Abstract
Conversion of pre-mRNAs into mature mRNAs includes several consecutive enzymatic modification steps that are carried out in the spliceosomes. Helicases have been shown to contribute to these catalytic processes both in yeast and in mammalian cells. Our results identify the mammalian protein MAH (matrix-associated helicase) as a new helicase present in the spliceosome complex. Sequence comparison describes MAH as the first higher eukaryotic member of the helicase superfamily I, with demonstrated enzymatic activity. Because MAH does not bind small nuclear ribonucleoproteins (snRNPs), it appears to be a non-snRNP binding factor of the splicing complex. In conclusion, our data suggest the involvement of MAH in processing of pre-mRNAs in mammalian cells.
Pre-mRNA maturation takes place in macromolecular complexes denoted the spliceosomes (1–3), which are characteristic structural components of the nuclease and high salt resistant part of the nucleus called nuclear matrix (4). The spliceosomes are composed of four small nuclear ribonucleoproteins (snRNPs) (5), and a large but as yet unidentified number of non-snRNP protein factors that do not bind small nuclear RNA (snRNA) (6). The snRNP particles consist of common proteins (also designated Sm proteins) that are associated with all snRNPs, and snRNP-specific proteins (7). The spliceosome proteins execute the precise cleavage and ligation steps generating mature mRNA that include the preparation of RNA for splicing, selection of splicing sites, catalysis of structural modifications of the RNA, ligation of spliced RNA ends, and production of mature RNA (7).
Helicases catalyze the unwinding of double-stranded DNA and RNA sequences by disrupting the hydrogen bonds between the two strands (8). They are thought to be involved both in DNA and RNA metabolism, including RNA splicing. Helicases are structurally characterized by the presence of several consensus sequence motifs that delimitate specific superfamilies (9–12). Several proteins of superfamily II with DEAD/H box typical for putative RNA helicases have been identified as splicing factors. These include members of the Prp family in Saccharomyces cerevisiae (13–17) as well as the mammalian proteins HRH1 (18), HEL117 (19), and U5-200kD (20). In contrast, involvement of superfamily I helicases in pre-mRNA splicing has not been described yet.
Here we report the association of a new mammalian superfamily I helicase called MAH (matrix-associated helicase) with the pre-mRNA splicing complex. Our data suggest that MAH is a non-snRNP binding factor of the spliceosomes that may be involved in the processing of pre-mRNAs in mammalian cells.
MATERIALS AND METHODS
Cell Culture.
Mouse A31 fibroblasts, HeLa, and 21PT human epithelial cells were maintained in Dulbecco’s modified Eagle’s medium, supplemented with 10% fetal bovine serum, 4 mM glutamine, 0.4 unit/ml penicillin, and 0.4 g/ml streptomycin.
Library Screening, Antibody Production, Protein Purification, and in Vitro Translation.
λgt11 cDNA libraries prepared from NIH 3T3 fibroblasts were used for expression screening with a concatemer 32P-labeled oligonucleotide probe (21) corresponding to the MT3 protein binding site of the mouse thymidine kinase promoter (−43 bp/−28 bp) (22, 23).
The recombinant C12 fragment of MAH (between amino acids 222–379) was expressed in the JM109 strain of Escherichia coli, using the pGEX-KG prokaryotic expression vector (24). Purification and adsorption to glutathione beads was carried out as described (25). The eluted C12 protein was used to produce a polyclonal rabbit antiserum (Hazleton Research Products, Denver, PA). Sonicated A31 cell extracts were then prepared and used for C12 antibody coupled purification of MAH by AminoLink column as suggested by the manufacturer (Pierce). To eliminate coelution of contaminating proteins, the column was washed with a linear gradient of 1–3 M KCl before elution with glycine buffer.
[35S]Methionine-labeled in vitro translated MAH protein was synthesized with the TnT-coupled reticulocyte lysate translation system (Promega) using full-length MAH cDNA as template.
ATPase and GTPase Assays.
ATPase activity was measured as described (26). The autoradiograms were evaluated by an imaging densitometer (model GS-700) and analyzed with molecular analysis software (Bio-Rad). The ATPase activity was expressed as the amount of ATP converted to ADP. GTPase activity was determined in analogous experiments using GTP instead of ATP.
Helicase Assays.
The standard reaction mixture (20 μl) contained 50 mM Tris (pH 7.8), 7 mM MgCl2, 5 mM DTT, 200 μg/ml bovine serum albumin, 1 mM ATP, various amounts of purified MAH protein, and 32P-labeled short double-stranded substrates (26, 27). The reaction mixtures were incubated at 30°C for 40 min, and the reaction was stopped by addition of 2.5 μl of a solution containing 40% glycerol, 40 mM EDTA, 1% SDS, and 0.25% bromophenol blue. Single- and double-stranded DNAs were separated on 12% nondenaturing polyacrylamide gels and the displacement of the radioactive oligonucleotide was analyzed by autoradiography.
Substrates for helicase assays were prepared as described (26). Briefly, substrate I, a 30-mer oligonucleotide, complementary to nucleotides between 6257–6287 from the multiple cloning site of mp18/pUC18, was 5′-end labeled with T4 polynucleotide kinase plus [γ-32P]ATP. Then it was equimolarly annealed to single-stranded M13 mp18 circular plasmid DNA (see Fig. 2C). Substrate II consists of substrate I linearized with HincII (see Fig. 2E). Substrate III consisted of the above 30-mer oligonucleotide first annealed to single-stranded M13 mp18 circular plasmid DNA and then digested with SphI, resulting in a partially double-stranded DNA with cohesive ends. The 3′ end of the 20-mer fragment was then radioactively labeled, using the Klenow fragment and [α-32P]dATP (see Fig. 2E).
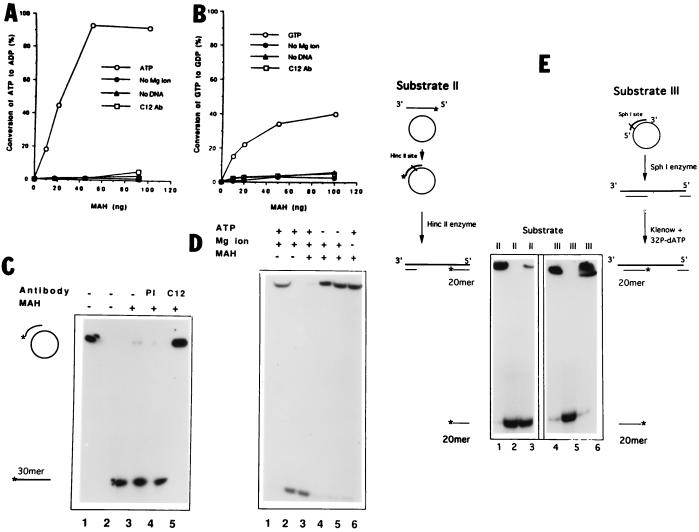
Figure 2.
(A and B) Purified MAH presents NTPase activity. MAH can hydrolyze both ATP (A) and GTP (B) in a DNA and Mg ion-dependent manner. Increasing concentrations of MAH were combined with 1 mM [2,8-3H]ATP or [2,8-3H]GTP at 30°C for 30 min. The reaction products were separated by thin layer chromatography, and the converted nucleotides were determined with autoradiography. The open circles indicate ATP or GTP hydrolysis in the presence of salmon sperm DNA and Mg ions. (C) MAH unwinds partially double-stranded DNA. Substrate I was created from circular M13 mp18 single-stranded plasmid DNA annealed with a 30-mer labeled oligonucleotide. Lanes: 1, native substrate I; 2, heat denatured substrate I without MAH protein (negative and positive controls, respectively); 3–5, 30 fmol substrate I combined with 40 ng MAH; 4–5, preincubation of MAH with preimmune serum (PI) or C12 antibody (C12). (D) MAH unwinding activity is ATP and Mg ion dependent. Lanes: 1 and 2, as in C; 3, MAH was combined with substrate I in the presence of Mg ion and ATP; 4 and 5, ATP was replaced with 1 or 2 mM ATP-γS, respectively; 6, Mg ions were omitted from the reaction mixture. (E) Polarity of MAH unwinding activity. Substrates II and III were created as described in Materials and Methods, with 5′- or 3′-end labeled short duplexes, respectively. MAH (40 ng) was combined with either substrate II (lane 3) or III (lane 6). Lanes: 1 and 4, native substrates as negative controls; 2 and 5, heat denatured substrates as positive controls.
All substrates were purified from unincorporated nucleotides in two steps. First, they were purified by gel filtration through a Quick Spin G-25 or G-50 column (Boehringer Mannheim), and then by diluting and reconcentrating twice in 2 ml TE buffer (10 mM Tris, pH 7.5/1 mM EDTA).
Immunocytochemistry.
HeLa or 21PT cells grown on coverslips were fixed in 2% paraformaldehyde for 10 min and permeabilized with 0.5% Triton X-100 for 5 min. In certain experiments the cells were permeabilized twice; first, the living cells were treated with 0.05% Triton X-100 on ice for 10 min before fixation, and then permeabilized again as above. Cells were then directly processed for antibody staining or before that treated with DNase I and RNase. Blocking and antibody treatments were carried out in PBS containing 1% BSA and 5% normal rabbit serum. Cells were incubated with the primary antibodies overnight at 4°C, and then stained with the appropriate secondary antibody conjugated with rhodamine or fluorescein isothiocyanate (FITC). Simultaneously, DNA was visualized by 4′,6′-diamidino-2-phenylindole (DAPI). Immunofluorescence was analyzed with LSM 410 confocal laser scanning microscope (Zeiss). The images were printed with Fujix Pictography 3000 color printer using Adobe Photoshop software.
Splicing Assay.
HeLa nuclear extracts were combined with in vitro-transcribed, capped, and uniformly 32P-labeled pre-mRNA substrates PIP85A (28) or β-globin (Promega) for in vitro splicing assay, essentially as described (29–31). Protein A beads were coated with the indicated antibodies, then combined with the splicing reaction mixture in IP100 buffer (32). Immunoprecipitated RNA was then recovered by elution with proteinase K buffer, ethanol precipitated, and resolved on polyacrylamide (1:30 bisacrylamide/acrylamide)/8 M urea gel.
RESULTS
Identification of a DNA Binding Protein with Putative Helicase Motifs.
We have cloned a gene, designated as MAH, by screening a cDNA expression library with an oligonucleotide probe corresponding to the MT3 protein binding element of the mouse thymidine kinase promoter. Searching the database, MAH was found to be identical to the mouse immunoglobulin switch region binding protein Smbp2 (33). Both the human and hamster counterparts of MAH/Smbp2 have already been cloned (34, 35), and it was also described that 45-kDa glial factor 1 is equal to the middle part of MAH/Smbp2 (36). Smbp2, its counterparts, and glial factor 1 have been shown to carry putative helicase motifs, although their possible helicase activity had not been yet demonstrated.
In addition to published data by others, our detailed database search revealed that MAH is homologous to members of the helicase superfamily I which includes the S. cerevisiae protein Upf1. Interestingly, Upf1 is an active DNA/RNA helicase and is involved in nonsense mRNA decay (37, 38). This homology with Upf1 and related proteins, and the presence of putative helicase motifs in MAH, suggested that MAH may have a general role in DNA/RNA metabolism.
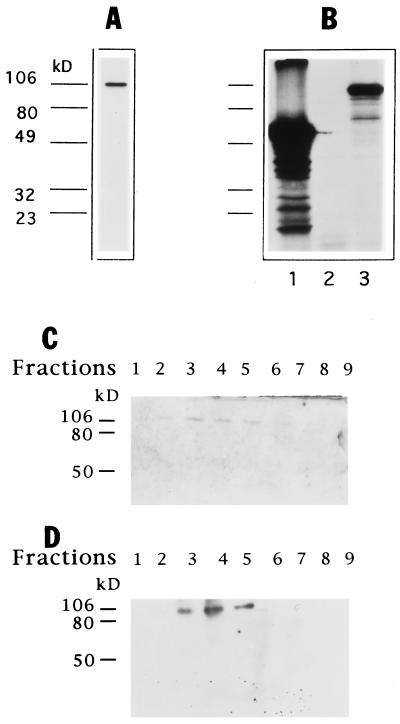
Therefore, first we wanted to know if MAH presents helicase activity. To this end, we expressed MAH in Sf9 cells with baculovirus and in E. coli; however, we did not yield enough protein for systematic studies because of the low expression level and the limited solubility, respectively. Then we raised a rabbit polyclonal antibody against a 157-amino acid long piece of MAH named C12. The C12 antibody identified a single polypeptide on Western blot, with an apparent molecular weight of 106 kDa (Fig. 1A), which is close to the calculated molecular weight of MAH (97 kDa), and equal to that of the in vitro-translated protein (Fig. 1B). The specificity of C12 was confirmed with using preimmune or protein A depleted sera that did not react with any protein on immunoblot (data not shown). Therefore, we used the C12 antibody to purify MAH protein from A31 cell extract by affinity column. The antibody-affinity purified MAH protein gave a single polypeptide band in the expected molecular weight range both on silver stained gel and on Western blot in fractions 2–6 (Fig. 1 C and D).
Figure 1.
(A) C12 antibody recognizes a single 106-kDa polypeptide on Western blot. A 50 μg A31 whole cell extract was separated on 10% polyacrylamide/SDS gel, then transferred to polyvinylidene difluoride membrane and hybridized with C12 antibody. (B) In vitro-translated MAH has 106-kDa molecular weight. In vitro-translated proteins were separated with 10% polyacrylamide/SDS gel electrophoresis and examined by autoradiography. Lanes: 1, luciferase (positive control); 2, no DNA in the reaction (negative control); 3, in vitro-translated product of the full length MAH cDNA. (C and D) Analysis of protein fractions collected from C12 affinity column by silver staining (C) and Western blot (D).
Purified MAH Is an Active Helicase.
After confirming the identity of the fraction, we used the pooled fractions of purified MAH protein to study its enzymatic activity. The ability to hydrolyze NTPs and unwind nucleic acids are two important features of a functional helicase. Therefore, to characterize MAH, we used the purified protein for measuring NTP hydrolytic and DNA unwinding activities in vitro. Consistent with the presence of type I helicase motifs (33), MAH hydrolyzed both ATP and GTP in a DNA and Mg ion-dependent manner (Fig. 2 A and B). The ATP/GTP hydrolysis activity was also identified with recombinant MAH obtained from prokaryotic expression. The Km values of ATP and GTP hydrolysis by MAH (90 and 37 μM, respectively) were similar to those of the yeast homologue Upf1 (37).
The unwinding activity was determined with an in vitro helicase assay using different substrates. A partially double-stranded DNA (substrate I) was efficiently unwound by MAH, resulting in the displacement of the radioactive strand (Fig. 2C). In contrast, MAH was inactive both with completely double-stranded linear DNA or with nicked circular DNA (data not shown). The specificity of the reaction was confirmed by preincubating MAH with C12 antibody which inhibited the unwinding (Fig. 2C, lane 5), whereas preimmune serum did not have any influence (Fig. 2C, lane 4). The helicase activity of MAH was dependent on ATP, because replacing ATP with its nonhydrolyzable analogue (ATP-γS) strongly interfered with dislocation of the 30-mer fragment (Fig. 2D, lanes 4 and 5). Besides ATP, MAH also required the presence of Mg ions in the reaction buffer (Fig. 2D, lane 6).
To determine the directionality of the unwinding activity, partially double stranded linear DNA substrates (substrates II and III) were created from M13 mp18 single-stranded DNA and end-labeled at different short duplex ends by 32P (Materials and Methods and Fig. 2E). Addition of MAH to these substrates resulted in displacement of the 20-mer from substrate II (Fig. 2E, lane 3), but not from substrate III (Fig. 2E, lane 6). Because double-stranded DNA with blunt ends did not serve as substrate for MAH in the unwinding reaction, the 20-mer fragment was released from substrate II only from the 3′ single-stranded end.
In summary, MAH displays helicase activity on a partially double-stranded DNA substrate in unipolarity to the 3′ → 5′ direction, as referred to the single-stranded DNA region.
MAH Is Localized to the Nuclear Matrix.
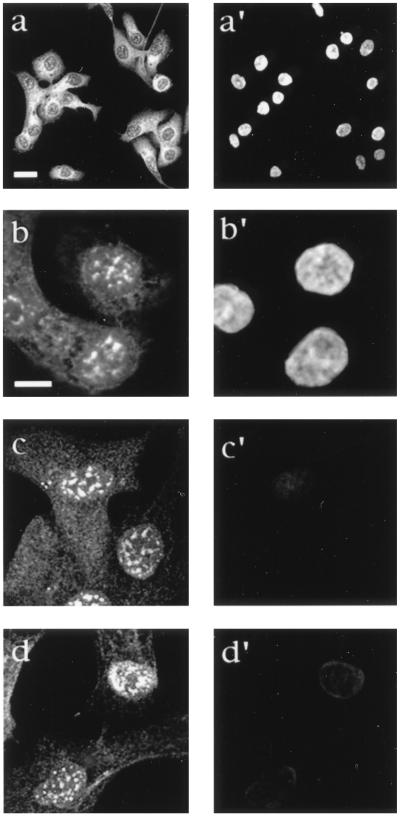
The homology with yeast proteins and the helicase activity suggest that MAH plays a role in RNA metabolic events. Because proteins involved in these events show characteristic cellular localization (39), we next determined the subcellular distribution of MAH by immunostaining. Incubation with C12 antibody stained the nuclei with prominent punctuate immunofluorescence, but also showed discrete foci in the cytoplasm with some perinuclear accumulation (Fig. 3 a and a′). In addition, all interphase cells were labeled uniformly, and no cell cycle-dependent redistribution of nuclear and cytoplasmic MAH was observed. MAH staining completely disappeared when the antiserum was depleted with purified MAH protein (data not shown), confirming the specificity of the reaction. Pretreatment of cells with 0.05% Triton-X-100 before fixation removed the cytoplasm, and the nuclear localisation of MAH became more visible (Fig. 3 b and b′). MAH proteins in the nuclei accumulated into bright speckles typical for proteins involved in mRNA splicing (39). The speckles were dispersed equally throughout the whole nucleus, but they were excluded from nucleoli. When cells were further extracted with high concentration of salts and digested with nucleases, the DNA content was released, resulting in the disappearance of DAPI staining. Interestingly, however, MAH staining was retained, suggesting the association of MAH with the nuclear matrix (Fig. 3 c and c′ and d and d′). This localization was further supported with biochemical fractionations when more than 50% of MAH remained in the salt and nuclease resistant fraction (data not shown).
Figure 3.
Subcellular localization of MAH. 21PT epithelial cells grown on coverslips were fixed by paraformaldehyde, then permeabilized with 0.5% Triton X-100 (a and a′) or permeabilized twice, first by 0.05% Triton X-100 before fixation, then after fixation as above (b and b′). After double permealization cells were treated with DNase I (c and c′) or DNAse I and RNase A (d and d′). The fixed samples were incubated with a 1:250 dilution of C12 antiserum, and subsequently with rhodamine-conjugated secondary antibody (a–d) followed by counterstaining with the DNA dye DAPI (a′–d′) before analysis with confocal laser beam microscopy.
In conclusion, MAH can be detected both in the cytoplasm and in the nucleus; moreover, the nuclear MAH is tightly bound to the nuclear matrix. Because of this association with the nuclear matrix, this protein has been named MAH (matrix-associated helicase).
MAH Is Colocalized with Splicing Factors.
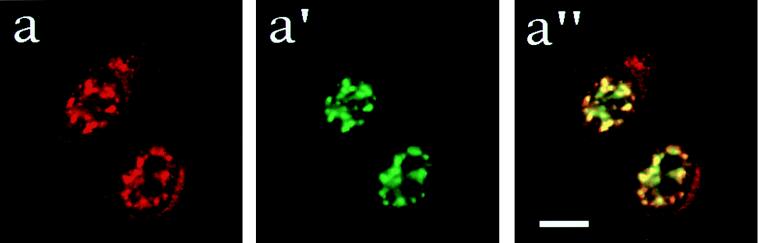
Because nuclear speckles are enriched in pre-mRNA splicing factors (39), and because MAH was distributed in speckles in the nucleus, we considered the possibility that MAH contributes to mRNA splicing. To support this hypothesis, we first made a direct comparison of the relative subnuclear locations of MAH and SC-35, a well-defined pre-mRNA splicing factor (40), by simultaneous staining with anti-MAH and anti-SC35 antibodies combined with FITC or rhodamine-conjugated secondary antibody treatments, respectively (Fig. 4 a, a′, and a"). Confocal images of cells stained with anti-MAH (Fig. 4a) and anti-SC35 (Fig. 4a′) antibodies showed similar staining pattern in the nucleus. Moreover, superimposition of the red and green images in the third column (Fig. 4a") identified the precise colocalization of anti-MAH and anti-SC35 stained speckles, represented by areas of yellow coloration. Identical results were obtained with antibodies against two other non-snRNP splicing factors, B1C8 and B4A11 (41), which, like MAH, are nuclear matrix antigens (data not shown). Thus it appears that MAH colocalizes with three different non-snRNP splicing factors described earlier, and is found in the same nuclear compartment.
Figure 4.
Colocalization of MAH with splicing factors. Fixed and permeabilized HeLa cells were stained with C12 (a), anti-SC35 (a′), or costained with C12 and anti-SC35 (a") primary antibodies. Then C12 antibody was detected with rhodamine conjugated secondary antibody (a and a"), whereas an FITC-conjugated secondary antibody was used to visualize anti-SC35 (a′ and a") antibodies. The staining pattern was examined with a confocal laser beam microscope.
Association of MAH with the pre-mRNA Splicing Complex.
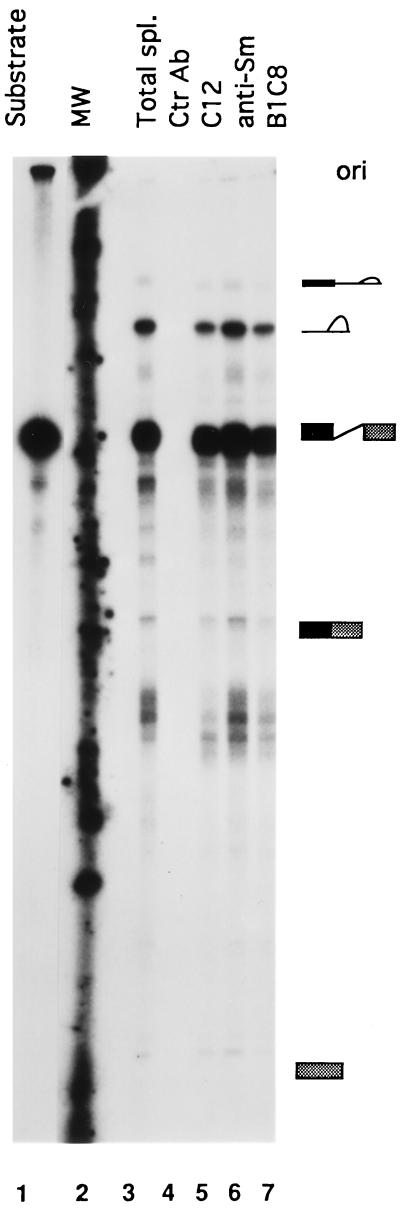
Colocalization with splicing factors suggests that MAH is present in spliceosomes and may be involved in pre-mRNA processing. Therefore, we investigated the interaction of MAH with pre-mRNA splicing complexes assembled in vitro. First, the PIP85A pre-mRNA splicing reaction mixture was immunoprecipitated with C12 antiserum, and then the RNAs were recovered from the immunoprecipitate and analyzed with urea/polyacrylamide gel electrophoresis. The C12 antiserum consistently immunoprecipitated all RNA products, including the precursor RNA, the lariat intermediates, the lariat and the final products (Fig. 5, lane 5). Importantly, the C12 antiserum was as efficient as the positive control Sm and B1C8 antibodies (lanes 5 versus 6 and 7). Because polyclonal sera may react with splicing factors, we used preimmune serum derived from the same animal as C12 and nonrelated anti-thymidylate synthase antiserum as controls. Neither the preimmune serum nor the anti-thymidylate synthase antiserum could immunoprecipitate pre-mRNA precursors or their processed products (lane 4 and data not shown). In addition, the protein A purified C12 antibody was as effective as the nonpurified C12 antibody in immunoprecipitating the splicing complexes (data not shown), further confirming the specificity. A similar pattern of immunoprecipitations was seen with the sequence nonrelated β-globin pre-mRNA complex, and again the C-12 antiserum appeared to be as efficient as the positive control anti-B1C8 (data not shown).
Figure 5.
MAH is present in the in vitro-assembled pre-mRNA splicing complex. HeLa nuclear extract was incubated with 32P-labeled PIP85A splicing substrate for 40 min at 30°C, and then equal volumes of splicing complexes were immunoprecipitated with the indicated antibodies. RNAs were separated on 15% polyacrylamide/urea gel. Lanes 1, in vitro-transcribed PIP85A pre-mRNA; 2, molecular weight marker, [γ-32P]ATP end-labeled 100 bp DNA ladder; 3, total splicing reaction without immunoprecipitation, half of assay reaction than in lanes 4–7. Ctr Ab, preimmune serum (lane 4); C12, anti-MAH antiserum (lane 5); Sm, anti-Sm antibody (lane 6); B1C8, anti-B1C8 antibody (lane 7). Ctr and C12 are equal to 150 μg IgG.  , pre-mRNA;
, pre-mRNA;  , lariat intermediate;
, lariat intermediate;  , lariat product;
, lariat product;  , spliced mRNA;
, spliced mRNA;  , intron.
, intron.
To analyze the possible direct interaction between MAH and the snRNAs of spliceosomes, HeLa nuclear extracts were immunoprecipitated with C12 antiserum, and coimmunoprecipitation of MAH with snRNAs was measured by Northern blot analysis. Unlike the anti-Sm antibody, the C12 antiserum did not immunoprecipitate detectable amount of snRNAs (data not shown), suggesting that MAH is a non-snRNP splicing factor.
In summary, these data together suggest the specific association of MAH with in vitro-assembled spliceosomes without direct association between MAH and snRNAs.
DISCUSSION
We have identified the mammalian helicase MAH as a new component of the pre-mRNA splicing complex. Involvement of helicases in pre-mRNA splicing has already been reported both in yeast and mammalian cells. In S. cerevisiae the DEAD/H box protein family members Prp2, Prp5, Prp16, Prp22, and Prp28 have been implicated in the splicing reactions (13–17). These enzymes modify the structure of pre-mRNAs and prepare them for the catalytic steps. They may also help for spliceosome disassembly by releasing snRNA from the snRNP particles (7).
The large number of putative helicases involved in yeast pre-mRNA splicing predicts that several helicases may function as splicing factors in mammalian cells. First, the ability of the human RNA helicase I (HRH1) to restore splicing activity of the temperature sensitive phenotype of the Prp22 yeast mutant has been demonstrated (18). Then another putative mammalian helicase, HEL117, has been cloned from rat and was suggested to participate in pre-mRNA splicing (19). Recently, U5–200kD and its yeast homologue Snu246 were shown to be the first putative RNA helicases as intrinsic components of snRNPs (20).
Comparison of sequences of the above listed superfamily II splicing helicases with MAH, however, did not describe a clear homology. On the other hand, our search of databases for MAH-related proteins revealed that MAH is homologous rather to a group of helicases designated helicase superfamily I (9). This protein group includes Upf1, an activator of nonsense mRNA decay (37, 38), Rent-1, which appears to be a mammalian functional homologue of Upf1 (42), Sen-1, a positive effector of tRNA splicing endonuclease (43), and Mov-10 with unknown function. Although the overall identity between MAH and these proteins does not exceed 30%, the helicase motif regions are much more similar, with an identity of up to 90%.
MAH presents especially strong structural as well as functional similarities to the S. cerevisiae protein Upf1, the most well-characterized member of this group of helicases. MAH and Upf1 have strongly homologous helicase motifs with 65% identity, and carry a DEAD/H box-like motif typical for RNA helicases which is DEcAq in MAH and DEstq in Upf1. Consistent with this, both MAH and Upf1 have similar NTP hydrolysis kinetics, affinity to single-stranded DNA and unwinding activity on partially double-stranded DNA. Upf1 was also shown to have RNA helicase activity (37). Our attempts with MAH to demonstrate RNA helicase activity have failed thus far, possibly due to the stringent sequence and/or structural requirements in RNA helices that are difficult to mimic in vitro in the absence of assembling spliceosome. For similar reasons, RNA helicase activity has not been yet demonstrated with the yeast RNA helicases of the Prp family involved in RNA splicing (7).
Disruption of the UPF1 gene results in stabilization of nonsense-containing mRNAs and leads to a nonsense suppression phenotype, suggesting the involvement of Upf1 in nonsense mRNA decay (44). This function is accompanied with a cytoplasmic localization of Upf1. Because MAH was found to be present in the cytoplasm with immunostaining, it might have a similar function in mammalian cells. In contrast to Upf1, a significant part of MAH is localized to the nuclear matrix. This nuclear MAH is colocalized with splicing factors, and the spliceosomes from nuclear extracts can be coimmunoprecipitated with MAH using an anti-MAH antibody. These data suggest that in addition to a possible cytoplasmic role, MAH as a helicase may contribute to the regulation of pre-mRNA splicing in the nucleus.
MAH resembles non-snRNP binding factors of the splicing complex because it does not associate with snRNAs, as determined by an immunoprecipitation assay combined with Northern blot analysis. Consistent with this, the molecular weight of MAH is significantly different from all snRNP binding proteins, except for two (102 and 110 kDa) present in the U5 snRNP particle (45). Although these two proteins have not been cloned yet, they do not seem to be identical with MAH, because the helicase activity in this complex is attributed to another already cloned helicase (U5-200kD) (20). In addition, MAH does not have any motifs typical for Sm proteins and cannot be immunoprecipitated with anti-Sm antibody (46, 47), confirming that MAH is not a common snRNA binding protein.
In conclusion, we have identified MAH as a non-snRNP binding factor present at least transiently in the mammalian splicing complex. We propose that MAH contributes to pre-mRNA splicing carried out in the nuclear matrix, most probably via its helicase activity. Because MAH is also present in the cytoplasm, it may have an additional function—possibly similar to that of the yeast homologue Upf1—in the metabolism of mRNAs.
Acknowledgments
We thank Benjamin J. Blencowe, Jeffrey A. Nickerson, and Bruce M. Spiegelman for kindly providing reagents. We are especially grateful to Benjamin J. Blencowe for valuable suggestions, Nuala O’Leary and Weimin Zhu for the excellent technical work, and Heide Ford for critical reading of the manuscript. This work was supported by grants from The Council for Tobacco Research (CTR 3415R1) and the National Institutes of Health (GM 24571).
ABBREVIATIONS
- MAH
matrix-associated helicase
- snRNP
small ribonucleoprotein
- snRNA
small nuclear RNA
- DAPI
4′,6′-diamidino-2-phenylindole
- FITC
fluorescein isothiocyanate
References
- 1.Brody J, Abelson J. Science. 1985;228:1344–1349. [Google Scholar]
- 2.Frendewey D, Keller N. Cell. 1985;42:355–367. doi: 10.1016/s0092-8674(85)80131-8. [DOI] [PubMed] [Google Scholar]
- 3.Grabowski P J, Seiler S, Sharp P A. Cell. 1985;42:345–353. doi: 10.1016/s0092-8674(85)80130-6. [DOI] [PubMed] [Google Scholar]
- 4.Berezney R, Coffey D S. Biochem Biophys Res Commun. 1974;60:1410–1417. doi: 10.1016/0006-291x(74)90355-6. [DOI] [PubMed] [Google Scholar]
- 5.Luhrmann R, Kastner B, Bach M. Biochim Biophys Acta. 1990;1087:265–292. doi: 10.1016/0167-4781(90)90001-i. [DOI] [PubMed] [Google Scholar]
- 6.Lamm G M, Lamond A I. Biochim Biophys Acta. 1993;1173:247–265. doi: 10.1016/0167-4781(93)90122-t. [DOI] [PubMed] [Google Scholar]
- 7.Kramer A. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 8.Matson S W, Kaiser-Rogers K A. Annu Rev Biochem. 1990;59:289–329. doi: 10.1146/annurev.bi.59.070190.001445. [DOI] [PubMed] [Google Scholar]
- 9.Koonin E V. Trends Biochem Sci. 1992;17:495–497. doi: 10.1016/0968-0004(92)90338-a. [DOI] [PubMed] [Google Scholar]
- 10.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. Nature (London) 1988;333:22. doi: 10.1038/333022a0. [DOI] [PubMed] [Google Scholar]
- 11.Hodgman T C. Nature (London) 1988;333:22–23. doi: 10.1038/333022b0. [DOI] [PubMed] [Google Scholar]
- 12.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. Nucleic Acids Res. 1988;17:4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Company M, Arenas J, Abelson J. Nature (London) 1991;349:478–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- 14.Chen J H, Lin R J. Nucleic Acids Res. 1990;18:6447. doi: 10.1093/nar/18.21.6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalbadie-McFarland G, Abelson J. Proc Natl Acad Sci USA. 1990;87:4236–4240. doi: 10.1073/pnas.87.11.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burgess S, Couto J R, Guthrie C. Cell. 1990;60:705–717. doi: 10.1016/0092-8674(90)90086-t. [DOI] [PubMed] [Google Scholar]
- 17.Strauss E J, Guthrie C. Genes Dev. 1991;5:629–641. doi: 10.1101/gad.5.4.629. [DOI] [PubMed] [Google Scholar]
- 18.Ono Y, Ohno M, Shimura Y. Mol Cell Biol. 1994;14:7611–7620. doi: 10.1128/mcb.14.11.7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sukegawa J, Blobel G. J Biol Chem. 1995;270:15702–15706. doi: 10.1074/jbc.270.26.15702. [DOI] [PubMed] [Google Scholar]
- 20.Lauber J, Fabrizio P, Teigelkamp S, Lane W S, Hartmann E, Luhrmann R. EMBO J. 1996;15:4001–4015. [PMC free article] [PubMed] [Google Scholar]
- 21.Singh H, LeBowitz J H, Baldwin A S, Jr, Sharp P A. Cell. 1988;52:415–423. doi: 10.1016/s0092-8674(88)80034-5. [DOI] [PubMed] [Google Scholar]
- 22.Fridovich-Keil J L, Gudas J M, Dou Q D, Bouvard I, Pardee A B. Cell Growth Diff. 1991;2:67–76. [PubMed] [Google Scholar]
- 23.Dou Q P, Fridovich-Keil J L, Pardee A B. Proc Natl Acad Sci USA. 1991;88:1157–1161. doi: 10.1073/pnas.88.4.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guan K L, Dixon J E. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 25.Smith D B, Johnson K S. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 26.Harosh I, Naumovski L, Friedberg E C. J Biol Chem. 1989;264:20532–20539. [PubMed] [Google Scholar]
- 27.Seo Y S, Lee S H, Hurwitz J. J Biol Chem. 1991;266:13161–13170. [PubMed] [Google Scholar]
- 28.Moore M J, Query C C, Sharp P A. In: The RNA World. Gesteland R, Atkins R J, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 303–357. [Google Scholar]
- 29.Barabino S M, Blencowe B J, Ryder U, Sproat B S, Lamond A I. Cell. 1990;63:293–302. doi: 10.1016/0092-8674(90)90162-8. [DOI] [PubMed] [Google Scholar]
- 30.Grabowski P J, Padgett R A, Sharp P A. Cell. 1984;37:415–427. doi: 10.1016/0092-8674(84)90372-6. [DOI] [PubMed] [Google Scholar]
- 31.Lamond A I, Konarska M M, Sharp P A. Genes Dev. 1987;1:532–543. doi: 10.1101/gad.1.6.532. [DOI] [PubMed] [Google Scholar]
- 32.Blencowe B J, Carmo-Fonseca M, Behrens S-E, Luhrmann R, Lamond A I. J Cell Sci. 1993;105:685–697. doi: 10.1242/jcs.105.3.685. [DOI] [PubMed] [Google Scholar]
- 33.Mizuta T R, Fukita Y, Miyoshi T, Shimizu A, Honjo T. Nucleic Acids Res. 1993;21:1761–1766. doi: 10.1093/nar/21.8.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shieh S Y, Stellrecht C M, Tsai M J. J Biol Chem. 1995;270:21503–21508. doi: 10.1074/jbc.270.37.21503. [DOI] [PubMed] [Google Scholar]
- 35.Fukita Y, Mizuta T R, Shirozu M, Ozawa K, Shimizu A, Honjo T. J Biol Chem. 1993;268:17463–70. [PubMed] [Google Scholar]
- 36.Kerr D, Khalili K. J Biol Chem. 1991;266:15876–15881. [PubMed] [Google Scholar]
- 37.Czaplinski K, Weng Y, Hagan K W, Peltz S W. RNA. 1995;1:610–623. [PMC free article] [PubMed] [Google Scholar]
- 38.Leeds P, Peltz S W, Jacobson A, Culbertson M R. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 39.Spector D L. Curr Opin Cell Biol. 1993;5:442–448. doi: 10.1016/0955-0674(93)90009-f. [DOI] [PubMed] [Google Scholar]
- 40.Fu, X. D., Mayeda, A., Maniatis, T. & Krainer, A. R. (1992) Proc. Natl. Acad. Sci. USA 11224–11228. [DOI] [PMC free article] [PubMed]
- 41.Blencowe B J, Nickerson J A, Issner R, Penman S, Sharp P A. J Cell Biol. 1994;127:593–607. doi: 10.1083/jcb.127.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perlick H A, Medghalchi S M, Spencer F A, Kendzior R J, Jr, Dietz H C. Proc Natl Acad Sci USA. 1996;93:10928–10932. doi: 10.1073/pnas.93.20.10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeMarini D J, Winey M, Ursic D, Webb F, Culbertson M R. Mol Cell Biol. 1992;12:2154–2164. doi: 10.1128/mcb.12.5.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He F, Peltz S W, Donahue J L, Rosbash M, Jacobson A. Proc Natl Acad Sci USA. 1993;90:7034–7038. doi: 10.1073/pnas.90.15.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Behrens S E, Luhrmann R. Genes Dev. 1991;5:1439–1452. doi: 10.1101/gad.5.8.1439. [DOI] [PubMed] [Google Scholar]
- 46.Hermann H, Fabrizio P, Raker V A, Foulaki K, Hornig H, Brahms H, Luhrmann R. EMBO J. 1995;14:2076–2088. doi: 10.1002/j.1460-2075.1995.tb07199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seraphin B. EMBO J. 1995;14:2089–2098. doi: 10.1002/j.1460-2075.1995.tb07200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]