Abstract
We investigated the mechanisms involved in imipenem resistance in 23 clinical strains of Acinetobacter baumannii. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis showed the presence of a 30-kDa protein in imipenem-intermediate A. baumannii (IIAB) and imipenem-resistant A. baumannii (IRAB) strains; this protein was almost undetectable in imipenem-susceptible A. baumannii (ISAB) strains. The 30-kDa protein was identified as an OXA-51-like carbapenemase using two-dimensional gel electrophoresis and mass spectrometry. Similar to other recent findings, blaOXA-51-like genes were found to exist in all 23 clinical strains; however, the transcript levels of blaOXA-51-like in the IIAB and IRAB were higher than in the ISAB strains using reverse transcriptase PCR (RT-PCR) and real-time RT-PCR assays. This change was due to the presence of an insertion sequence, ISAba1, upstream of blaOXA-51-like in the IIAB and IRAB strains that was not present in the ISAB strains. The introduction of blaOXA-66 (a blaOXA-51-like gene), identified in ISAB ab1254 and IRAB ab1266, into Escherichia coli TOP10 cells resulted in 3.95-fold and 7.90-fold elevations in resistance to imipenem, respectively. Furthermore, when ISAB ab8 and ISAB ab1254 and their in vitro-selected imipenem-resistant mutants ISAB ab8(r) and ISAB ab1254(r) were compared, the results showed no change in the blaOXA-66/blaOXA-51-like gene sequences, in expression of the gene, and in the outer membrane protein profiles. However, there was a four- to eightfold reduction in imipenem resistance upon adding carbonyl cyanide m-chlorophenylhydrazone. Taken together, these results suggest that the OXA-66/OXA-51-like carbapenemase contributes to intrinsic resistance to imipenem; however, drug export by an efflux pump may be more important and/or occur more frequently in imipenem-resistant A. baumannii. Furthermore, this is the first report of a Taiwanese strain of an OXA-66/OXA-51-like carbapenemase that confers imipenem resistance in A. baumannii.
Acinetobacter baumannii accounts for a large percentage of nosocomial infections including pneumonia, bacteremia, skin infections, wound infections, and urinary tract infections (2, 19). Increasingly, multidrug resistance strains of A. baumannii have become common in hospitals worldwide and especially in intensive care units (10, 12, 30) and burn units (2, 31, 32, 40). The types of resistance include many commonly used antibiotics such as aminoglycosides, fluoroquinolones, and β-lactams, although carbapenems are the most used antimicrobial drugs. However, an increasing number of recent studies reported the emergence of clinical A. baumannii strains that are resistance to imipenem (7, 9, 27).
The mechanism of resistance to carbapenems in A. baumannii has mostly been ascribed to the acquisition of carbapenemases (1, 26) or to synergistic effects between β-lactamases with an ability to hydrolyze carbapenems and decreased expression of certain penicillin-binding proteins (13, 14). The carbapenemases in A. baumannii known to be involved in carbapenem resistance are the class B metallo-beta-lactamases and the class D carbapenem-hydrolyzing oxacillinases (OXA type) (38, 39, 42). Recently, the number of OXA-type carbapenemases has increased substantially, and they have been divided into eight distantly related groups (39). Of these OXA-type carbapenemases, the OXA-51-like group consists of a large number of closely related variants from a wide range of geographical origins (4, 5). Furthermore, there is increasing evidence that A. baumannii encodes a naturally occurring carbapenemase gene, blaOXA-51-like, that is intrinsic to the species (15, 22, 37, 41). Similar to most of the other OXA-type carbapenemases, the OXA-51-like enzymes show weak carbapenemase activity; however, the presence of the insertion sequence ISAba1 upstream of the blaOXA-51-like gene may provide a promoter that allows the overproduction of carbapenemase, and this results in carbapenem resistance (36). A few reports have demonstrated that a loss or decreased expression of an outer membrane protein or porin may be involved in resistance to carbapenems (3, 6, 11, 20, 24, 33, 35). However, there is less research supporting the association of increased expression of an outer membrane protein, which may form a component of an efflux pump, with carbapenem resistance (17, 21, 34).
In this study, we investigated the outer membrane proteins of clinical strains of A. baumannii isolated from three hospitals at different times in Taiwan. Using sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), two-dimensional gel electrophoresis (2-DE), and mass spectrometry, we have been able to identify an OXA-66/OXA-51-like carbapenemase that is overexpressed in imipenem-intermediate and imipenem-resistant strains of A. baumannii. We also show here that the overexpression of OXA-66 is able to elevate resistance to imipenem in Escherichia coli. In addition to carbapenemase, we speculate that another additional mechanism(s) may be involved in imipenem resistance in A. baumannii.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Twenty-three strains of A. baumannii were randomly selected from many clinical isolates that had been collected at three hospitals in Taiwan over a long period. The collection dates were 1993 (3 strains), 1994 (2 strains), 2001 (6 strains), and 2003 (12 strains) (Table 1). These strains were confirmed as being A. baumannii using PCR-based ribosomal DNA restriction analysis (data not shown) and using A. baumannii (ATCC 19606) as a reference strain. Escherichia coli DH5α and TOP10 were used as hosts for the cloning and expression experiments, respectively. The plasmid pGEM-T Easy (Promega) was used as the cloning vector. Plasmid pBAD/Myc-His was used as the expression vector. E. coli and A. baumannii cultures were routinely grown in Luria broth and tryptic soy broth, respectively at 37°C for 18 h.
TABLE 1.
Antibiotic susceptiblities of the A. baumannii strains used in this studya
| Strain | MIC (μg ml−1)a
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAZ | CTX | FEP | PIP | TZPb | ATM | IMP | MEM | CIP | AMK | GEN | SXT | |
| ab3 | 48 | >256 | 6 | >256 | >256 | 24 | 1 | 0.38 | 0.125 | 3 | 256 | >32 |
| ab4 | 32 | >256 | 16 | >256 | >256 | 24 | 1 | 0.5 | 0.25 | 8 | 256 | >32 |
| ab8 | 48 | >256 | 24 | >256 | >256 | 24 | 1 | 0.5 | 0.38 | 12 | >256 | >32 |
| ab12 | 48 | >256 | 16 | >256 | >256 | 16 | 1 | 0.5 | 0.25 | >256 | >256 | >32 |
| ab20 | 24 | >256 | 24 | >256 | >256 | 12 | 1.5 | 0.75 | 3 | 128 | >256 | >32 |
| ab1308 | >256 | >256 | >256 | >256 | >256 | 64 | 2 | 0.75 | >32 | >256 | >256 | >32 |
| ab1254 | >256 | >256 | 32 | >256 | >256 | 32 | 3 | 0.75 | >32 | >256 | >256 | >32 |
| ab1316 | >256 | >256 | >256 | >256 | >256 | 24 | 3 | 1.5 | >32 | >256 | >256 | >32 |
| ab1297 | >256 | >256 | 32 | >256 | >256 | 32 | 8 | 32 | >32 | >256 | >256 | >32 |
| ab1265 | >256 | >256 | 24 | >256 | >256 | 32 | >32 | >32 | >32 | >256 | >256 | >32 |
| ab1266 | >256 | >256 | 32 | >256 | >256 | 48 | >32 | >32 | >32 | >256 | >256 | >32 |
| abh9 | >256 | >256 | >256 | >256 | >256 | 24 | 32 | >32 | >32 | >256 | >256 | >32 |
| abh11 | >256 | >256 | 48 | >256 | >256 | 32 | 32 | >32 | >32 | >256 | >256 | >32 |
| abh13 | >256 | >256 | >256 | >256 | >256 | 16 | 32 | >32 | >32 | >256 | >256 | >32 |
| abh14 | >256 | >256 | 256 | >256 | >256 | 24 | 32 | >32 | >32 | >256 | >256 | >32 |
| abh17 | >256 | >256 | 48 | >256 | >256 | 64 | 32 | 16 | >32 | >256 | >256 | >32 |
| abh18 | >256 | >256 | 256 | >256 | >256 | 16 | 32 | 32 | >32 | >256 | >256 | >32 |
| abh20 | >256 | >256 | 256 | >256 | >256 | 16 | 32 | 32 | >32 | >256 | >256 | >32 |
| abh26 | >256 | >256 | 256 | >256 | >256 | 16 | 32 | 32 | >32 | >256 | >256 | >32 |
| abh29 | >256 | >256 | 96 | >256 | >256 | 16 | 32 | >32 | >32 | >256 | >256 | >32 |
| abh36 | >256 | >256 | 256 | >256 | >256 | 12 | 32 | >32 | >32 | >256 | >256 | >32 |
| abh37 | >256 | >256 | 128 | >256 | >256 | 32 | 32 | 32 | >32 | >256 | >256 | >32 |
| abh39 | >256 | >256 | 256 | >256 | >256 | 32 | 32 | 32 | >32 | >256 | >256 | >32 |
Antibiotic abbreviations: PIP, piperacillin; TZP, piperacillin-tazobactam; CAZ, ceftazidime; CTX, cefotaxime; FEP, cefepime; ATM, aztreonam; IMP, imipenem; MEM, meropenem; CIP, ciprofloxacin; AMK, amikacin; GEN, gentamicin; SXT, trimethoprim-sulfamethoxazole.
Contains 4 μg ml−1 of tazobactam.
Multistep selection resistance.
The generation of imipenem-resistant mutants from the imipenem-susceptible A. baumannii (ISAB) strain ab8 and ISAB strain ab1254 was done according to procedures described previously by Pankuch et al. (28). Briefly, tubes containing antibiotic concentrations that were three doubling dilutions above and three doubling dilutions below the MIC were used. Inocula were adjusted to achieve a final concentration of 5 × 105 CFU ml−1 in each tube. The tubes were incubated at 37°C for 24 h. For each subsequent daily passage, 10 μl inocula was taken from the first tube containing a subinhibitory drug concentration and subcultured in the next-passage tubes containing each diluted drug. Daily subculturing was done until the MIC was more than 64 μg/ml. Mutants from the last subculturing tube were inoculated onto drug-free LB plates. Twenty resistant clones were subcultured on drug-free medium for 10 days, and they were then frozen at −80°C. Mutants resistant to imipenem, designated ISAB ab8R(r) and ISAB ab1254(r), were chosen to examine their blaOXA-66/blaOXA-51-like gene sequences and the expression of this gene. Furthermore, in these strains, the outer membrane protein profiles were analyzed together with efflux pump function.
Antibiotics for susceptibility testing.
The antibiotics used for the Etest were amikacin, gentamicin, ceftazidime, cefotaxime, cefepime, aztreonam, piperacillin, piperacillin-tazobactam, imipenem, meropenem, ciprofloxacin, and trimethoprim-sulfamethoxazole. Carbonyl cyanide m-chlorophenylhydrazone (CCCP), imipenem, and meropenem were obtained from Sigma-Aldrich Corp. (St. Louis, MO) in order to assess efflux pump function. The MICs were determined by the broth dilution method as recommended by the Clinical and Laboratory Standards Institute (formerly National Committee for Clinical Laboratory Standards) (25) or by Etest strips (AB Biodisk, Solna, Sweden) according to the manufacturer's recommendations.
Preparation of bacterial outer membrane.
The outer membrane fraction was prepared according to methods described previously by Molloy et al. (23). Briefly, the bacterial cells were disrupted by ultrasonic disintegration with a Vibra-cell VCX 600 (Sonics & Materials, Inc., CT), and the unbroken cells were removed by centrifugation at 2,500 × g for 10 min. The supernatant was added to an equal volume of ice-cold 0.1 M sodium carbonate (pH 11) and stirred slowly on ice overnight. The carbonate-treated membranes were collected by ultracentrifugation in a Beckman 55.2 Ti rotor at 115,000 × g for 1 h at 4°C. The supernatant was discarded, and the membrane pellets were rinsed with 2 ml 50 mM Tris-HCl (pH 7.3) and resuspended in the same buffer for SDS-PAGE. Alternatively, the rinsed membrane pellets were resuspended in 2D lysis buffer {7 M urea, 2 M thiourea, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propaneslulfonate (CHAPS), 40 mM Tris base, 0.5% (vol/vol) Triton X-100, 30 mM dithiothreitol, and 0.5% (vol/vol) Biolytes 3 to 10} for 2-DE. The protein concentration was determined using the Bradford method (3a).
SDS-PAGE.
The fraction containing the outer membrane protein as described above was analyzed by SDS-PAGE using 12% (wt/vol) polyacrylamide gels. All samples were incubated for 5 min in a boiling water bath before they were subjected to electrophoresis. The resolved proteins were visualized by silver staining.
2-DE.
The procedure for 2-DE was done according to methods described previously by Molloy et al. (23). In brief, a sample of the outer membrane fraction was loaded onto an 18-cm (pH 4 to 7) IPG (Amersham Pharmacia Biotec, Uppsala, Sweden). Isoelectric focusing (IEF) was conducted for a total of 50,000 V h. For the second dimension, the IPG was embedded onto a 12% SDS-PAGE gel with 0.5% (wt/vol) agarose, 25 mM Tris base, 192 mM glycine, 0.1% SDS, and a few grains of bromophenol blue. The running buffer for the SDS-PAGE consisted of 25 mM Tris base, 192 mM glycine, and 0.1% SDS. The gels were run at 14°C and 50 mA for 9 h. Protein spots were visualized using silver staining. Spot differences were confirmed by three independent experiments.
Peptide sequencing and protein identification.
In order to identify specific proteins, tryptic in-gel digestion of the protein spots of interest was followed by tandem mass spectrometry performed as described previously by Kristensen et al. (18) by using a Q-TOF instrument (Micromass, Manchester, United Kingdom) at the National Yang-Ming University Genome Center. Peptide masses were queried against the entries for microorganisms in the NCBInr database using the MASCOT program (http://www.matrixscience.com).
Cloning and sequencing of blaOXA-66.
To obtain the blaOXA-51-like gene from clinical strains of A. baumannii, primers OXA-F and OXA-R (Table 2) were designed based on the DNA sequences of members of OXA-51-like family. ISAB ab1254 or imipenem-resistant A. baumannii (IRAB) ab1266 genomic DNA was used as the template to amplify the blaOXA-51-like gene using PCR. The PCR product from ISAB ab1254 or IRAB ab1266 was then ligated into the pGEM-T Easy vector (Promega) according to the manufacturer's instructions. The DNA sequences of both inserts from the resulting plasmids, pGEM-oxa1254 and pGEM-oxa1266, were determined using an Applied Biosystems sequencer (ABI 3100). Sequence similarity was estimated using BLAST (http://www.ncbi.nlm.nih.gov).
TABLE 2.
Sequences of primers designed for this study
| Primer | Sequence | Reference or source |
|---|---|---|
| OXA-F | 5′-CCATGGCAATGAACATTAAAGCACTCTTAC-3′ | This study |
| OXA-R | 5′-CTATAAAATACCTAATTGTTC-3′ | This study |
| OXA-RTF | 5′-GATTTAGCTCGTCGTATTGGA-3′ | This study |
| OXA-RTR | 5′-AAGCGTTTTATTAGCTAGCTTG-3′ | This study |
| 16sRNA-F | 5′-CAGCTCGTGTCGTGAGATGT-3′ | 17 |
| 16sRNA-R | 5′-CGTAAGGGCCATGATGACTT-3′ | 17 |
| ISAb-F1 | 5′-AGTTGCACTTGGTCGAATGAA-3′ | This study |
| ISAb-F2 | 5′-TTGAAAATACGCGCTTGACAGA-3′ | This study |
| ISAb-F3 | 5′-CTCTGTACACGACAAATTTCAC-3′ | This study |
| OXA/IS-R | 5′-CCATAGCTTTGTTGAGTTTGG-3′ | This study |
Cloning and sequencing of the ISAba1 gene upstream of blaOXA-66/blaOXA-51-like.
To determine whether the ISAba1 gene was present upstream of the blaOXA-66/blaOXA-51-like genes and the ISAba1 gene's orientation, two primer sets, ISAb-F1 and OXA/IS-R and ISAb-F2 and OXA/IS-R (Table 2), were used for PCR. These primers were designed based on the sequence of the ISAba1 gene and based on the sequence of blaOXA-66/blaOXA-51-like. After the orientation was determined, ISAb-F3 (Table 2), based on the 3′-end sequence of ISAba1, and OXA/IS-R were used with PCR to obtain a complete copy of the ISAba1 gene and part of the adjacent blaOXA-66/blaOXA-51-like gene from imipenem-intermediate A. baumannii (IIAB) ab1297, IRAB ab1266, and IRAB abh9. These three PCR products were cloned, sequenced, and analyzed using the same procedure as described above for the cloning and sequencing of blaOXA-66.
Expression of blaOXA-66 in E. coli.
The result of sequence analyses indicated that both blaOXA-51-like genes from ISAB ab1254 and IRAB ab1266 are indeed blaOXA-66 genes. The complete coding sequences of the blaOXA-66 genes from ISAB ab1254 and IRAB ab1266 were obtained using NcoI/PstI double digestions of pGEM-oxa1254 and pGEM-oxa1266, respectively. The 831-bp NcoI-PstI fragments containing the blaOXA-66 from ISAB ab1254 and IRAB ab1266 were then directionally inserted into pBAD/Myc-His that had been cut with NcoI/PstI to construct pBAD/Myc-His-oxa66-1254 and pBAD/Myc-His-oxa66-1266, respectively. Plasmids pBAD/Myc-His, pBAD/Myc-His-oxa66-1254, and pBAD/Myc-His-oxa66-1266 were then introduced into E. coli TOP10 cells. The transformed cells containing the above-described three plasmids were grown at 37°C in LB medium containing 50 μg of ampicillin per ml. When the cultures had reached an optical density at 600 nm of 0.5, the cells were plated onto a Mueller-Hinton agar plate containing 50 μg of ampicillin per ml for the imipenem susceptibility test using the Etest method.
RT-PCR and real-time RT-PCR.
Total RNA was isolated from 1 × 109 cells of A. baumannii using an RNeasy Mini kit (QIAGENE) according to the manufacturer's instructions. The concentrations and the quality of the RNA in each sample were determined by measuring their absorbance at 260 nm. All RNAs were adjusted to a concentration of 100 ng μl−1. The detection of A. baumannii 16S rRNA was carried out using previously described primers (17), and it was employed as an internal control. The reverse transcriptase PCR (RT-PCR) protocol was done using the SuperScript first-strand synthesis system (Invitrogen Corporation, Carlsbad, CA). Briefly, the reaction mixtures contained 500 ng total RNA and appropriate amounts of reagents and primers. These were first incubated at 42°C for 50 min to synthesize cDNA and then heated to 70°C for 15 min to inactivate the reverse transcriptase. Next, they were subjected to 25 thermal cycles (95°C for 30 s, 56°C for 60 s, and 72°C for 90 s) to allow PCR amplification. Real-time RT-PCR was performed using a capillary real-time thermal cycle (LightCycler; Roche Diagnostics), and results were normalized against the 16S rRNA mRNA. Amplification was carried out in a 10-μl final volume containing 1 μl from the LightCycler FastStart DNA Master SYBR green kit (Roche Diagnostics), 1 μl of primer mix (5 nM each), 1.2 μl MgCl2 (4 mM final concentration), 1.8 μl H2O, and 5 μl cDNA. The following run protocol was used: denaturation at 95°C for 10 min, followed by 30 cycles consisting of 10 s at 95°C for denaturation, 5 s at 56°C for annealing, and 10 s at 72°C for elongation. The primers used to perform the RT-PCR and real-time RT-PCR experiments are described in Table 2.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been submitted to the GenBank nucleotide sequence database under the accession numbers DQ987478 for blaOXA-66 from ISAB ab1254, DQ987479 for blaOXA-66 from IRAB ab1266, EF433474 for ISAba1and part of blaOXA-51-like from IIAB ab1297, EF433475 for ISAba1 and part of blaOXA-66 from IRAB ab1266, and EF433476 for ISAba1 and part of blaOXA-51-like from IRAB abh9.
RESULTS
Susceptibility testing.
Antibiotic susceptibility testing revealed that the 23 clinical A. baumannii strains were resistant to cefotaxime, ceftazidime, piperacillin, trimethoprim-sulfamethoxazole, and gentamicin and were intermediate or resistant to aztreonam; however, the addition of tazobactam did not reduce the MICs of piperacillin. The strains isolated in 2001 and 2003 were resistant to all antibiotics tested except imipenem (Table 1). Overall, three strains from 1993, two strains from 1994, and three out of six strains from 2001 were both imipenem susceptible (ISAB) and meropenem susceptible, one out of six strains from 2001 was imipenem intermediate (IIAB) but meropenem resistant, and all 12 strains from 2003 and two out of six strains from 2001 were both imipenem resistant (IRAB) and meropenem resistant (Table 1).
Association between imipenem resistance and overexpression of the OXA-51-like carbapenemase in multidrug-resistant clinical strains of A. baumannii.
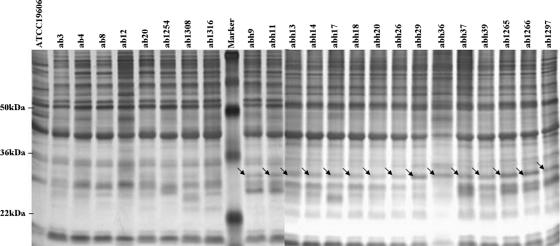
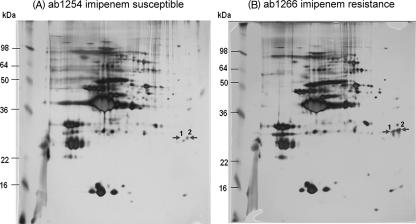
The outer membrane proteins from 23 clinical strains of A. baumannii were analyzed by SDS-PAGE. The expression level of a unique protein band at about 30 kDa was significantly higher in the IIAB and IRAB strains than in the ISAB strains (Fig. 1). Based on the pulsed-field gel electrophoresis banding pattern using ApaI restriction enzyme digestion (see Fig. S1 in the supplemental material), the most genetic closely related strains, ISAB ab1254 and IRAB ab1266, together with ISAB ab1316 and IRAB abh9, were selected for further analysis by 2-DE. The levels of expression of two spots, spots 1 and 2 (near pI 7.0), were higher in the IRAB strains than in the ISAB strains, and this is exemplified in Fig. 2 by strains ab1254 (A) and ab1266 (B). These two spots from the 2-DE were cut out, washed, reduced, alkylated, digested in gel with trypsin, extracted from the gel, and subjected to peptide mass fingerprinting by tandem mass spectrometry. The results revealed that these two spots were made up of peptides in reasonable agreement with predicted values for an OXA-51-like carbapenemase, with a relative molecular mass 30.548 kDa and a pI value of 9.07; the protein coverage was between 19% and 32%. Similar results were obtained for ISAB ab1316 and IRAB abh9 (data not shown).
FIG. 1.
Overexpression of a 30-kDa outer membrane protein band in imipenem-resistant clinical strains of A. baumannii. Outer membrane fractions of the various strains, each containing 10 μg protein, were analyzed by SDS-PAGE on 12% polyacrylamide gels and silver stained. Lanes 1 to 9, ISAB ATCC 19606, ab3, ab4, ab8, ab12, ab20, ab1254, ab1308, and ab1316, respectively; lanes 11 to 24, IRAB abh9, abh11, abh13, abh14, abh17, abh18, abh20, abh26, abh29,abh36, abh37, abh39, ab1265, and ab1266, respectively; lane 25, IIAB ab1297; and lane 10, molecular mass markers. The molecular masses of three protein markers are indicated in the left margin. The arrows indicate the 30-kDa protein band.
FIG. 2.
2-DE analysis of the outer membrane proteins comparing ISAB ab1254 (A) and IRAB ab1266 (B). The molecular masses of six protein markers are indicated in the left margin. The protein spots that were significantly changed are indicated by arrows.
Cloning and sequencing of the blaOXA-51-like gene.
In order to identify the exact type of OXA-51-like carbapenemase in these organisms, the gene encoding this enzyme was obtained by PCR amplification using OXA-F as the forward primer and OXA-R as the reverse primer (Table 2). These primers were designed based on an alignment of the nucleotide sequences of blaOXA-51 and its closely related variants. An approximately 800-bp PCR product was obtained from both ISAB ab1254 and IRAB ab1266. The two PCR products were cloned into the pGEM-T Easy vector to construct pGEM-oxa1254 and pGEM-oxa1266, respectively. DNA sequence analysis of the inserts of both plasmids, pGEM-oxa1254 and pGEM-oxa1266, revealed an 825-bp open reading frame encoding a 274-amino-acid protein (Fig. 3). DNA sequence alignment showed that the blaOXA-51-like gene from ISAB ab1254 is 100% identical to a blaOXA-66 gene of A. baumannii deposited in the GenBank databank. Only one base change was present in the blaOXA-66 gene sequence of IRAB ab1266 relative to ISAB ab1254. This was a C-to-G change at nucleotide position 499, and this resulted in a leucine (CTG)-to-valine (GTG) change at amino acid position 167 (Fig. 3). Furthermore, an 825-bp PCR product was also obtained using OXA-F and OXA-R from all other 21 tested clinical strains of A. baumannii (data not shown). Thus, the gene encoding OXA-66 or a very closely related variant (OXA-51-like) carbapenemase seems to exist in all clinical A. baumannii strains analyzed in this study.
FIG. 3.
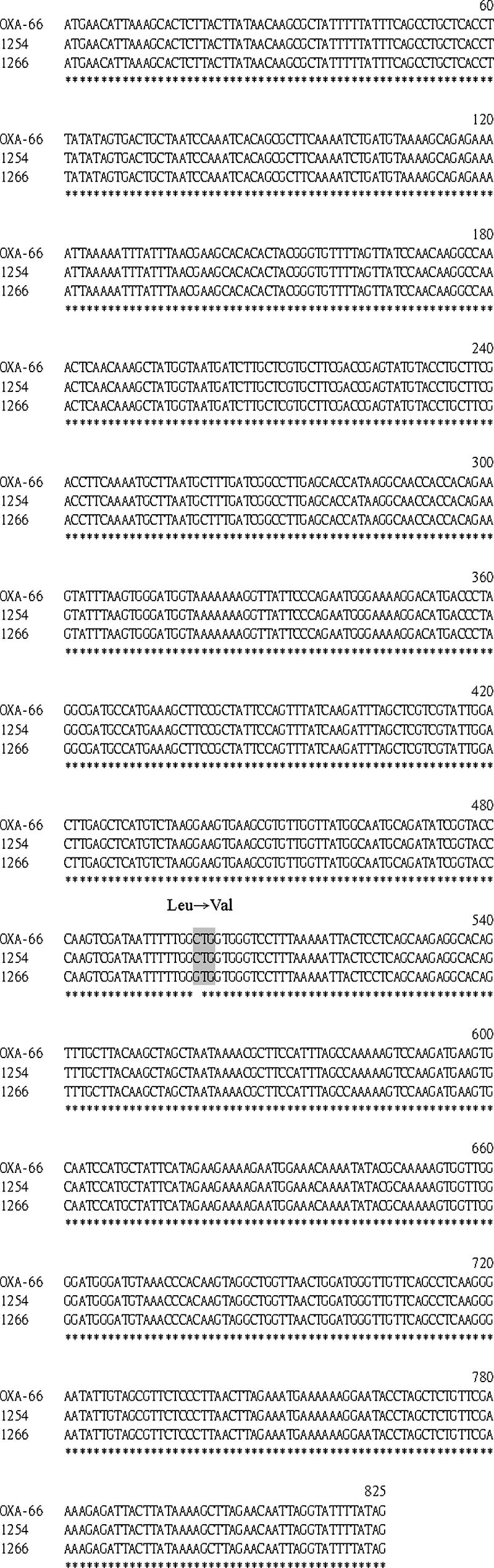
blaOXA-66 gene sequence alignment of three A. baumannii strains (1266 and 1254 [this study] and OXA-66 [reported in the GenBank database]). The corresponding strain is indicated at the left of each line. Numbers on the top of each three-line group indicate the relative posi tions within the open reading frame. Identity is indicated below the alignment by an asterisk. The point mutation (C499G), leading to amino acid substitution Leu167Val in the deduced protein sequence of the blaOXA-66 gene of the IRAB 1266 strain, is highlighted by gray shading.
RT-PCR and real-time RT-PCR analysis of the expression of blaOXA-66 and its closely related variants (blaOXA-51-like) in the ISAB, IIAB, and IRAB strains.
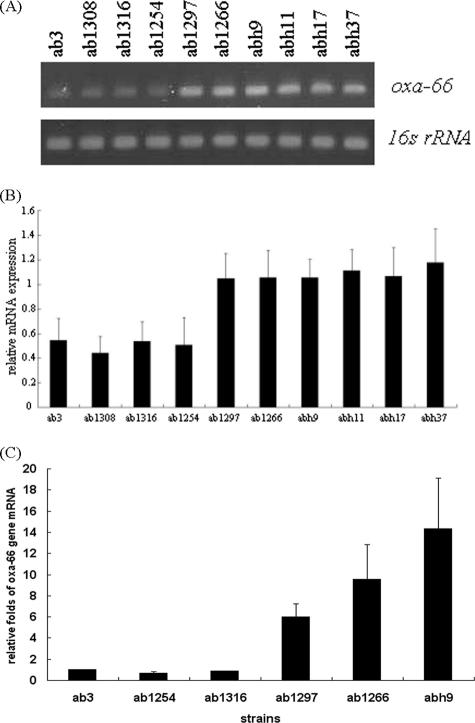
The blaOXA-66 or blaOXA-51-like gene is present in all clinical strains tested, and therefore, we determined whether there were differences in gene expression at the mRNA level across the various A. baumannii strains. To test this hypothesis, the synthetic oligonucleotides OXA-RTF and OXA-RTR (Table 2) were used as a primer set for the semiquantification of the mRNA levels by RT-PCR. As shown in Fig. 4A, the mRNA levels of the blaOXA-66/blaOXA-51-like gene were higher in IRAB ab1266, abh9, abh11, abh17, and abh37 and in IIAB ab1297 than in ISAB ab3, ab1308, ab1316, and ab1254. The results of three independent experiments showed that the relative mRNA level of blaOXA-66/blaOXA-51-like was about twofold higher in the IIAB and IRAB strains than in the ISAB strains (Fig. 4B). The increase was found to be even higher when real-time RT-PCR was used. This approach showed that the relative mRNA levels had increased by 5.98-fold ± 1.22-fold in IIAB ab1297 and 9.5-fold ± 3.25-fold and 14.3-fold ± 4.83-fold in IRAB ab1266 and abh9, respectively (Fig. 4C). Therefore, it is likely that the higher production of OXA-66 or OXA-51-like carbapenemase in these strains is a result of higher levels of mRNA. Thus, this would seem to mean that the production of the OXA-66 or OXA-51-like carbapenemase is regulated at the transcriptional level in the IIAB and IRAB strains.
FIG. 4.
Quantification of the transcript levels of the blaOXA-66 genes from different clinical strains of A. baumannii (ISAB ab3, ab1308, ab1316, and ab1254; IIAB ab1297; and IRAB ab1266, abh9, abh11, abh17, and abh37). (A) RT-PCR analysis of mRNA expression. (B) Relative expression (n-fold) determined from data in A. (C) Relative expression (n-fold) determined by real-time RT-PCR. The relative expression (n-fold) was measured against the expression level of ISAB strain ab3. The expression of the 16S rRNA gene was used as the internal control. Each bar represents the average value of three independent experiments, and the error bars represent the standard deviations.
Detection of ISAba1 upstream of blaOXA-66/blaOXA-51-like.
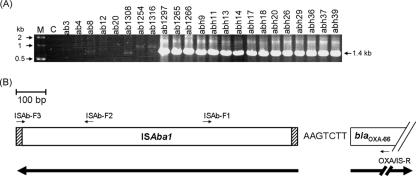
Next, it was necessary to investigate whether, as suggested in recent reports, an insertion sequence, ISAba1, was present upstream of the blaOXA-66/blaOXA-51-like gene. To do this, the synthetic oligonucleotides ISAb-F1 and ISAb-F2 were used as the forward primers and OXA/IS-R was used as the reverse primer (Table 2) for a PCR assay. A 540-bp PCR product was obtained from the IIAB and IRAB strains but not from the ISAB strains when ISAb-F1 and OXA/IS-R were used as the primer set. However, the PCR amplifications were found to be negative when ISAb-F2 and OXA/IS-R were used as the primer set (data not shown). The result indicated that an insertion sequence, ISAba1, may be present upstream of blaOXA-66/blaOXA-51-like with a reverse orientation (Fig. 5B). In order to obtain a full copy of the ISAba1 adjacent to blaOXA-66/blaOXA-51-like, another synthetic primer, ISAb-F3, which is based on the 3′-end sequence of ISAba1 (Table 2), and OXA/IS-R were used together, and they produced a 1.4-kb PCR product from IIAB ab1297, IRAB ab1266, and IRAB abh9 (Fig. 5A). The three 1.4-kb PCR products were cloned into the pGEM-T Easy vector to construct pGEM-IS/OXA1297, pGEM-IS/OXA1266, and pGEM-IS/OXAh9. DNA sequence analysis of the inserts of pGEM-IS/OXA1297, pGEM-IS/OXA1266, and pGEM-IS/OXAh9 revealed an insertion sequence seven bases (AAGTCTT) upstream of the blaOXA-66/blaOXA-51-like gene (Fig. 5B). DNA sequence alignment showed that these insertion sequences are 100% identical to the ISAba1 gene of A. baumannii deposited in the GenBank databank. Furthermore, a 1.4-kb PCR product was also obtained using ISAb-F3 and OXA/IS-R from all other 12 IRAB strains tested (Fig. 5A). Thus, ISAba1 upstream of blaOXA-66/blaOXA-51-like may provide a strong promoter sequence that is able to enhance mRNA levels and thus increase carbapenemase production; as a result, this would confer imipenem resistance to A. baumannii.
FIG. 5.
Detection of ISAba1 upstream of blaOXA-66/blaOXA-51-like from different clinical strains of A. baumannii. (A) PCR results for the amplification of DNA fragments containing partial blaOXA-66/blaOXA-51-like and its upstream ISAba1. Lanes 3 to 10, ISAB strains ab3, ab4, ab8, ab12, ab20, ab1308, ab1254, and ab1316; lane 11, IIAB strain ab1297; lanes 12 to 25, IRAB strain ab1265, ab1266, abh9, abh11, abh13, abh14, abh17, abh18, abh20, abh26, abh29,abh36, abh37, and abh39. Lane C represents a negative control (no A. baumannii chromosomal DNA). Lane M represents DNA size markers, with molecular masses (in kilodaltons) indicated by the arrows in the left margin. The arrow in the right margin indicates a 1.4-kb DNA fragment that was amplified by PCR. (B) Schematic map of ISAba1 and blaOXA-66/blaOXA-51-like in IIAB ab1297, IRAB ab1266, and IRAB abh9. The heavy arrows indicate the transcription orientations of the ISAba1 and blaOXA-66/blaOXA-51-like genes. These two genes are separated by seven bases, namely, AAGTCTT. The dashed areas represent the terminal inverted repeat sequences of ISAba1. The positions of the primers are indicated and are referred to in Table 2.
Expression of the blaOXA-66 genes from ISAB 1254 and IRAB 1266 in E. coli.
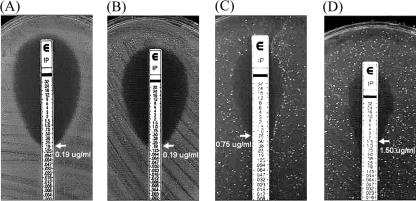
In order to evaluate the role of OXA-66 carbapenemase in imipenem resistance, the DNA fragments containing the blaOXA-66 gene from pGEM-oxa1254 and pGEM-oxa1266 were subcloned into the expression vector pBAD/Myc-His to construct pBAD/Myc-His-oxa66-1254 and pBAD/Myc-His-oxa66-1266, respectively. Plasmids pBAD/Myc-His, pBAD/Myc-His-oxa66-1254, and pBAD/Myc-His-oxa66-1266 were transformed into E. coli TOP10 to obtain the recombinant strains TOP10/pBAD/Myc-His, TOP10/pBAD/Myc-His-oxa66-1254, and TOP10/pBAD/Myc-His-oxa66-1266. Susceptibilities of these three recombinant strains to imipenem were determined by Etest. As shown in Fig. 6, the resistance to imipenem increased by 3.95-fold when the blaOXA-66 gene from strain ab1254 was introduced into E. coli TOP10 cells (TOP10/pBAD/Myc-His-oxa66-1254 versus TOP10/pBAD/Myc-His, 0.75 μg ml−1 versus 0.19 μg ml−1, respectively). Furthermore, the resistance to imipenem increased by 7.90-fold when the blaOXA-66 gene from strain ab1266 was introduced into E. coli TOP10 (TOP10/pBAD/Myc-His-oxa-66-1266 versus TOP10/pBAD/Myc-His, 1.5 μg ml−1 versus 0.19 μg ml−1, respectively). However, resistance to meropenem was not affected when the blaOXA-66 gene from either strain ab1254 or ab1266 was introduced into E. coli TOP10 cells (data not shown). Thus, the increased imipenem resistance effect of the OXA-66 carbapenemase occurs not only in A. baumannii but also in E. coli.
FIG. 6.
Effect of the introduction of the blaOXA-66 gene from A. baumannii into E. coli TOP10 cells on imipenem susceptibility. (A) TOP10 only. (B) TOP/pBAD/Myc-His. (C) TOP10/pBAD/Myc-His-oxa66-1254. (D) TOP/pBAD/Myc-His-oxa66-1266. The imipenem susceptibility tests were determined by Etest. The expression of OXA-66 carbapenemase was induced by the addition of 0.002% arabinose to the agar plate. The MICs are indicated by arrows.
Study of selected imipenem resistance mutants.
In order to examine any differences in the blaOXA-66/blaOXA-51-like gene sequence and gene expression, in the outer membrane proteins profiles, or in efflux pump function between imipenem-susceptible strains and their imipenem-resistant isogenic strains, in vitro selection of the imipenem-resistance mutants ISAB ab8(r) and ISAB ab1254(r) was carried out from ISAB ab8 and ISAB ab1254, respectively. The results showed that there were no differences in the blaOXA-66/blaOXA-51-like gene sequence, in the levels of gene expression of this beta-lactamase, and in their outer membrane protein profiles when ISAB ab8 and ISAB ab8(r) or ISAB ab1254 and ISAB ab1254(r) were compared (data not shown). However, there was a four- to eightfold reduction in imipenem resistance and a twofold reduction in meropenem resistance with ISAB ab8(r) and ISAB ab1254(r) when CCCP was added. This suggested that a putative efflux mechanism may play a role in the imipenem and/or meropenem resistance of these selected mutants but not the clinical strains (Table 3). Thus, increased activity of a drug export system may be important and/or occur frequently when IRAB strains are selected.
TABLE 3.
Multistep resistance selection with imipenem from two clinical strains of A. baumannii
| A. baumannii strain | Initial MICa before exposure to drug
|
MICa of resistant mutants (fold reduction)
|
||||
|---|---|---|---|---|---|---|
| IMP | MEM | IMP | IMP + CCCPb | MEM | MEM + CCCPc | |
| ab8 | 1 | 0.5 | 256 | 64 (4) | 8 | 4 (2) |
| ab1254 | 3 | 0.75 | 128 | 16 (8) | 6 | 3 (2) |
Determination of MIC using the broth dilution method. IMP, imipenem; MEM, meropenem.
The concentration of CCCP is 24 μg/ml.
The concentration of CCCP is 2 μg/ml.
DISCUSSION
In A. baumannii, carbapenem resistance is due mostly to carbapenemases (1, 26) or to synergistic effects between carbapenemases that hydrolyze carbapenems and decreased expression of certain penicillin-binding proteins or other outer membrane proteins (13, 14). There are few studies that have examined drug resistance mediated by an efflux pump(s). They include the efflux system AdeABC, which has been identified as mediating multidrug resistance, and the efflux system AbeM, which has been suggested to be involved in drug resistance (17, 21, 34). In this study, we used a combination of 2-DE and mass spectrometry and identified the overexpression of an OXA-66/OXA-51-like carbapenemase, which is thought to be periplasmic, in the outer membrane fraction of clinical imipenem-resistant strains of A. baumannii.
Our results showed that blaOXA-66/blaOXA-51-like genes exist in all clinical strains of A. baumannii that were tested in this study. This result agrees with recent reports that have demonstrated that an OXA-51-like oxacillinase naturally occurs in A. baumannii; it has been suggests that this protein is an intrinsic class D carbapenemase (15, 22, 37, 41). The expression level of the OXA-66 carbapenemase was found to be much higher in IRAB ab1266 than in ISAB 1254 (Fig. 2). Like with another OXA-14 class D beta-lactamase (8), it may be possible to explain the resistance because a high level of OXA-66 carbapenemase is able to convert the enzyme from a monomeric to a dimeric form; this form is more active in the bacterial periplasm. This would confer imipenem resistance to A. baumannii strains. As shown in Fig. 4, the production of this beta-lactamase was up-regulated at the transcriptional level. Like other recent reports, the up-regulation seems to have resulted from the presence of an upstream insertion sequence, ISAba1 (Fig. 5B), which provides a strong promoter (36). Similar regulation by ISAba1 has also been found for A. baumannii blaampC, blaOXA-23, and blaOXA-58 gene expression (16, 29, 36). The successful expression in E. coli of the genes thought to confer carbapenem resistance in A. baumannii strongly supports the idea that these genes are central to carbapenem resistance in this species. We also identified a C-to-G change at nucleotide position 499, which resulted in a leucine-to-valine change at amino acid residue 167 of the OXA-66 or OXA-51-like carbapenemase in several IRAB strains tested (data not shown). However, the elevation of resistance in E. coli TOP10 cells was not significantly difference when the two genes were compared (3.95-fold for ab1254 versus 7.90-fold for ab1266) (Fig. 5). Thus, the cloned OXA-66 enzyme from either strain shows carbapenemase activity that is able to confer reduced susceptibility to imipenem but not to meropenem. This suggests that this beta-lactamase has hydrolytic activity specific to imipenem. This is supported by a previous result where the OXA-69 carbapenemase (a member of OXA-51 family) was shown to confer low levels of imipenem resistance to E. coli and was also found to be associated with full imipenem resistance (16). Therefore, it is necessary to sequence more clinical strains to determine the importance of the leucine-to-valine change at amino acid residue 167 in IRAB strains isolated from Taiwan. If confirmed, the role of amino acid residue 167 in carbapenemase activity would then need to be further studied because residue 167 is not among the various known conserved motifs that have been predicted by multiple sequence alignment for class D beta-lactamase (39). Although it has been reported that class B metallo-beta-lactamases (IMP or VIM) are involved in some A. baumannii outbreaks in the Far East (38, 42), we were unable to detect any such metallo-beta-lactamases using the Etest MBL strip on our A. baumannii clinical strains (data not shown). Thus, our data support the hypothesis that an OXA-66 or OXA-51-like carbapenemase plays a key role in A. baumannii imipenem resistance in our tested strains.
The result for the in vitro selection of mutants showed no significant differences in the DNA sequences of the blaOXA-66/blaOXA-51-like genes between ISAB ab8 and ISAB 1254 strains and their isogenic mutants, ISAB ab8(r) and ISAB ab1254(r). Furthermore, no changes in gene expression were detected either. This suggests that this intrinsic gene is not induced by exposure to imipenem but that resistance is affected mainly by the presence of ISAba1 upstream. In addition, there were no differences in the outer membrane protein profiles. These results were supported by the fact that the mRNA levels of the already known imipenem resistance-associated porins or outer membrane proteins, such as CarO, 33- to 36-kDa protein, and 43-kDa protein (3, 6, 11, 20, 24, 33, 35), did not show any significant differences between ISAB ab8 and ISAB ab8 (r) or ISAB ab1254 and ISAB ab1254(r) by RT-PCR. However, we could not exclude the hypothesis that decreased drug permeability was associated with imipenem resistance, and this resistance mechanism is found quite often as a drug resistance mechanism in bacteria. Nonetheless, adding CCCP did reduce imipenem resistance by four- to eightfold and meropenem resistance by twofold when ISAB ab8(r) and ISAB ab1254(r) were exposed to this drug. This suggests that an efflux mechanism is possibly involved in the imipenem and meropenem resistance of these selected mutants of A. baumannii (Table 3). Since there was no significant difference in mRNA levels for the expression of the adeB and abeM genes (data not shown), efflux transporters other than AdeB and AbeM are suggested to be associated with imipenem resistance between ISAB ab8 and ISAB ab8(r) or ISAB ab1254 and ISAB ab1254(r).
In conclusion, this is the first report of a class D carbapenemase, OXA-66/OXA-51-like enzyme, that contributes to imipenem resistance in clinical strains of A. baumannii isolated from Taiwan. Also, the results of this study confirm that this naturally occurring beta-lactamase plays an important role in A. baumannii imipenem resistance. However, increased efflux pump activity as a mechanism giving imipenem resistance may be more important and/or occur frequently in A. baumannii. Thus, it is necessary to identify the efflux pump(s), other than AdeB and AbeM, that may be affected and which, as a result, may be correlated with imipenem resistance in our clinical strains of A. baumannii. Finally, whether other class D carbapenemases, such as OXA-23, OXA-24, or OXA-58, are associated with imipenem resistance also needs to be investigated in our clinical strains of A. baumannii.
Supplementary Material
Acknowledgments
We thank Bor-Shen Hu and Kwok-Woon Yu for providing us with some of the A. baumannii strains used in this study. We also thank Ya-Luen Chen for assistance with preparation of manuscript.
This work was supported in part by a grant from the Ministry of Education of the Republic of China, Aim for the Top University Plan.
Footnotes
Published ahead of print on 27 August 2007.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Amyes, S. G. B., and H. K. Young. 1996. Mechanism of antibiotic resistance in Acinetobacter spp.—genetics of resistance, p. 185-223. In E. Bergogene-Berezin, M. L. Joly-Guillou, and K. J. Towner (ed.), Acinetobacter: microbiology, epidemiology, infections, management, 1996. CRC Press, New York, NY.
- 2.Bergogene-Berezin, E., and K. J. Towner. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9:148-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bou, G., G. Cervero, M. A. Dominguez, C. Quereda, and J. Martinez-Beltran. 2000. Characterization of a nosocomial outbreak caused by a multiresistant Acinetobacter baumannii strain with a carbapenem-hydrolyzing enzyme: high-level carbapenem resistance in A. baumannii is not due solely to the presence of β-lactamases. J. Clin. Microbiol. 38:3299-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Brown, S., and S. G. B. Amyes. 2005. The sequences of seven class D beta-lactamases isolated from carbapenem-resistant Acinetobacter baumannii from four continents. Clin. Microbiol. Infect. 11:326-329. [DOI] [PubMed] [Google Scholar]
- 5.Brown, S., and S. Amyes. 2006. OXA beta-lactamases in Acinetobacter: the story so far. J. Antimicrob. Chemother. 57:1-3. [DOI] [PubMed] [Google Scholar]
- 6.Clark, R. B. 1996. Imipenem resistance among Acinetobacter baumannii: association with reduced expression of a 33-36 kDa outer membrane protein. J. Antimicrob. Chemother. 38:245-251. [DOI] [PubMed] [Google Scholar]
- 7.Corbella, W., A. Montero, M. Pujol, M. A. Dominguez, J. Ayats, M. J. Argerich, F. Garrigosa, J. Ariza, and F. Gudiol. 2000. Emergence and rapid spread of carbapenem resistance during a large and sustained hospital outbreak of multiresistant Acinetobacter baumannii. J. Clin. Microbiol. 38:4086-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danel, F., J. M. Frere, and D. M. Livermore. 2001. Evidence of dimerisation among class D beta-lactamases: kinetics of OXA-14 beta-lactamase. Biochim. Biophys. Acta 1546:132-142. [DOI] [PubMed] [Google Scholar]
- 9.Del Mar Tomas, M., M. Cartelle, S. Pertega, A. Beceiro, P. Llinares, D. Canle, F. Molina, R. Villanueva, J. M. Cisneros, and G. Bou. 2005. Hospital outbreak caused by a carbapenem-resistant strain of Acinetobacter baumannii: patient prognosis and risk-factors for colonization and infection. Clin. Microbiol. Infect. 11:540-546. [DOI] [PubMed] [Google Scholar]
- 10.Denton, M., M. H. Wilcox, P. Parnell, D. Green, V. Keer, P. M. Hawkey, I. Evans, and P. Murphy. 2004. Role of environmental cleaning in controlling outbreak of Acinetobacter baumannii on a neurosurgical intense care unit. J. Hosp. Infect. 56:106-110. [DOI] [PubMed] [Google Scholar]
- 11.Dupont, M., J. M. Pages, D. Lafitee, A. Siroy, and C. Bollet. 2006. Identification of an OprD homologue in Acinetobacter baumannii. J. Proteome Res. 4:2386-2390. [DOI] [PubMed] [Google Scholar]
- 12.El Shafe, S. S., M. Alishaq, and M. Leni Garcia. 2004. Investigation of an outbreak of multidrug-resistant Acinetobacter baumannii in trauma intensive care unit. J. Hosp. Infect. 56:101-105. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez-Cuenca, F., L. Martinez-Martinez, M. C. Conejo, J. A. Ayala, E. J. Perea, and A. Pascual. 2003. Relationship between beta-lactamase production, outer membrane protein and penicillin-binding protein profiles on the activity of carbapenems against clinical isolates of Acinetobacter baumannii. J. Antimicrob. Chemother. 51:565-574. [DOI] [PubMed] [Google Scholar]
- 14.Gehrlein, M., H. Leying, W. Cullmann, S. Wendt, and W. Opferkuch. 1991. Imipenem resistance in Acinetobacter baumannii is due to altered penicillin-binding proteins. Chemotherapy 37:405-412. [DOI] [PubMed] [Google Scholar]
- 15.Heritier, C., L. Poirel, P.-E. Fournier, J.-M. Claverie, D. Raoult, and P. Nordmann. 2005. Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:4174-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heritier, C., L. Poirel, and P. Nordmann. 2006. Cephalosporinase over-expression resulting from insertion of ISAba1 in Acinetobacter baumannii. Clin. Microbiol. Infect. 12:123-130. [DOI] [PubMed] [Google Scholar]
- 17.Higgins, P. G., H. Wisplinghoff, D. Stefanik, and H. Seifert. 2004. Selection of topoisomerase mutations and overexpression of adeB mRNA transcripts during an outbreak of Acinetobacter baumannii. J. Antimicrob. Chemother. 54:821-823. [DOI] [PubMed] [Google Scholar]
- 18.Kristensen, D. B., K. Imamura, Y. Miyamoto, and K. Yoshizato. 2000. Mass spectrometric approaches for the characterization of proteins on a hybrid quadrupole time-of-flight (Q-TOF) mass spectrometer. Electrophoresis 21:430-439. [DOI] [PubMed] [Google Scholar]
- 19.Levi, I., and E. Rubinstein. 1996. Acinetobacter infections—overview of clinical features, p. 101-115. In E. Bergogene-Berezin, M. L. Joly-Guillou, and K. J. Towner (ed.), Acinetobacter: microbiology, epidemiology, infections, management, 1996. CRC Press, New York, NY.
- 20.Limansky, A. S., M. A. Mussi, and A. M. Viale. 2002. Loss of a 29-kilodalton outer membrane protein in Acinetobacter baumannii is associated with imipenem resistance. J. Clin. Microbiol. 40:4776-4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magnet, S., P. Courvalin, and T. Lambert. 2001. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob. Agents Chemother. 45:3375-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merkier, A. K., and D. Centron. 2006. blaOXA-51-type beta-lactamase genes are ubiquitous and vary within a strain in Acinetobacter baumannii. Int. J. Antimicrob. Agents 28:110-113. [DOI] [PubMed] [Google Scholar]
- 23.Molloy, M. P., B. R. Herbert, M. B. Slade, T. Rabilloud, A. S. Nouwens, K. L. Williams, and A. A. Gooley. 2000. Proteomic analysis of the Escherichia coli outer membrane. Eur. J. Biochem. 267:2871-2881. [DOI] [PubMed] [Google Scholar]
- 24.Mussi, M. A., A. S. Limansky, and A. M. Viale. 2005. Acquisition of resistance to carbapenems in multidrug-resistant clinical strains of Acinetobacter baumannii: natural insertional inactivation of a gene encoding a member of a novel family of beta-barrel outer membrane proteins. Antimicrob. Agents Chemother. 49:1432-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 7th ed. NCCLS document M7-A7. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 26.Nordmann, P., and L. Poirel. 2002. Emerging carbapenemases in gram-negative aerobes. Clin. Microbiol. Infect. 8:321-331. [DOI] [PubMed] [Google Scholar]
- 27.Nordmann, P. 2004. Acinetobacter baumannii, the nosocomial pathogen par excellence. Pathol. Biol. 52:301-303. [DOI] [PubMed] [Google Scholar]
- 28.Pankuch, G. A., S. A. Jueneman, T. A. Davis, M. R. Jacobs, and P. C. Appelbaum. 1998. In vitro selection of resistance to four β-lactams and azithromycin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2914-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poirel, L., and P. Nordmann. 2006. Genetic structures at the origin of acquisition and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-58 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 50:1442-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rello, J., and E. Diaz. 2003. Pneumonia in the intensive care unit. Crit. Care Med. 31:2544-2551. [DOI] [PubMed] [Google Scholar]
- 31.Roberts, S. A., R. Findlay, and S. D. Lang. 2001. Investigation of an outbreak of multi-drug resistant Acinetobacter baumannii in an intensive care burns unit. J. Hosp. Infect. 48:228-232. [DOI] [PubMed] [Google Scholar]
- 32.Sengupta, S., P. Humar, A. M. Ciraj, and P. G. Shivananda. 2001. Acinetobacter baumannii—an emerging nosocomial pathogen in the burns unit Manipal, India. Burns 27:140-144. [DOI] [PubMed] [Google Scholar]
- 33.Siroy, A., V. Molle, C. Lemaitre-Guillier, D. Vallenet, M. Pestel-Caron, A. J. Cozzone, T. Jouenne, and E. De. 2005. Channel formation by CarO, the carbapenem resistance-associated outer membrane protein of Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:4876-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su, X.-Z., J. Chen, T. Mizushima, T. Kuroda, and T. Tsuchiya. 2005. AbeM, an H+-coupled Acinetobacter baumannii multidrug efflux pump belonging to the MATE family of transporters. Antimicrob. Agents Chemother. 49:4362-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas, M. M., A. Beceiro, A. Perez, D. Velasco, R. Moure, R. Villanueva, J. Martinoz-Beltran, and G. Bou. 2005. Cloning and functional analysis of the gene encoding the 33- to 36-kilodalton outer membrane protein associated with carbapenem resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:5172-5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turton, J. F., M. E. Ward, N. Woodford, M. E. Kaufmann, R. Pike, D. M. Livermore, and T. L. Pitt. 2006. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 258:72-77. [DOI] [PubMed] [Google Scholar]
- 37.Turton, J. F., N. Woodford, J. Glover, S. Yarde, M. E. Kaufmann, and T. L. Pitt. 2006. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 44:2974-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walsh, T. R., M. A. Toleman, L. Poirel, and P. Nordmann. 2005. Metallo-β-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18:306-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walther-Rasmussen, J., and N. Hoiby. 2006. OXA-type carbapenemases. J. Antimicrob. Chemother. 57:373-383. [DOI] [PubMed] [Google Scholar]
- 40.Wong, T. H., B. H. Tan, M. L. Ling, and C. Song. 2002. Multi-resistant Acinetobacter baumannii on a burn unit-clinical risk factors and prognosis. Burns 28:349-357. [DOI] [PubMed] [Google Scholar]
- 41.Woodford, N., M. J. Ellington, J. M. Coelho, J. F. Turton, M. E. Ward, S. Brown, S. G. B. Amyes, and D. Livermore. 2006. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27:351-353. [DOI] [PubMed] [Google Scholar]
- 42.Yong, D., Y. S. Choi, K. H. Roh, C. K. Kim, Y. H. Park, J. H. Yum, K. Lee, and Y. Chong. 2006. Increasing prevalence and diversity of metallo-β-lactamases in Pseudomonas spp., Acinetobacter spp., and Enterobacteriaceae from Korea. Antimicrob. Agents Chemother. 50:1884-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.