were not different compared to control at two or five days post-SCI (P > 0.05, Fig. 3A–C).
Figure 2.
Western blotting for PSMD11 Protein levels of PSMD11 increased ∼20% five days post-SCI compared to healthy controls (P < 0.001) and compared to two days post-SCI (P < 0.05). A representative blot (4 controls, 4 SCI patients day 2 (D2 Leg), and 3 SCI patients Day 5 (D5 Leg)) is below the graph and the control value has been assigned the value 100%. Values represent means ± standard error (s.e.m.). *P < 0.05 versus control and two days post-SCI. Arrow indicates approximate molecular weight (kDa) of the band of interest, according to protein standard.
Figure 3.
Western blotting for E3 ligases A–C, Western blotting results for the E3 ligases (A: Atrogin-1; B: MURF1; C: UBE3C). Protein levels for Atrogin-1, MURF1, and UBE3C were not different at two (D2 Leg) or five (D5 Leg) days post-SCI compared to healthy controls (P > 0.05). Representative blots are below each graph and in each case, the control value has been assigned the value 100%. Values represent means ±s.e.m.
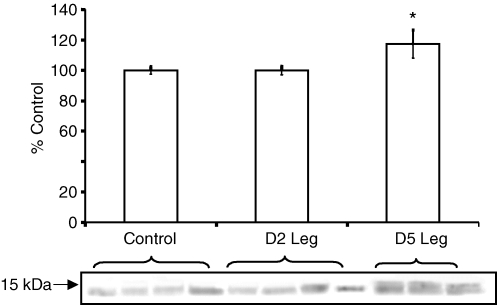
Metallothioneins
Western blotting for metallothionein I and II showed that protein levels were not changed two days post-SCI, but by five days post-SCI, protein levels increased compared to healthy controls (∼17%, P < 0.05) and compared to two days post-SCI (∼18%, P < 0.05, Fig. 4).
Figure 4.
Western blotting for metallothionein I and II Metallothionein protein levels were not changed two days post-SCI (D2 Leg), but by five days post-SCI (D5 Leg), protein levels increased ∼17% compared to healthy controls (P < 0.05) and ∼18% five days post-SCI compared to two days post-SCI (P < 0.05). A representative blot is below the graph and the control value has been assigned the value 100%. Values represent means ±s.e.m.*P < 0.05 versus control and two days post-SCI.
Protease inhibitor
Although there was a significant increase in gene expression for SLPI at two and five days post-SCI, we were unable to detect an increase in this protein via Western blotting between healthy controls and biopsies taken two and/or five days post-SCI (Fig. 5).
Figure 5.
Western blotting for SLPI Protein levels for SLPI were not different at two (D2 Leg) or five (D5 Leg) days post-SCI compared to healthy controls (P > 0.05). A representative blot is below the graph and the control value has been assigned the value 100%. Values represent means ±s.e.m.
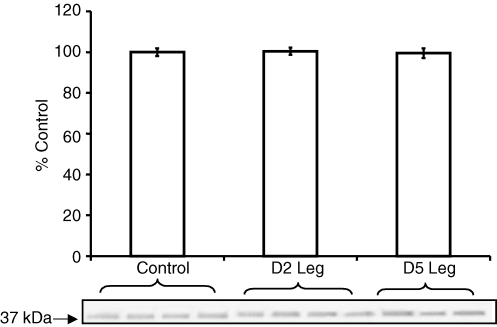
Loading control
In addition to Western blotting for each protein of interest, membranes were also incubated in a rabbit polyclonal primary antibody against GAPDH. Results of the Western blotting analysis revealed that protein levels for GAPDH were the same in muscle samples from healthy controls and SCI patients (Fig. 6).
Figure 6.
Western blotting for GAPDH GAPDH was used as a loading control each time a protein was analysed using Western blotting. To do this, membranes were stripped and reprobed for antibodies specific for GAPDH. Representative blot illustrates that equal amounts of protein were loaded per lane, and that protein levels for GAPDH were not affected by SCI. The control value has been assigned the value 100%. Values represent means ±s.e.m.
Immunohistochemistry (IHC)
Although our original rationale was to use IHC analysis to confirm alterations in protein levels observed via Western blotting, we only documented alterations in protein levels of metallothionein and PSMD11 via Western blotting analysis. Therefore, in addition to metallothionein and PSMD11, we decided to perform IHC on a subset of protein products to identify if the location of protein products of genes of interest within the myofibril changed as a result of the SCI, offering insight to their mechanism of action in response to SCI.
Ubiquitin proteasome pathway
Western blotting analysis for PSMD11 revealed a 20% increase five days post-SCI. IHC analysis not only confirmed this increase via increased staining for PSMD11 (Fig. 7A–C), staining for PSMD11 appears to be localized to the peripheral region of the cell at five days post-SCI (Fig. 7C).
Figure 7.
Immunohistochemistry for PSMD11 A (control), B (two days post-SCI) and C (five days post-SCI) are representative images of muscle biopsies stained for nuclei (blue), dystrophin (green) and PSMD11 (red). The last panel in each row (A–C) is the merged image for nuclei, dystrophin and PSMD11. White arrows indicate the most intense region of staining for PSMD11. Scale bar equals 50 μm.
We also performed IHC on Atrogin-1, since this gene had the highest level of expression among the E3 ligases in the microarray and qRT-PCR analysis. IHC images stained for Atrogin-1 in muscle samples from five days post-SCI appeared to show increased staining in the peripheral region of the cell. Figure 8A–C depicts staining for Atrogin-1 and dystrophin in muscle samples from control (Fig. 8A), two days post-SCI (Fig. 8B), and five days post-SCI (Fig. 8C). Control samples show strong staining for dystrophin, with little staining for Atrogin. By two days post-SCI, however, there is increased staining for Atrogin-1 within the fibre, and by five days post-SCI, this staining for Atrogin appears to move to the peripheral region of the cell. Moreover, in the unmerged images, white arrows indicate instances where staining for Atrogin-1 and dystrophin are identical. These data indicate that the membrane region is one of the first targets of proteolytic activity through tagging via the E3 ligase, Atrogin-1. Additionally, considering Atrogin-1 functions to tag proteins and insert them into the proteolytic core, our finding that staining for PSMD11, a component of the proteolytic core, also increases in a similar temporal and spatial fashion, is noteworthy.
Figure 8.
Immunohistochemistry for Atrogin-1 A (control), B (two days post-SCI) and C (five days post-SCI) are representative images of muscle biopsies stained for nuclei (blue), dystrophin (green) and Atrogin-1 (red). The last panel in each row (A–C) is the merged image for nuclei, dystrophin and Atrogin-1. White arrows indicate the most intense region of staining for Atrogin-1. Scale bar equals 50 μm.
Metallothioneins
In agreement with Western blotting analysis for metallothionein proteins showing an increase in metallothionein protein levels at five days post-SCI, IHC revealed increased staining for metallothionein in muscle samples taken five days post-SCI compared to those taken from healthy controls and two days post-SCI (Fig. 9). Similar to our IHC results for PSMD11 and Atrogin-1, metallothionein staining increased along the periphery of the cell when comparing images from muscle biopsies taken from healthy controls to those taken two days post-SCI and to those taken five days post-SCI.
Figure 9.
Immunohistochemistry for metallothionein I and II Merged representative images of muscle biopsies (control, two days post-SCI, and five days post-SCI, respectively) stained for nuclei (blue) and metallothionein (green). White arrows indicate the most intense region of staining for metallothionein. Scale bar equals 50 μm.
Protease inhibitor
Although we were unable to document differences in protein levels of SLPI between healthy controls and patients two and five days post-SCI using Western blotting, IHC analysis suggests that at five days post-SCI, staining for SLPI increases along the peripheral region of the cell (Fig. 10C) compared to two-days post-SCI (Fig. 10B) and healthy controls (Fig. 10A), where staining for SLPI is almost undetectable.
Figure 10.
Immunohistochemistry for SLPI A (control), B (two days post-SCI) and C (five days post-SCI) are representative images of muscle biopsies stained for nuclei (blue), dystrophin (green) and SLPI (red). The last panel in each row (A–C) is the merged image for nuclei, dystrophin and SLPI. White arrows indicate the most intense region of staining for SLPI at five days post-SCI. Scale bar equals 50 μm.
Discussion
The overall aim of the present investigation was to identify genes and proteins involved in the early stages of the muscle atrophy programme following SCI. We used microarray analysis, qRT-PCR, Western blotting and IHC to pursue several genes and proteins thought to play a significant role in the regulation of muscle protein loss at two and five days post-SCI in humans. Consistent with our hypothesis, analysis of muscle biopsies two and five days post-SCI compared to healthy controls revealed: (1) an increase in transcripts (by qRT-PCR) that encode essential components of the ubiquitin proteasome pathway (Atrogin-1, UBE3C and PSMD11), and this increase was greater five days post-SCI; (2) an increase in PSMD11 protein at five days post-SCI; and (3) an increase in staining (via IHC) for PSMD11 five days post-SCI, and this increase was localized to the peripheral region of the cell. We also hypothesized that there would be an increase in protein products of Atrogin-1, MURF1 and UBE3C by five days post-SCI, due to the progressive increase in mRNA for these components from two to five days post-SCI. However, we did not document a change in protein products for Atrogin-1, MURF1 or UBE3C at two or five days post-SCI, possibly suggesting that translation of protein products of the E3 ligases may follow a slower time course than transcriptional activation. Additionally, skeletal muscle protein turnover is affected by the SCI, thus synthesis and degradation of proteins is altered.
The preliminary microarray analysis was used to characterize the expression of novel genes thought to be involved in signalling pathways that regulate the loss of muscle protein, or increase their activity in response to the atrophic stimulus, in the first days following SCI. The microarray analysis revealed that at two and five days post-SCI, there was a dramatic increase in the expression of several transcripts involved in metallothionein function (MT2A, MT1A, MT1E, MT1F, MT1G, MT1H, MT1M, MT1R, and MT1X) and protease inhibition (SLPI). We were able to confirm increases in gene expression two and five days post-SCI using qRT-PCR analysis on a subset of the metallothioneins (MT1A, MT1F, and MT1H) and SLPI. We also identified increases in metallothionein protein levels five days post-SCI using Western blotting and IHC, and an increase in staining for SLPI proteins along the periphery of the myofibre at five days post-SCI using IHC. Thus, through the combined use of microarray technology, qRT-PCR, Western blotting and IHC, we were able to identify fundamental increases in gene expression and in some cases, respective protein products, in a class of genes and their protein products not previously shown to increase during the early stages of muscle atrophy. We discuss these alterations and implications below. When interpreting our data, however, it is critical that the reader considers limitations in the study design, including age ranges of the different groups, and the influence of different muscle types (e.g. biceps versus quadriceps) on alterations in skeletal muscle gene and protein levels.
Ubiquitin proteasome pathway
The role of the ubiquitin proteasome pathway in skeletal muscle is to tag and degrade proteins for destruction (Gomes et al. 2001; Taillandier et al. 2004). Four molecules are essential to the functioning of the ubiquitin-proteasome pathway: the ubiquitin-activating enzyme (E1), which activates the ubiquitin molecule; the ubiquitin-conjugating enzyme (E2), a ubiquitin carrier protein; and the ubiquitin ligase (E3), which catalyses transfer of ubiquitin to a protein substrate, marking the substrate for degradation by the fourth component, the 26S proteasome, a proteolytic core where proteins are degraded. The ubiquitin ligase (E3) is the key enzyme required for identifying and attaching ubiquitin to proteins to be degraded, and the specificity of the ubiquitin proteasome system is attributed to the E2 and E3 ligases (Mitch et al. 1999). Because the proteasome is an early regulator of muscle atrophy (Medina et al. 1991; Mitch et al. 1999; Bodine et al. 2001; Jagoe et al. 2002; Taillandier et al. 2003, 2004; Li et al. 2004; Krawiec et al. 2005; Whitman et al. 2005) whose components are induced in response to denervation (Medina et al. 1991; Bodine et al. 2001; Batt et al. 2006), it is an appealing target for interventions designed to attenuate muscle atrophy in the days following SCI. However, most studies have been done in animal models, and little data exist regarding alterations in this pathway in humans following SCI at both the transcript and protein levels. We discuss below the implications of our observed alterations in gene expression and protein levels for these essential components of the ubiquitin proteasome pathway in humans following SCI.
Our analysis of skeletal muscle biopsies from patients two and five days post-SCI show increased gene expression for the ubiquitin ligases, MURF1 and Atrogin-1. In contrast to these findings, we previously showed that 48 h of immobilization did not alter the expression of MURF1 and Atrogin-1 (Urso et al. 2006). While it is conceivable that discrepancies among these models in the level of gene expression for MURF1 and Atrogin-1 is a direct result of the rate of protein degradation, Bodine et al. (2001) have previously shown in a rat model that denervation, immobilization, and unweighting resulted in similar increases in mRNA expression for MURF1 and Atrogin-1 at two days after the treatment began. Thus, the discrepancy between the results from our previous and current study in humans, and those from Bodine et al. (Bodine et al. 2001; Urso et al. 2006) in rats is unexplained, but may suggest that adaptive responses to inactivity in skeletal muscle are not identical between humans and animals. In humans, although two days of immobilization in our previous study did not result in a significant increase in MURF1 expression, longer-term immobilization (2 weeks) did result in a significant 62% and 34% increase in Atrogin-1 and MURF1 expression, respectively (Jones et al. 2004). These data in humans suggest that SCI is a more potent and earlier atrophic stimulus compared with immobilization.
Two and five days post-SCI, we documented a 2.4- and 2.5-fold increase, respectively, in another E3 ligase, UBE3C, that is highly expressed in skeletal muscle. The effect of muscle atrophy on UBE3C expression is not well characterized, but in studies where UBE3C was examined, expression was not found to be altered by atrophic stimuli. For example: (1) our previous work examining global changes in gene expression in response to two days of immobilization failed to document an increase in this E3 ligase (Urso et al. 2006); (2) using the differential display approach, Bodine et al. (2001) did not detect differences in the expression of UBE3C in response to three days of denervation, immobilization, or unweighting in rats, although genes included in the analysis were only those with at least a 3-fold increase in expression; and (3) Jones et al. (2004) did not document alterations in UBE3C using a targeted approach to analyse gene expression after two weeks of immobilization in humans. These authors (Jones et al. 2004) argued that their findings indicate that UBE3C is not a general marker of skeletal muscle atrophy. Lecker et al. (2004) demonstrated that while MURF1 and Atrogin-1 were consistently induced, UBE3C was not expressed under conditions of muscle wasting including fasted mice and rats with cancer cachexia. Based on our data showing a concomitant increase in gene expression in the biceps muscles of paraplegic patients two days post-SCI, we suggest that this increase may be the result of confounding stresses associated with SCI (e.g. nutritional, pharmacological, physiological), and not a direct indicator of the effects of denervation on the molecular response within the muscle.
Although the present findings provide evidence that there is an overall increase in gene expression for the E3 ligases, MURF1, Atrogin-1 and UBE3C two and five days post-SCI, and an increase in staining for Atrogin-1 protein about the periphery of the cell, it is interesting that we did not document an increase in gene expression for other components of the pathway, specifically the E1-activating enzymes and E2-conjugating enzymes using microarray analysis, as reported by Nikawa et al. (2004). We also did not find an increase in gene expression of the E1 and E2 enzymes in our previous two-day immobilization protocol in humans according to microarray analysis (Urso et al. 2006). Similarly, Bodine et al. (2001) reported increases in gene expression for only the E3 ligases three days after immobilization, hindlimb suspension or denervation. Thus, our results and others' (Bodine et al. 2001; Welle et al. 2003; Di Giovanni et al. 2004; Chen et al. 2005; Khal et al. 2005) show dramatic increases in gene expression of the E3 ligases in various atrophy models in humans and animals, but no change in the E1 and E2 transcripts.
We were surprised to find that despite increases in gene expression for MURF1, Atrogin-1 and UBE3C, we were unable to document increases in respective protein products. Until recently, however, research has focused primarily on alterations in gene expression for ubiquitin proteasome pathway components (Bodine et al. 2001; Ikemoto et al. 2001; Jones et al. 2004; Taillandier et al. 2004; Khal et al. 2005; Krawiec et al. 2005), and measurements of respective protein products are lacking. Bodine et al. (2001) measured protein levels of MURF1 at one and three days following immobilization in the rat, and reported that protein levels increased by three days post-immobilization. We did not detect alterations in MURF1 protein levels at two or five days post-SCI in the present study. The one study we are aware of in humans that measured both MURF1 and Atrogin-1 gene expression and protein did so in patients experiencing muscle atrophy due to amyotrophic lateral sclerosis (ALS/Lou Gehrig's disease), a rapidly progressive neurological disease that attacks neurons responsible for controlling skeletal muscle. The authors reported that levels of gene expression and protein products were increased for Atrogin-1 in all patients who had been diagnosed with ALS at least three months previously, but there was no change in gene expression for MURF1 and respective protein products (Léger et al. 2006). We were surprised to find in the present study, that despite a 5.6- to 6.0-fold increase in expression of the gene for MURF1 in the days post-SCI, protein levels remained unaltered. Since we also failed to document an increase in Atrogin-1 protein, despite increases in gene expression, we suggest that expression of the E3 ligases, particularly MURF1 and Atrogin-1, is tightly controlled at the level of translation. Results from Léger et al. (2006) suggest that for translation of the Atrogin-1 gene product to occur, expression of Atrogin-1 must not only reach a certain magnitude, but must occur for an extended period of time as evidenced by lack of change observed in the present study after only five days of altered transcription. Atrogin-1 is an essential component in the pathway leading to muscle protein degradation (Attaix et al. 1998; Bodine et al. 2001; Gomes et al. 2001), and, mice who are unable to make the Atrogin-1 protein, maintain muscle mass following denervation (Bodine et al. 2001). Thus, because the Atrogin-1 protein plays a critical role in regulating muscle cell size, acute increases in Atrogin-1 gene expression may not be adequate to increase Atrogin-1 protein product, particularly as early as five days post-SCI. However, although we did not detect an increase in Atrogin-1 protein product using Western blotting analysis despite documenting increases in staining for Atrogin-1 about the periphery of the cell at five days post-SCI, it is possible that increases in gene expression result in a translocation of the protein, rather than an overall increase in cellular protein content. Indeed, the possibility exists that when performing protein isolation techniques for the Western blotting analysis, membrane proteins were discarded. This may explain why we did not measure an increase in protein levels when performing Western blotting, yet did detect an increase about the periphery when performing IHC.
In addition to the increases in the E3 ligases, we also documented increased gene expression for PSMD11 at two (2.7-fold) and five days (3.2-fold) post-SCI using qRT-PCR. PSMD11 is a gene that encodes a subunit of the 26S proteolytic core and is responsible for modulating activation of proteolysis (Bassaglia et al. 2005). There is a general consensus that gene expression for components of the 26S proteolytic core, including PSMD11, increases in response to an atrophic stimulus (Jones et al. 2004; Khal et al. 2005). Bodine et al. (2001) documented a >3-fold increase in the 26S proteasome subunit three days post-denervation, immobilization, and unweighting in rats. In an earlier study (Urso et al. 2006), we did not find an increase in any components of the 26S proteasome after two days of limb immobilization, but Jones et al. (2004) reported a 26% increase in the 20S alpha-subunit following two weeks of immobilization in humans. The discrepancy between our findings in humans in response to immobilization and those from Bodine et al. (2001) support our previous statement that adaptive responses to inactivity in skeletal muscle may not be identical between humans and animals. Additionally, the observed increases in PSMD11 gene expression after SCI (present study) and two weeks of immobilization (Jones et al. 2004), but not two days of immobilization (Urso et al. 2006) supports our hypothesis that SCI is a more potent and earlier atrophic stimulus compared with immobilization.
We also documented an increase in protein levels for PSMD11 five days post-SCI. To our knowledge, previous disuse atrophy models have not examined protein levels of PSMD11 or other components of the 26S proteasome, although some reports show concomitant increases in 26S proteasome subunit gene expression and protein products in various muscle-wasting conditions (Kumamoto et al. 2000; Khal et al. 2005). It appears from those results, as well as results from our present study, that translation of proteins essential to the 26S proteasome is not only induced within a shorter time period after the atrophic stimulus, but that translation is less dependent on the magnitude of mRNA expression, compared to the E3 ligases. We base this conclusion on the modest fold-change in PSMD11 mRNA expression at five days post-SCI (3.2-fold), yet there was a 20% increase in protein levels, whereas there was no detectable change in Atrogin-1 and MURF1 protein levels according to Western blot analysis, despite more robust fold-changes in gene expression (5.6- to 12.9-fold). Because the 26S proteasome is the critical site for proteolysis in the muscle, it serves as the rate-limiting step for muscle protein degradation during the course of atrophy (Bassaglia et al. 2005). In contrast, the various E3 ligases tag specific proteins for degradation via the 26S proteasome complex and do not exert the same rate-limiting effects on the ubiquitin proteasome pathway.
In addition to the observed increase in PSMD11 protein at five days post-SCI, we observed via IHC that at five days post-SCI, there was strong staining for this protein along the peripheral region of the cell (Fig. 7). Previous IHC analysis of proteasome subunit distribution in Duchenne muscular dystrophy (Kumamoto et al. 2000) and healthy skeletal muscle (Bassaglia et al. 2005) has shown sarcomeric distribution of this protein, with stronger staining in dystrophic muscle compared to faint staining patterns in healthy controls. We are unaware of any studies that have explored the distribution of this protein during atrophy, but we suggest that the increased staining for this protein along the peripheral region of the cell suggests that the first proteins to be tagged for degradation following SCI are those involved in membrane/extracellular matrix integrity. We have previously shown after two days of immobilization, that there is decreased gene expression and protein levels for components of the extracellular matrix (Urso et al. 2006), and IHC revealed decreased staining for collagen III and collagen IV. Although our preliminary microarray analysis on a subset of SCI patients showed a 2.5- and 5.0-fold decrease in gene expression for collagen III two and five days post-SCI, we were unable to document alterations in collagen III protein content using Western blotting analysis. In addition, collagen IV gene expression was actually upregulated according to the microarray analysis at two days post-SCI, with no change at five days post-SCI. We did not document any changes at the protein level in the SCI patients. We hypothesize that, in response to immobilization (Urso et al. 2006), decreased membrane/extracellular matrix integrity at the protein level is one of the first adaptations in the muscle atrophy programme stimulating proteolysis in skeletal muscle. We suggest that we do not see this same alteration within two days of SCI because our immobilization model allowed weight bearing, and extracellular matrix turnover is strongly influenced by changes in both physical activity and mechanical load (Kjaer, 2004). Therefore, we suggest that in the SCI patients, the reduction in mechanical load following injury slowed the rate of extracellular matrix degradation. This may also explain why increases in protein products and peripheral staining for the proteasome subunit did not occur until five days post-SCI.
Metallothioneins
One aspect of this study was to use microarray technology to acquire information about novel genes whose expression is altered during the initiation of the atrophy programme in human skeletal muscle following SCI. Therefore, we chose to perform additional analyses on a subset of metallothionein genes found to be upregulated in our microarray dataset. We were able to confirm, using qRT-PCR, increased gene expression at both two and five days post-SCI of three metallothionein genes, MTIA, MTIF and MTIH (Table 2). We also show that metallothionein protein levels increase at five, but not two days post-SCI. IHC confirmed these results, with increased staining for metallothionein along the peripheral regions by five days post-SCI.
Increased metallothionein expression in skeletal muscle has been reported previously in various atrophy models in animals (Kondo et al. 1992; Lecker et al. 2004) and humans (Urso et al. 2006). For example, Kondo et al. (1992) found an increase in metallothionein gene expression following four days of ankle-joint immobilization in the rat. We were the first to explore metallothionein gene expression and protein levels in response to two days of immobilization in humans. We found that two days of immobilization resulted in a significant increase in gene expression in several metallothionein transcripts (1.8–2.3-fold), but no change in protein levels (Urso et al. 2006), similar to results of the current study after two days of SCI. It should be noted that the increase in gene expression as a result of SCI is more robust compared to the immobilization intervention, supporting our previous statement that SCI is a more potent stimulator of the atrophy programme in humans than immobilization. Moreover, by five days post-SCI, protein levels of metallothionein had increased.
Although there are no data on atrophy-induced expression of the metallothioneins in humans, previous work in humans has focused on the effects of an acute bout of exercise on metallothionein gene expression in skeletal muscle, and show robust increases in mRNA for metallothionein in response to an exercise stimulus (Penkowa et al. 2005; Scheede-Bergdahl et al. 2005). Metallothionein protein levels have also been shown to increase 24 h after an acute bout of exercise (Penkowa et al. 2005), and eight weeks after the start of an exercise intervention in healthy populations (Scheede-Bergdahl et al. 2005). This increased expression of metallothionein gene expression and respective protein products in response to physiological stressors, including intense exercise and muscle atrophy, is thought to serve several purposes in skeletal muscle. Metallothioneins are antioxidant proteins, and in response to oxidative stress, glucocorticoids, catecholamines, and matrix metalloprotease secretion, metallothioneins inhibit reactive oxygen species and protect against DNA damage and degradation (Kondo et al. 1992; Apostolova et al. 1999; Lecker et al. 2004; Penkowa et al. 2005). Metallothioneins are also induced in response to inflammatory stimuli and provide anti-inflammatory and anti-apoptotic functions to protect cells in response to trauma and pathological conditions (Kondo et al. 1992; Apostolova et al. 1999; Lecker et al. 2004; Penkowa et al. 2005). A role has been proposed for the metallothioneins, in skeletal muscle, to attenuate the infiltration of inflammatory cytokines in response to trauma and disuse (Kondo et al. 1992; Lecker et al. 2004). Increased staining along the peripheral region of the cell supports this role, as increased metallothionein activity along the membrane will counteract infiltration of macrophages and cytokines that induce proteolytic signalling in muscle.
A number of reports on the expression of ubiquitin proteasome genes during muscle atrophy have also documented a concomitant increase in metallothionein gene expression in animals (Kondo et al. 1992; Lecker et al. 2004; Nikawa et al. 2004). This increase in metallothionein expression, particularly in the first few days following the atrophic stimulus, may attenuate the overall rate of muscle atrophy by reducing DNA damage, reactive oxygen species-mediated toxicity, and/or the inflammatory response. These factors may be responsible for the induction of proteolytic pathways, including the ubiquitin proteasome pathway. In the present study, despite robust increases in metallothionein gene expression within two days of SCI, protein levels did not increase until five days post-SCI. Thus, should treatment regimens that manipulate metallothionein expression be explored, interventions should focus on decreasing this delay between increases in transcription and eventual translation of the metallothionein protein, so that beneficial effects of metallothionein protein induction will occur sooner in the atrophy programme, possibly attenuating the overall rate of atrophy.
Protease inhibitor
The microarray analysis revealed that another gene, SLPI, not previously associated with skeletal muscle atrophy in humans, was upregulated 21- and 44-fold two and five days post-SCI, respectively. SLPI is a member of the serine protease group of proteins that inhibit protein-degrading enzymes, such as proteases involved in the ubiquitin proteasome pathway. We chose to confirm this alteration in gene expression because this upregulation was so robust, and SLPI has been shown to function in tissue by controlling protease activity (Ma et al. 2004; Sehnert et al. 2004). Additionally, since previous work has shown increased expression of skeletal muscle ubiquitin in response to inflammation (DeJong et al. 2005), we speculate that induction of SLPI may inhibit proteolysis in skeletal muscle by either attenuating the inflammatory response or binding to proteases and rendering them inactive.
Although we were able to confirm alterations in gene expression using qRT-PCR technology (Table 2), we did not show concomitant increases in protein levels of SLPI at two or five days post-SCI. Despite this, we performed IHC using a primary antibody targeted against SLPI to determine the location of this protein in healthy muscle and to identify if this location was altered by SCI. Our motivation for this analysis was that identifying the intracellular location of SLPI might help to ascertain the functional interactions of SLPI with target molecules in skeletal muscle. We first performed IHC using single antibody staining for SLPI. IHC (Fig. 10) revealed that staining for SLPI was not detectable in skeletal muscle samples from healthy controls, and in muscle biopsies taken two days post-SCI staining was faint, with sparse staining in the peripheral region of the cell. However, by five days post-SCI, there was considerable staining for SLPI in the peripheral region of the cell (Fig. 10).
These results are novel because to our knowledge, we are the first to explore SLPI gene expression and protein products in skeletal muscle in response to an atrophy stimulus. Our previous work examining global changes in gene expression after two days of limb immobilization did not detect alterations in SLPI (Urso et al. 2006). Similarly, SLPI was not identified as being upregulated after three days of immobilization, denervation or unweighting in rats (Bodine et al. 2001). Moreover, we explored comprehensive data sets from atrophy models previously discussed in this paper that used microarray or cDNA array analysis to identify global changes in gene expression (Bodine et al. 2001; Ikemoto et al. 2001; Jagoe et al. 2002; Welle et al. 2003; Lecker et al. 2004; Batt et al. 2006) and found that SLPI expression was not induced in any of these models discussed. Therefore, we used a public expression profiling resource to explore gene expression profiles among various disease conditions (see http://pepr.cnmcresearch.org). We were able to query the SLPI refseq transcript (203021_at; http://www.genome.ucsc.edu) across 13 muscle disease groups, and we found that this transcript was significantly induced only in patients with acute quadriplegic myopathy (AQM), a muscle disease characterized by profound atrophy in response to systemic illness or exposure to corticosteroids or neuroblocking agents (Di Giovanni et al. 2004). The induction of SLPI in this model was in stark contrast to other chronic atrophy conditions, including the previously discussed ALS (Léger et al. 2006), where SLPI expression was similar to that in healthy controls. These data indicate that induction of SLPI is a unique response in humans to conditions that are characterized by rapid and severe atrophy, as in SCI and AQM. Because SLPI is a serine protease inhibitor which functions to block protease activity and subsequent destruction of muscle tissue, induction of SLPI may be dependent upon the magnitude of protease activity, explaining why previous work in less severe models of muscle atrophy has not documented increases in SLPI.
Additional work is warranted to explore the relationship between SLPI and protease activity. More importantly, it is critical to understand if SLPI induction is indeed beneficial in skeletal muscle in attenuating proteolytic activity. The selective nature of SLPI activity in inhibiting proteases, particularly proteases associated with degradation of the membrane/extracellular matrix makes it an ideal target for therapeutic intervention. Currently, serine protease inhibitors are under experimentation in clinical trials to attenuate the progression of HIV by inhibiting proteases essential for viral replication (Lin et al. 2004; Ma et al. 2004).
Summary
Transcriptional activity of the ubiquitin proteasome pathway, specifically the E3 ligases and the proteasome subunit, was profoundly increased two and five days post-SCI. We showed that protein levels for the proteasome subunit increase in the days following SCI and that staining for the proteasome subunit increased in the peripheral region of the cell. Similarly, Atrogin-1, the E3 ligase responsible for tagging proteins for destruction, increased in the peripheral region of the cell. This work also demonstrated that in the first days following SCI, there was an increase in metallothionein gene expression and respective protein products, and that localization of these proteins, which are thought to exert protective effects in skeletal muscle, increased in the peripheral region of the cell. These results lend evidence to our proposition that the membrane/extracellular matrix may be an early target for degradation in the days following SCI. Finally, our data showed that gene expression for a protease inhibitor, not previously associated with the muscle atrophy programme (SLPI), increased dramatically two and five days post-SCI, and staining for the SLPI protein increased in the peripheral region of the cell, similar to the other genes explored in this human SCI model. In summary, the changes in gene and mRNA expression and the associated changes in protein and localization of these proteins that we report here, provide insight into specific signalling pathways in the muscle atrophy programme in the days following SCI, probably contributing to losses in muscle mass in humans. These data suggest that the primary signalling networks activated in the days post-SCI are those involved in protein degradation (ubiquitin proteasome pathway), and those which may function to protect skeletal muscle from rapid degradation (metallothioneins, SLPI). These alterations are important biomarkers for future work investigating ways to manipulate these pathways post-SCI to attenuate losses in muscle mass.
Acknowledgments
This study was supported by a research grant from the Christopher Reeve Foundation. We acknowledge Dr Rongye Shi for her expert assistance in performing expression profiling; Dr Milena Penkowa who generously supplied the Metallothionein (MT-I/II) antibody; and Dr Lawrence Schwartz who assisted with protocol development and data interpretation. The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the US Army or the Department of Defense. Any citations of commercial organizations and trade names in this report do not constitute an official Department of the Army endorsement of approval of the products or services of these organizations.
References
- Apostolova MD, Ivanova IA, Cherian MG. Metallothionein and apoptosis during differentiation of myoblasts to myotubes: protection against free radical toxicity. Toxicol Appl Pharmacol. 1999;159:175–184. doi: 10.1006/taap.1999.8755. [DOI] [PubMed] [Google Scholar]
- Attaix D, Aurousseau E, Combaret L, Kee A, Larbaud D, Ralliere C, Souweine B, Taillandier D, Tilignac T. Ubiquitin-proteasome-dependent proteolysis in skeletal muscle. Reprod Nutr Dev. 1998;38:153–165. doi: 10.1051/rnd:19980202. [DOI] [PubMed] [Google Scholar]
- Bassaglia Y, Cebrian J, Covan S, Garcia M, Foucrier J. Proteasomes are tightly associated to myofibrils in mature skeletal muscle. Exp Cell Res. 2005;302:221–232. doi: 10.1016/j.yexcr.2004.08.038. [DOI] [PubMed] [Google Scholar]
- Batt J, Bain J, Goncalves J, Michalski B, Plant P, Fahnestock M, Woodgett J. Differential gene expression profiling of short and long term denervated muscle. FASEB J. 2006;20:115–117. doi: 10.1096/fj.04-3640fje. [DOI] [PubMed] [Google Scholar]
- Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- Booth FW, Gollnick PD. Effects of disuse on the structure and function of skeletal muscle. Med Sci Sports Exerc. 1983;15:415–420. [PubMed] [Google Scholar]
- Chen YW, Hubal MJ, Hoffman EP, Thompson PD, Clarkson PM. Molecular responses of human muscle to eccentric exercise. J Appl Physiol. 2003;95:2485–2494. doi: 10.1152/japplphysiol.01161.2002. [DOI] [PubMed] [Google Scholar]
- Chen YW, Nagaraju K, Bakay M, McIntyre O, Rawat R, Shi R, Hoffman EP. Early onset of inflammation and later involvement of TGFα in Duchenne muscular dystrophy. Neurology. 2005;65:826–834. doi: 10.1212/01.wnl.0000173836.09176.c4. [DOI] [PubMed] [Google Scholar]
- Chen YW, Zhao P, Borup R, Hoffman EP. Expression profiling in the muscular dystrophies: identification of novel aspects of molecular pathophysiology. J Cell Biol. 2000;151:1321–1336. doi: 10.1083/jcb.151.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJong CH, Busquets S, Moses AG, Schrauwen P, Ross JA, Argiles JM, Fearon KC. Systemic inflammation correlates with increased expression of skeletal muscle ubiquitin but not uncoupling proteins in cancer cachexia. Oncol Rep. 2005;14:257–263. [PubMed] [Google Scholar]
- Di Giovanni S, Molon A, Broccolini A, Melcon G, Mirabella M, Hoffman EP, Servidei S. Constitutive activation of MAPK cascade in acute quadriplegic myopathy. Ann Neurol. 2004;55:195–206. doi: 10.1002/ana.10811. [DOI] [PubMed] [Google Scholar]
- Dudley GA, Castro MJ, Rogers S, Apple DF., Jr A simple means of increasing muscle size after spinal cord injury: a pilot study. Eur J Appl Physiol Occup Physiol. 1999;80:394–396. doi: 10.1007/s004210050609. [DOI] [PubMed] [Google Scholar]
- Dupont-Versteegden EE, Houle JD, Gurley CM, Peterson CA. Early changes in muscle fiber size and gene expression in response to spinal cord transection and exercise. Am J Physiol Cell Physiol. 1998;275:C1124–C1133. doi: 10.1152/ajpcell.1998.275.4.C1124. [DOI] [PubMed] [Google Scholar]
- Goldspink DF. The effects of denervation on protein turnover of rat skeletal muscle. Biochem J. 1976;156:71–80. doi: 10.1042/bj1560071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci U S A. 2001;98:14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman EPAT, Palma J, Webster T, Hubbell E, Warrington JA, Spira A, Wright G, Buckley J, Triche T, Davis R, Tibshirani R, Xaio W, Jones W, Tomkins R, West M. Expression profiling – best practices for data generation and interpretation in clinical trials. Nat Rev Genet. 2004;5:229–237. doi: 10.1038/nrg1297. [DOI] [PubMed] [Google Scholar]
- Hornberger TA, Hunter RB, Kandarian SC, Esser KA. Regulation of translation factors during hindlimb unloading and denervation of skeletal muscle in rats. Am J Physiol Cell Physiol. 2001;281:C179–C187. doi: 10.1152/ajpcell.2001.281.1.C179. [DOI] [PubMed] [Google Scholar]
- Ikemoto M, Nikawa T, Takeda S, Watanabe C, Kitano T, Baldwin KM, Izumi R, Nonaka I, Towatari T, Teshima S, Rokutan K, Kishi K. Space shuttle flight (STS-90) enhances degradation of rat myosin heavy chain in association with activation of ubiquitin-proteasome pathway. FASEB J. 2001;15:1279–1281. doi: 10.1096/fj.00-0629fje. [DOI] [PubMed] [Google Scholar]
- Jagoe RT, Lecker SH, Gomes M, Goldberg AL. Patterns of gene expression in atrophying skeletal muscles: response to food deprivation. FASEB J. 2002;16:1697–1712. doi: 10.1096/fj.02-0312com. [DOI] [PubMed] [Google Scholar]
- Jones SW, Hill RJ, Krasney PA, O'Conner B, Peirce N, Greenhaff PL. Disuse atrophy and exercise rehabilitation in humans profoundly affects the expression of genes associated with the regulation of skeletal muscle mass. FASEB J. 2004;18:1025–1027. doi: 10.1096/fj.03-1228fje. [DOI] [PubMed] [Google Scholar]
- Khal J, Hine AV, Fearon KC, Dejong CH, Tisdale MJ. Increased expression of proteasome subunits in skeletal muscle of cancer patients with weight loss. Int J Biochem Cell Biol. 2005;37:2196–2206. doi: 10.1016/j.biocel.2004.10.017. [DOI] [PubMed] [Google Scholar]
- Kjaer M. Role of extracellular matrix in adaptation of tendon and skeletal muscle to mechanical loading. Physiol Rev. 2004;84:649–698. doi: 10.1152/physrev.00031.2003. [DOI] [PubMed] [Google Scholar]
- Kondo H, Miura M, Nakagaki I, Sasaki S, Itokawa Y. Trace element movement and oxidative stress in skeletal muscle atrophied by immobilization. Am J Physiol Endocrinol Metab. 1992;262:E583–E590. doi: 10.1152/ajpendo.1992.262.5.E583. [DOI] [PubMed] [Google Scholar]
- Krawiec BJ, Frost RA, Vary TC, Jefferson LS, Lang CH. Hindlimb casting decreases muscle mass in part by proteasome-dependent proteolysis but independent of protein synthesis. Am J Physiol Endocrinol Metab. 2005;289:E969–E980. doi: 10.1152/ajpendo.00126.2005. [DOI] [PubMed] [Google Scholar]
- Kumamoto T, Fujimoto S, Ito T, Horinouchi H, Ueyama H, Tsuda T. Proteasome expression in the skeletal muscles of patients with muscular dystrophy. Acta Neuropathol. 2000;100:595–602. doi: 10.1007/s004010000229. [DOI] [PubMed] [Google Scholar]
- Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, Price SR, Mitch WE, Goldberg AL. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004;18:39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- Léger BVL, Sorarù G, Hespel P, Derave W, Goblet C, D'Ascenzio C, Angelini C, Russell AP. Human skeletal muscle atrophy in amyotrophic lateral sclerosis reveals a reduction in Akt and an increase in atrogin-1. FASEB J. 2006;20:583–585. doi: 10.1096/fj.05-5249fje. [DOI] [PubMed] [Google Scholar]
- Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues, and standard error application. Genome Biol. 2001;8:32.31–32.32.11. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HH, Kedar V, Zhang C, McDonough H, Arya R, Wang DZ, Patterson C. Atrogin-1/muscle atrophy F-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex. J Clin Invest. 2004;114:1058–1071. doi: 10.1172/JCI22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AL, Johnson DA, Stephan KT, Yeh CK. Salivary secretory leukocyte protease inhibitor increases in HIV infection. J Oral Pathol Med. 2004;33:410–416. doi: 10.1111/j.1600-0714.2004.00218.x. [DOI] [PubMed] [Google Scholar]
- Ma G, Greenwell-Wild T, Lei K, Jin W, Swisher J, Hardegen N, Wild CT, Wahl SM. Secretory leukocyte protease inhibitor binds to annexin II, a cofactor for macrophage HIV-1 infection. J Exp Med. 2004;200:1337–1346. doi: 10.1084/jem.20041115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney DJ, Carey K, Fu MH, Snow R, Cameron-Smith D, Parise G, Tarnopolsky MA. Real-time RT-PCR analysis of housekeeping genes in human skeletal muscle following acute exercise. Physiol Genomics. 2004;18:226–231. doi: 10.1152/physiolgenomics.00067.2004. [DOI] [PubMed] [Google Scholar]
- Medina R, Wing SS, Haas A, Goldberg AL. Activation of the ubiquitin-ATP-dependent proteolytic system in skeletal muscle during fasting and denervation atrophy. Biomed Biochim Acta. 1991;50:347–356. [PubMed] [Google Scholar]
- Mitch WE, Bailey JL, Wang X, Jurkovitz C, Newby D, Price SR. Evaluation of signals activating ubiquitinproteasome proteolysis in a model of muscle wasting. Am J Physiol Cell Physiol. 1999;276:C1132–C1138. doi: 10.1152/ajpcell.1999.276.5.C1132. [DOI] [PubMed] [Google Scholar]
- Nikawa T, Ishidoh K, Hirasaka K, Ishihara I, Ikemoto M, Kano M, Kominami E, Nonaka I, Ogawa T, Adams GR, Baldwin KM, Yasui N, Kishi K, Takeda S. Skeletal muscle gene expression in space-flown rats. FASEB J. 2004;18:522–524. doi: 10.1096/fj.03-0419fje. [DOI] [PubMed] [Google Scholar]
- Penkowa M, Keller P, Keller C, Hidalgo J, Giralt M, Pedersen BK. Exercise-induced metallothionein expression in human skeletal muscle fibres. Exp Physiol. 2005;90:477–486. doi: 10.1113/expphysiol.2004.029371. [DOI] [PubMed] [Google Scholar]
- Raffaello A, Laveder P, Romualdi C, Bean C, Toniolo L, Germinario E, Megighian A, Danieli-Betto D, Reggiani C, Lanfranchi G. Denervation in murine fast-twitch muscle: short term physiological changes and temporal expression profiling. Physiol Genomics. 2006;25:60–74. doi: 10.1152/physiolgenomics.00051.2005. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Li C, Ellis B, Wong WH. Feature extraction and normalization algorithms for high-density oligonucleotide gene expression array data. J Cell Biochem Suppl. 2001;37:120–125. doi: 10.1002/jcb.10073. [DOI] [PubMed] [Google Scholar]
- Scheede-Bergdahl C, Penkowa M, Hidalgo J, Olsen DB, Schjerling P, Prats C, Boushel R, Dela F. Metallothionein-mediated antioxidant defense system and its response to exercise training are impaired in human type 2 diabetes. Diabetes. 2005;54:3089–3094. doi: 10.2337/db12-wd10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnert B, Cavcic A, Bohm B, Kalden JR, Nandakumar KS, Holmdahl R, Burkhardt H. Antileukoproteinase: modulation of neutrophil function and therapeutic effects on anti-type II collagen antibody-induced arthritis. Arthritis Rheum. 2004;50:2347–2359. doi: 10.1002/art.20339. [DOI] [PubMed] [Google Scholar]
- Sevetson BRKX, Lawrence JC. Increasing cAMP attenuates activation of mitogen-activated protein kinase. Proc Natl Acad Sci U S A. 1993;90:10305–10309. doi: 10.1073/pnas.90.21.10305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taillandier D, Aurousseau E, Combaret L, Guezennec CY, Attaix D. Regulation of proteolysis during reloading of the unweighted soleus muscle. Int J Biochem Cell Biol. 2003;35:665–675. doi: 10.1016/s1357-2725(03)00004-9. [DOI] [PubMed] [Google Scholar]
- Taillandier D, Combaret L, Pouch MN, Samuels SE, Bechet D, Attaix D. The role of ubiquitin-proteasome-dependent proteolysis in the remodelling of skeletal muscle. Proc Nutr Soc. 2004;63:357–361. doi: 10.1079/PAR2004358. [DOI] [PubMed] [Google Scholar]
- Tang H, Cheung WM, Ip FC, Ip NY. Identification and characterization of differentially expressed genes in denervated muscle. Mol Cell Neurosci. 2000;16:127–140. doi: 10.1006/mcne.2000.0864. [DOI] [PubMed] [Google Scholar]
- Urso ML, Scrimgeour A, Chen YW, Thompson PD, Clarkson PM. Analysis of mRNA and protein in human skeletal muscle after 48h immobilization. J Appl Physiol. 2006;101:1136–1148. doi: 10.1152/japplphysiol.00180.2006. [DOI] [PubMed] [Google Scholar]
- Welle S, Brooks AI, Delehanty JM, Needler N, Thornton CA. Gene expression profile of aging in human muscle. Physiol Genomics. 2003;14:149–159. doi: 10.1152/physiolgenomics.00049.2003. [DOI] [PubMed] [Google Scholar]
- Whitman SA, Wacker MJ, Richmond SR, Godard MP. Contributions of the ubiquitin-proteasome pathway and apoptosis to human skeletal muscle wasting with age. Pflugers Arch. 2005;450:437–446. doi: 10.1007/s00424-005-1473-8. [DOI] [PubMed] [Google Scholar]