Abstract
Here we report the nucleotide sequence of pCTX-M3, a highly conjugative plasmid that is responsible for the extensive spread of the gene coding for the CTX-M-3 extended-spectrum β-lactamase in clinical populations of the family Enterobacteriaceae in Poland. The plasmid belongs to the IncL/M incompatibility group, is 89,468 bp in size, and carries 103 putative genes. Besides blaCTX-M-3, it also bears the blaTEM-1, aacC2, and armA genes, as well as integronic aadA2, dfrA12, and sul1, which altogether confer resistance to the majority of β-lactams and aminoglycosides and to trimethoprim-sulfamethoxazole. The conjugal transfer genes are organized in two blocks, tra and trb, separated by a spacer sequence where almost all antibiotic resistance genes and multiple mobile genetic elements are located. Only blaCTX-M-3, accompanied by an ISEcp1 element, is placed separately, in a DNA fragment previously identified as a fragment of the Kluyvera ascorbata chromosome. On the basis of sequence analysis, we speculate that pCTX-M3 might have arisen from plasmid pEL60 from plant pathogen Erwinia amylovora by acquiring mobile elements with resistance genes. This suggests that plasmids of environmental bacterial strains could be the source of those plasmids now observed in bacteria pathogenic for humans.
Bacterial plasmids constitute an important part of the so-called horizontal gene pool, the pool of genes that can be acquired by various bacterial strains by means of horizontal gene transfer (56). The ability of plasmids to spread by conjugation greatly enhances their impact on the genetic plasticity, metabolic potential, and environmental adaptability of bacteria, which in the case of plasmids carrying virulence or antimicrobial resistance genes is a source of serious clinical and epidemiological problems (see references 35, 56, and 61 and references therein).
Among the genes that are often located in conjugative plasmids are those coding for extended-spectrum β-lactamases (ESBLs), which in large part are responsible for the resistance of the members of the family Enterobacteriaceae to newer β-lactams (11, 24, 35). The epidemiology of ESBLs has recently been dominated by the extremely rapid and worldwide spread of organisms producing enzymes of the CTX-M family, both in nosocomial environments and in the community (10, 36, 45). To date, over 60 CTX-M-type β-lactamases have been identified, showing the intensive evolution of this group (http://www.lahey.org/studies/webt.stm). blaCTX-M genes are derivatives of the chromosomal β-lactamase genes of the genus Kluyvera (17, 29, 43, 46, 49). In transmissible plasmids found in clinical isolates, they usually reside in fragments of Kluyvera sp. chromosomes in association with either ISEcp1-like insertion sequences (30, 34, 37, 47) or CR1 elements inserted in sul1-type class 1 integrons (3, 19, 41, 50, 58). The role of ISEcp1-like sequences in the mobilization of Kluyvera genes and details of this transposition process have been demonstrated recently (35, 47, 48); moreover, these elements are also frequently responsible for the high-level expression of blaCTX-M genes (10, 13, 47).
In Poland, the first gene of the blaCTX-M type, blaCTX-M-3, was identified in 1996 in clinical isolates of Citrobacter freundii and Escherichia coli from one of the hospitals in Warsaw. It was associated with a large plasmid that could be easily transferred to E. coli via conjugation and that conferred resistance to penicillins, cephalosporins, and aztreonam, as well as aminoglycosides, gentamicin, and tobramycin (25). Later, the rapid spread of CTX-M-3-producing organisms was observed in the same center (44) and all over the country (7, 8), and it has mainly been attributed to the horizontal transmission of plasmids. Of the several types of plasmids with blaCTX-M-3 genes identified by restriction fingerprinting, one family of similar molecules in particular has been widely disseminated into multiple locales and multiple species, including E. coli, Klebsiella pneumoniae, Klebsiella oxytoca, Enterobacter cloacae, C. freundii, Morganella morganii, Serratia marcescens, and Salmonella enterica (7, 8). The most prevalent variant of that family was the plasmid first observed in 1996 in C. freundii isolates, in which CTX-M-3 had been originally identified (25). In Taiwan, where CTX-M-3 is the most common ESBL, it has been encoded by large, transmissible IncI1-like plasmids (37), while in Spain the dissemination of CTX-M-3 and other CTX-M-1-like β-lactamases is ascribed to the broad-host-range plasmids of the IncA/C2, IncL/M, and IncN groups (42).
In this paper we report the sequence of the pCTX-M3 plasmid originating from C. freundii isolate 2526 from the Praski Hospital in Warsaw (25). The results of our study demonstrate the physical link between the blaCTX-M-3 gene, the IncL/M replicon, and an efficient conjugal transfer system. We also discuss the possible evolution of pCTX-M3 from a putative ancestor pEL60 from a plant pathogen, Erwinia amylovora (22).
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli A15(pCTX-M3), a transconjugant of C. freundii 2526 (25), was the initial source of pCTX-M3 in this study. Clinical isolates K. pneumoniae 179, S. marcescens 12, and E. coli 279 were described previously (44); and the other clinical strains or transconjugants used in the study included E. coli 2112, E. coli 3624, E. coli 1145, E. coli 8350, E. coli 1775, K. pneumoniae 9172, and K. pneumoniae 2113, which were also described previously (7). E. coli DH5α (27) was used as a host strain in all cloning experiments. In the matings, E. coli DH5α(pCTX-M3) and Agrobacterium tumefaciens UBAPF(pCTX-M3) (39) were used as the donors and E. coli JE2571 Rifr (leu thr thi lacY thy pil fla Rifr [12]) and DH5α(pACYC184) were used as the recipients, respectively. The shotgun library was prepared in vector pCR4BluntTOPO (Invitrogen, Carlsbad, CA). Plasmid pMI3, the pUC19 derivative with the bla gene replaced by cat of pACYC184 (the AvaII-NarI fragment of pUC19 blunt ended by use of the Klenow fragment and ligated with FnuDII-digested pACYC184), was used as the vector for cloning of the IncL/M and IncI1 oriT sequences. Plasmid pLG221, a ColIb-P9 derivative (14), was used as an IncI1 helper plasmid in oriT mobilization experiments.
Plasmid conjugal transfer.
One-milliliter volumes of cultures of the donor and recipient strains (109 CFU of each strain per ml) grown in Luria-Bertani (LB) broth (Biocorp, Warsaw, Poland) were mixed and incubated for 30 min at 37°C. Conjugation was stopped by vigorous vortexing for 30 s and placing the mating mixture on ice. Transconjugants were selected on LB agar (Biocorp) supplemented with cefotaxime (5 μg/ml; Polfa Tarchomin, Warsaw, Poland) and rifampin (50 μg/ml; Polfa Tarchomin). Solid mating was performed likewise, with an additional step of filtering of the mating mixture through a sterile HA 45-μm-pore-size filter (Millipore, Billerica, MA), which was then incubated on an LB agar plate. In the control experiments, the frequencies of spontaneous mutation of both the donor and the recipient to the phenotype of the transconjugants were measured.
DNA cloning and sequencing.
Plasmid DNA was isolated by the alkaline lysis method, and cloning procedures were performed by standard protocols (53). The pCTX-M3 DNA for construction of a shotgun library was purified by CsCl-ethidium bromide gradient ultracentrifugation (53). The shotgun library was prepared in the pCR4BluntTOPO plasmid with the use of a TOPO shotgun subcloning kit (Invitrogen). The plasmid was sequenced by using a combination of shotgun sequencing and primer walking methods. BigDye (version 2.0; Applied Biosystems, Foster City, CA) chemistry on an ABI377 automated sequencer (Applied Biosystems) was used. All enzymes used for cloning were from MBI Fermentas, Vilnius, Lithuania.
Plasmid sequence assembly and analysis.
The assembly of the sequence was performed with the use of the phred/phrap/consed software suite (20, 21), and the consed software autofinish function was used for gap closing and finishing of the sequence. The following programs were used for bioinformatic analysis: for gene identification, the Glimmer program (version 2.0) trained on E. coli long open reading frames (ORFs) was used (18, 52); for homology searches, the BLAST (version 2.0) program was used (2); and for alignment construction, the ClustalW (version 1.8) program was used (57).
PCR detection of repA and traU genes and mapping of blaCTX-M-3-containing regions.
Primers repAu (5′-CACTTTGGCCGGTCAGAGAT-3′) and repAd (5′-CGATCATCTTCCAGAGGCG-3′) were used for the amplification of repA, and primers traUC (5′-ATCTGACGCATCATTGCGCC-3′) and traUN (5′-TGTAGGGCTTACGTCGCAGG-3′) were used for the amplification of traU. The presence of ISEcp1 and its distance to blaCTX-M-3 were analyzed by PCR with primers ALA-3 and ALA-4, as described previously (6). The 3′ flank of blaCTX-M-3 was mapped with primer P1A (7), which anneals to the more distal part of blaCTX-M-3, and primer IRR (5′-GCGCACGTAGGTCCCAGG-3′), which anneals to the very 3′ end of the Kluyvera ascorbata DNA present in pCTX-M3 (49).
Nucleotide sequence accession number.
The complete nucleotide sequence of the pCTX-M3 plasmid has been submitted to the GenBank database under accession no. AF550415.
RESULTS AND DISCUSSION
Overall sequence analysis.
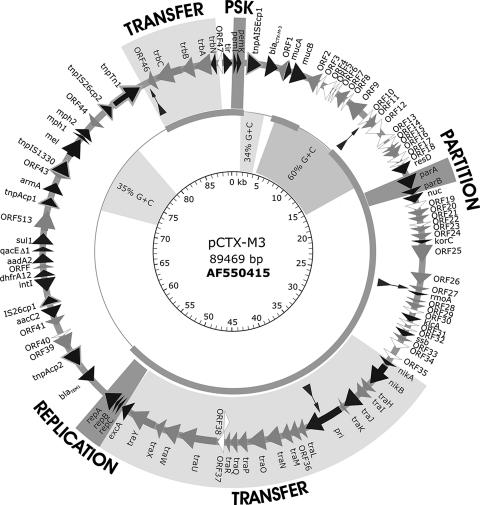
The complete nucleotide sequence of plasmid pCTX-M3 was verified by comparison of in silico-generated restriction maps with the experimental ones. The plasmid is a circular molecule of 89,468 bp and contains 103 putative genes (Fig. 1), of which 22 (21.4%) have no homologs in public databases, 44 (42.7%) have homologs of unknown functions, and the remaining 37 (35.9%) have homologs with known functions. In sixty-seven percent, the plasmid consists of a known backbone, described before in plasmid pEL60 from the plant pathogen E. amylovora (21), enriched with two additional regions that contain all pCTX-M3 antimicrobial resistance genes and mobile genetic elements (see below). The mean G+C content of pCTX-M3 is 51%, but it varies from 32% to 65%, depending on the sequence block (Fig. 1). The high G+C content (ca. 60%) is characteristic of a region located at position 3.2 to 14.0 kb, with the sequence of pCTX-M3-specific orf8 (positions 9639 to 9911) displaying the highest G+C content (65%). Two fragments of exceptionally low G+C content, 34% and 35%, are present at positions 1.3 to 3.2 kb and 72.0 to 80.0 kb, respectively, and are located inside the two regions with resistance genes and mobile elements. Thus, pCTX-M3 probably evolved from an environmental plasmid similar to pEL60 through the stepwise integration of mobile genetic elements associated with antimicrobial resistance genes (see below).
FIG. 1.
Overview of the pCTX-M3 sequence. ORFs are shown by arrows: pCTX-M3-specific ORFs are in white, ORFs homologous to genes of known function are in black, and ORFs homologous to genes of unknown function are in gray. Functional sequence blocks are highlighted with shades of gray bands and are indicated on the outside: replicon, stabilizing cassettes (PSK and partition), and conjugal transfer system. The middle circle with a thick line indicates the pCTX-M3 sequence identical to that of plasmid pEL60. The inner circle is a schematic representation of blocks of low and high G+C contents. Arrows inside the outer circle indicate the positions in which pEL60 has additional sequences that are absent from pCTX-M3.
Plasmid backbone.
Only one replication region was identified in the sequence of pCTX-M3. This replication cassette (positions 54341 to 55979) was confirmed to be identical to that of the prototypical IncL/M replicon of pMU407.1 (4, 5). The segregational stability of the plasmid is ensured by an active partition system encoded by parAB genes (position 17203 to 18633) related to those of IncI1 plasmid ColIb-P9 (parA, 64% similarity; parB, 34% similarity). The presence of a single replicon in pCTX-M3, as well as the functionality of the parAB system, were confirmed in a separate analysis (39). Two other regions possibly involved in stable plasmid maintenance were identified; and these included (i) region pemIK (positions 757 to 1348), which is identical to that of IncFII plasmid R100 and which encodes a postsegregational killing system (59), and (ii) the resD gene, which codes for a putative resolvase (positions 16229 to 16972) that is most similar to the resolvase of pYVe8081 (GenBank accession no. AF074611; 76% similarity). pCTX-M3 also bears mucAB genes (positions 4698 to 6435), which are members of the umuDC-like family of UV resistance genes (28).
Conjugal transfer system.
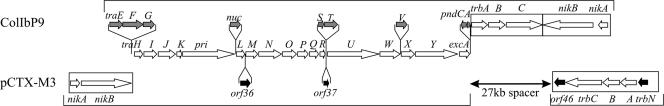
Genes coding for the pCTX-M-3 conjugal transfer system are organized in two blocks, tra (positions 31620 to 54603) and trb (positions 89418 to 83109). The system is related to that of IncI1 plasmid ColIb-P9 in terms of the amino acid sequences of specific proteins (30% to 60%) and of a conserved gene order (Fig. 2). Nevertheless, significant differences are observed between the two systems. First, the oriT region (oriT, nikAB) in ColIb-P9 is located downstream of the trb cluster, and they are both convergently transcribed, while in pCTX-M3 this region is a part of the tra operon. Second, the trb operon in the IncI1 system is placed next to tra and is transcribed in the same direction, whereas in pCTX-M3, trb lies 28.5 kb apart from the tra genes and is transcribed in the opposite direction. Third, some genes present in one of the plasmids are missing from the other; e.g., pCTX-M3 lacks traEFG, the three initial genes of the tra operon, and traV, which was found to be essential for the conjugal transfer of ColIb-P9 (33). In contrast, ColIb-P9 lacks a counterpart of orf36, which in pCTX-M3 separates traL and traM, and the traST genes of ColIb-P9 are replaced in pCTX-M3 by a single gene, orf37. Interestingly, no homology between orf37 and either traS or traT was detected. It is noteworthy that pil genes have not been found in pCTX-M3; thus, its host should not produce thin pili, which in IncI1 carriers enable liquid mating (31, 32). Therefore, it should be underlined that pCTX-M3 is able to transfer in liquid medium (7, 25, 44) at the same frequency as it does on solid support, reaching a value of 0.1 transconjugants/donor (the frequency of spontaneous mutations of donor and recipient strains to the transconjugant phenotype was estimated to be lower than 10−8).
FIG. 2.
Comparison of regions coding for the conjugal transfer systems of pCTX-M3 and IncI1 plasmid ColIb-P9. ORFs are presented as arrows: those present in both plasmids are not filled, and ORFs present only in pCTX-M3 or ColIb-P9 are black and gray, respectively, and are shown under or above the scheme of the common structure region.
As shown recently by Mierzejewska et al. (39), the broad host range of pCTX-M3 is not limited to the family Enterobacteriaceae but also includes members of the classes Alpha-, Beta-, and Gammaproteobacteria. We managed to transfer pCTX-M3 back from a soil bacterium, A. tumefaciens (a member of the class Alphaproteobacteria), to E. coli at a frequency of 10−5 per donor cell. This demonstrated the functionality of the transfer system in an Agrobacterium sp. and confirmed the possibility that environmental bacteria are a reservoir of pCTX-M3 ancestors.
oriT identification.
We have identified a region of pCTX-M3 (positions 31616 to 31721) that resembles oriT of IncI1 plasmid ColIb-P9 (23). The 105-bp fragment, which contains the putative oriTpCTX-M3, was cloned into the pMI3 vector to yield the construct pOriT, which was tested for mobilization by pCTX-M3 in both liquid and solid matings. The plasmid was mobilized at a frequency of 0.1 transconjugant/donor cell in both cases, demonstrating the full functionality of the cloned oriT. In parallel, oriTIncI1 of IncI1 plasmid pLG221 (GenBank accession no. AP005147; positions 67061 to 66944) cloned in pMI3 (pOriTIncI1) was mobilized by pLG221 at a frequency of 2.9 × 10−4. Moreover, pOriT (oriTpCTX-M3) was mobilized by pLG221 and pOriTIncI1 was mobilized by pCTX-M3 at frequencies of 5.9 × 10−5 and 8.7 × 10−6, respectively. The results indicate that the mobilization of oriTpCTX-M3 by a heterologous transfer system (IncI1) is possible, albeit at low frequency, thus suggesting an even increased possibility of plasmid dissemination in microbial populations.
The 27-kb replicon-trb region.
A 27-kb region separating the replicon and the trb operon (positions 55980 to 83109; Fig. 1) is one of the two segments that differentiate plasmid pCTX-M3 from plasmid pEL60 from E. amylovora (22). It contains almost all of the pCTX-M3 mobile elements and antimicrobial resistance genes, including blaTEM-1, which codes for the broad-spectrum β-lactamase TEM-1; aadA2, aacC2, and armA, which confer resistance to aminoglycosides; and dfrA12 and sul1, which are responsible for resistance to trimethoprim-sulfamethoxazole.
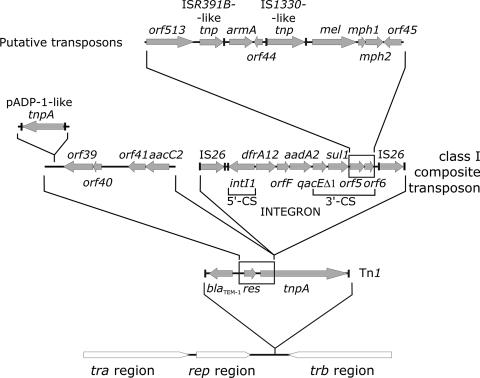
A fragment of transposon Tn1 containing the blaTEM-1 gene is located (positions 56790 to 57990) next to the replicon. The direct repeat (DR) created by the Tn1 insertion and the Tn1 left inverted repeat (IRL) are present directly at the boundary with the replicon. The second part of Tn1, which includes the truncated tnp gene, the right inverted repeat, and the second DR (positions 80415 to 83069), is found at the other end of the 27-kb insert, close to the trb genes. Tn1 was disrupted by other mobile elements (see below), which removed a part of its tnp and the whole res gene. A putative insertion sequence (IS; positions 58454 to 60070) is located behind blaTEM-1, with complete inverted repeats (IRs) and DRs. Its tnp reveals 97% similarity to that in plasmid pADP-1 (38). This IS is followed by orf39 to orf41 and the aacC2 gene, and the entire segment from Tn1 to aacC2 is almost identical to a fragment of recently described plasmid pU302L of S. enterica serovar Typhimurium (15).
The first of two IS26 copies present in pCTX-M3 (positions 63800 to 64622) and a class 1 integron (positions 64640 to 69064) are located downstream of the aacC2 gene. This integron bears three gene cassettes in its variable region, namely, dfrA12, orfF, and aadA2. An identical array of gene cassettes was found in an integron of plasmid pLEW517 from E. coli (60) and in several others (16) (GenBank accession no. AF188331). The pCTX-M3 integron contains a complete 5′ conserved sequence (5′-CS) with the terminal IR (IRi), whereas its 3′-CS is truncated 24 bp after the sul1 stop codon due to the insertion of a CR1 element with orf513 (positions 71184 to 71218) (54). Downstream of CR1 there is a putative IS homologous to ISR391B (9) with truncated IRL (positions 71219 to 72092), the aminoglycoside resistance gene armA, an IS1330-like element (positions 73766 to 75094), and genes reported before to be associated with resistance to macrolides: mel (1) and mph1 and mph2 (40, 55). This region ends with the second copy of IS26 (positions 79595 to 80417). The whole segment flanked by two IS26 copies might be considered a putative large composite transposon, and a similar structure was found before in pMUR050, an IncN group plasmid (26). It differs from that in pCTX-M3 only by the lack of two integronic gene cassettes (dfrA12 and orfF) and by a peculiar structure of the integron's 5′-CS, where the integrase gene intI1 is truncated, duplicated, and surrounded by two IS26 copies. pMUR050 is of “animal” origin (it was isolated from an E. coli strain from a diarrheic pig), and it has been implicated in the conjugative dissemination of the armA gene (26).
The mosaic structure of the 27-kb replicon-trb region clearly suggests that it arose from multiple insertions (Fig. 3). As both DRs created by the Tn1 insertion are present at boundaries of the region, the first event had to be the Tn1 transposition. The subsequent integration events disrupted Tn1 by removing 156 initial codons of tnp and the whole res gene but leaving blaTEM-1 intact. These events caused the acquisition of several mobile elements, some of which were acquired together with other resistance genes, including those present in the class 1 integron. It is impossible to determine if all these elements were acquired by pCTX-M3 in a series of separate events or as bigger modules that were preformed earlier. Since no DRs were found on the flanks of the IS26 elements, the composite transposon-like structure emerged in pCTX-M3 rather not by transposition but by another recombination mechanism.
FIG. 3.
Organization of the 27-kb region of the replicon and the trb genes. The partial pCTX-M3 sequence is drawn schematically out of scale. Arrows, ORFs; black lines, the sites of integration of particular sequence segments; black frames, hypothetical fragments of Tn1 and the integron that are not observed in pCTX-M3, probably due to integration events; vertical bars, IRs of the respective mobile elements and IRi of the integron; gray blocks, the regions of the plasmid backbone.
The ISEcp1- and blaCTX-M-3-containing region.
ISEcp1 (positions 1457 to 3112) and blaCTX-M-3 (positions 3161 to 4151) are the only mobile element and resistance gene, respectively, that are located outside the 27-kb replicon-trb region of pCTX-M3. The presence of ISEcp1 at a distance 128 bp upstream from the blaCTX-M-3 and the blaCTX-M-15 genes in pCTX-M3 and a similar plasmid, respectively, has been reported earlier (6). The pCTX-M3 fragment placed directly downstream from ISEcp1 was identified by Rodríguez et al. (49) to be a chromosomal fragment from a K. ascorbata strain. Apart from the 128-bp spacer and the entire coding region of blaCTX-M-3, it contains 343 bp of a Kluyvera orf477 that, together with 204 bp of the pCTX-M3 sequence, constitutes an orf1 of unknown function. The 3′ end of the insert is terminated by an 18-bp sequence which is a part of orf477 that most probably mimicked the right IR of ISEcp1. The duplication of a pentanucleotide (TGCAG), the ISEcp1 target sequence (positions 1452 to 1456), was found next to this boundary (positions 4508 to 4512) and most probably was generated in the one-ended transposition event that mobilized blaCTX-M-3 from the K. ascorbata chromosome (48).
Cooccurrence of the blaCTX-M-3 gene with the IncL/M replicon and the conjugal transfer system.
Multiple plasmids purified from Polish CTX-M-3 producers were compared before by PstI restriction fingerprinting and were split into several types, with the highly predominant type A (pCTX-M3 family). Several pCTX-M3-like plasmid variants and the plasmid of type B were nonconjugative (7, 8, 44). We decided to check whether these differences could be due to differences in the conjugal transfer genes and whether the blaCTX-M-3 gene has always been linked to the IncL/M replicon and located in the same context as it is in pCTX-M3. The repA gene was chosen as an IncL/M marker, and the traU gene was chosen as a marker of the transfer system. repA is essential for IncL/M plasmid replication (4, 5), whereas traU codes for one of the most conserved and essential proteins of the transfer system (25a, 51), The results of PCR tests carried out with several pCTX-M3-like variants, as well as plasmids of types B, C, and D, are shown in Table 1. All plasmids of types pCTX-M3, C, and D contained the repA and traU genes, whereas the plasmid of type B had neither of these. PCR mapping of the blaCTX-M-3 locus revealed that all of the types of plasmids carried the same fragment of the K. ascorbata chromosome described above. The results demonstrated clearly the major role of conjugative IncL/M plasmids in the spread of the blaCTX-M-3 gene in populations of the family Enterobacteriaceae in Poland and suggested the possible transfer of the blaCTX-M-3-containing element to other plasmids.
TABLE 1.
Cooccurrence of blaCTX-M-3, IncL/M traU, and repA
| Isolatea | Plasmid type/variant by PstI fingerprintinga | Occurrence
|
|||
|---|---|---|---|---|---|
| Matingb | traUc | repAc | blaCTX-M-3 locusd | ||
| C. freundii 2526* | A1-pCTX-M3 | + | ++ | ++ | + |
| K. pneumoniae 179 | A4 | − | ++ | ++ | + |
| S. marcescens 12 | A5 | − | ++ | ++ | + |
| E. coli 2112* | A11 | + | ++ | ++ | + |
| K. pneumoniae 2113* | A12 | + | ++ | ++ | + |
| E. coli 3624* | A18 | + | ++ | ++ | + |
| E. coli 1145* | A33 | + | ++ | ++ | + |
| K. pneumoniae 9172 | A34 | − | ++ | ++ | + |
| E. coli 279 | B | − | − | − | + |
| E. coli 8350* | C | + | + | ++ | + |
| E. coli 1775* | D | + | ++ | ++ | + |
Clinical isolates and their plasmid fingerprints were described previously (6, 24, 45); in cases marked with an asterisk, plasmids were purified from transconjugants for this analysis.
+ and −, positive and negative mating results, respectively.
++, intense band of a PCR product; +, weak band, −, no band present.
Defined as ISEcp1, blaCTX-M-3, (128 bp from ISEcp1), and orf477 (373 bp) (50).
Conclusions.
We have sequenced plasmid pCTX-M3, which seems to be in large part responsible for the rapid dissemination of CTX-M-3-producing microorganisms in Poland (7, 8, 25, 44). The blaCTX-M-3 gene observed in the country most probably emerged by a single ISEcp1-mediated mobilization from the K. ascorbata genome (49), and our results suggest that it could have been transmitted to other plasmids of the same or different replicon types. The pCTX-M3 plasmid bears the IncL/M type of replicon, which enables replication in a broad range of hosts (39) and which codes for the conjugal transfer system that is most similar to that of IncI1 plasmids. Despite the lack of the pil genes, pCTX-M3 is able to transfer with a high efficiency both in liquid and on solid media, and the transfer system is also functional in A. tumefaciens. Moreover, the oriT sequence of pCTX-M3 may serve as a transfer origin both for its cognate IncL/M system and for the heterologous IncI1 system. The backbones of pCTX-M3 and pEL60, a plasmid from bacterial species pathogenic for plants, reveal extended identity, with differences clustered in two regions packed with mobile genetic elements and antibiotic resistance genes. Our results allow us to speculate that an ancestral plasmid similar to pEL60 might have originated from an environmental bacterium and that the plant bacterial community may be the source of plasmids utilized by species pathogenic for human in their rapid adaptation to quickly changing clinical environments.
Acknowledgments
This work was started and conducted by Piotr Cegłowski until he passed away in 2004. We are grateful to Christopher Thomas of University of Birmingham (Birmingham, United Kingdom) for his help and discussions on the evolution of the region separating tra and trb. We thank Angela Thomas (University of Leicester, Leicester, United Kingdom) for plasmid pLG221.
This work was supported by a grant from the Polish State Committee for Scientific Research (grant 2 P04A 024 26 to J.B. and M.G.), by the EU grant within 5 FP project no. ICA-CT-2000-70010 (to P.C.), and by the 6 FP EU grant 6 PCRD LSHM-CT-2003-503-335 (COBRA; to A.B. and M.G.).
Footnotes
Published ahead of print on 13 August 2007.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambrose, K. D., R. Nisbet, and D. S. Stephens. 2005. Macrolide efflux in Streptococcus pneumoniae is mediated by a dual efflux pump (mel and mef) and is erythromycin inducible. Antimicrob. Agents Chemother. 49:4203-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arduino, S. M., P. H. Roy, G. A. Jacoby, B. E. Orman, S. A. Pineiro, and D. Centron. 2002. blaCTX-M-2 is located in an unusual class 1 integron (In35) which includes Orf513. Antimicrob. Agents Chemother. 46:2303-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Athanasopoulos, V., J. Praszkier, and A. J. Pittard. 1995. The replication of an IncL/M plasmid is subject to antisense control. J. Bacteriol. 177:4730-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Athanasopoulos, V., J. Praszkier, and A. J. Pittard. 1999. Analysis of elements involved in pseudoknot-dependent expression and regulation of the repA gene of an IncL/M plasmid J. Bacteriol. 181:1811-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baraniak, A., J. Fiett, W. Hryniewicz, P. Nordmann, and M. Gniadkowski. 2002. Ceftazidime-hydrolysing CTX-M-15 extended-spectrum β-lactamase (ESBL) in Poland. J. Antimicrob. Chemother. 50:393-396. [DOI] [PubMed] [Google Scholar]
- 7.Baraniak, A., J. Fiett, A. Sulikowska, W. Hryniewicz, and M. Gniadkowski. 2002. Countrywide spread of CTX-M-3 extended-spectrum β-lactamase-producing microorganisms of the family Enterobacteriaceae in Poland. Antimicrob. Agents Chemother. 46:151-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baraniak, A., E. Sadowy, W. Hryniewicz, and M. Gniadkowski. 2002. Two different extended-spectrum β-lactamases (ESBLs) in one of the first ESBL-producing Salmonella isolates in Poland. J. Clin. Microbiol. 40:1095-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boltner, D., C. MacMahon, J. T. Pembroke, P. Strike, and A. M. Osborn. 2002. R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J. Bacteriol. 184:5158-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonnet, R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bradford, P. A. 2001. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14:933-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bradley, D. E. 1980. Determination of pili by conjugative bacterial drug resistance plasmids of incompatibility groups B, C, H, J., K, M, V, and X. J. Bacteriol. 141:828-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao, T., T. Lambert, and P. Courvalin. 2002. ColE1-like plasmid pIP843 of Klebsiella pneumoniae encoding extended-spectrum β-lactamase CTX-M-17. Antimicrob. Agents Chemother. 46:1212-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chatfield, L. K., E. Orr, G. J. Boulnois, and B. M. Wilkins. 1982. DNA primase of plasmid ColIb-P9 is involved in conjugal DNA synthesis in donor and recipient bacteria. J. Bacteriol. 152:1188-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, C.-Y., G. W. Nace, B. Solow, and P. Fratamico. 2007. Complete nucleotide sequences of 84.5- and 3.2-kb plasmids in the multi-antibiotic resistant Salmonella enterica serovar Typhimurium U302 strain G8430. Plasmid 57:29-43. [DOI] [PubMed] [Google Scholar]
- 16.Chiu, C.-H., P. Tang, C. Chu, S. Hu, Q. Bao, J. Yu, Y.-Y. Chou, H.-S. Wang, and Y.-S. Lee. 2005. The genome sequence of Salmonella enterica serovar Cholerasuis, a highly invasive and resistant zoonotic pathogen. Nucleic Acids Res. 33:1690-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Decousser, J. W., L. Poirel, and P. Nordmann. 2001. Characterization of a chromosomally encoded extended-spectrum class A β-lactamase from Kluyvera cryocrescens. Antimicrob. Agents Chemother. 45:3595-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delcher, A. L., D. Karmon, D. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Conza, J., J. A. Ayala, P. Power, M. Mollerach, and G. Gutkind. 2002. Novel class 1 integron (InS21) carrying blaCTX-M-2 in Salmonella enterica serovar Infantis. Antimicrob. Agents Chemother. 46:2257-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ewing, B., and P. Green. 1998. Basecalling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 21.Ewing, B., L. Hillier, M. Wendl, and P. Green. 1998. Basecalling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 22.Foster, G. C., G. C. McGhee, A. L. Jones, and G. W. Sundin. 2004. Nucleotide sequence, genetic organization, and distribution of pEU30 and pEL60 from Erwinia amylovora. Appl. Environ. Microbiol. 70:7539-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furuya, N., T. Nisioka, and T. Komano. 1991. Nucleotide sequence an functions of the oriT operon in IncI1 plasmid R64. J. Bacteriol. 173:2231-2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gniadkowski, M. 2001. Evolution and epidemiology of extended-spectrum β-lactamases (ESBLs) and ESBL-producing microorganisms. Clin. Microbiol. Infect. 7:597-608. [DOI] [PubMed] [Google Scholar]
- 25.Gniadkowski, M., I. Schneider, A. Palucha, R. Jungwirth, B. Mikiewicz, and A. Bauernfeind. 1998. Cefotaxime-resistant Enterobacteriaceae isolates from a hospital in Warsaw, Poland: identification of a new CTX-M-3 cefotaxime-hydrolyzing β-lactamase that is closely related to the CTX-M-1/MEN1 enzyme. Antimicrob. Agents Chemother. 42:827-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Gołebiewski, M. 2004. Complete nucleotide sequence of pCTX-M3 plasmid and analysis of its genes responsible for conjugational transfer. Ph.D. thesis. Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Warsaw, Poland.
- 26.Gonzalez-Zorn, B., A. Catalan, J. A. Escudero, L. Dominguez, T. Teshager, C. Porrero, and M. A. Moreno. 2005. Genetic basis for dissemination of armA. J. Antimicrob. Chemother. 56:583-585. [DOI] [PubMed] [Google Scholar]
- 27.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 28.Ho, C., O. I. Kulaeva, A. S. Levine, and R. Woodgate. 1993. A rapid method for cloning mutagenic DNA repair genes: isolation of umu-complementing genes from multidrug resistance plasmids R391, R446b, and R471a. J. Bacteriol. 175:5411-5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Humeniuk, C., G. Arlet, V. Gautier, P. Grimont, R. Labia, and A. Philippon. 2002. β-Lactamases of Kluyvera ascorbata, probable progenitors of some plasmid encoded CTX-M types. Antimicrob. Agents Chemother. 46:3045-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karim, A., L. Poirel, S. Nagarajan, and P. Nordmann. 2001. Plasmid-mediated extended-spectrum β-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol. Lett. 201:237-241. [DOI] [PubMed] [Google Scholar]
- 31.Komano, T., N. Funayama, S.-R. Kim, and T. Nisioka. 1990. Transfer region of IncI1 plasmid R64 and role of shufflon in R64 transfer. J. Bacteriol. 172:2230-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Komano, T., S.-R. Kim, and T. Yoshida. 1995. Mating variation by DNA inversions of shufflon in plasmid R64. Adv. Biophys. 31:181-193. [DOI] [PubMed] [Google Scholar]
- 33.Komano, T., T. Yoshida, K. Narahara, and N. Furuya. 2000. The transfer region of IncI1 plasmid R64: similarities between R64 tra and Legionella icm/dot genes. Mol. Microbiol. 35:1348-1359. [DOI] [PubMed] [Google Scholar]
- 34.Lartigue, M.-F., L. Poirel, and P. Nordmann. 2004. Diversity of genetic environments of blaCTX-M genes. FEMS Microbiol. Lett. 234:201-207. [DOI] [PubMed] [Google Scholar]
- 35.Livermore, D. M. 2005. Minimising antibiotic resistance. Lancet Infect. Dis. 5:450-459. [DOI] [PubMed] [Google Scholar]
- 36.Livermore, D. M., R. Canton, M. Gniadkowski, P. Nordmann, G. M. Rossolini, G. Arlet, J. Ayala, T. Coque, I. Kern-Zdanowicz, F. Luzzaro, L. Poirel, and N. Woodford. 2007. CTX-M: changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 59:165-174. [DOI] [PubMed] [Google Scholar]
- 37.Liu, S. Y., L. H. Su, Y.L. Yeh, C. Chu, J.C. Lai, and C. H. Chiu. 2007. Characterisation of plasmids encoding CTX-M-3 extended-spectrum β-lactamase from Enterobacteriaceae isolated at a university hospital in Taiwan. Int. J. Antimicrob. Agents 29:440-445. [DOI] [PubMed] [Google Scholar]
- 38.Martinez, B., J. Tomkins, L. P. Wackett, R. Wing, and M. J. Sadowsky. 2001. Complete nucleotide sequence and organization of the atrazine catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP. J. Bacteriol. 183:5684-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mierzejewska, J., A. Kulinska, and G. Jagura-Burdzy. 2007. Functional analysis of replication and stability regions of broad-host-range conjugative plasmid pCTX-M3 from the IncL/M incompatibility group. Plasmid 57:95-107. [DOI] [PubMed] [Google Scholar]
- 40.Nogouchi, N., A. Emura, H. Matsuyama, K. O'Hara, M. Sasatsu, and M. Kono. 1995. Nucleotide sequence and characterization of erythromycin resistance determinant that encodes macrolide 2′-phosphotransferase I in Escherichia coli. Antimicrob. Agents Chemother. 39:2359-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Novais, A., R. Cantón, A. Valverde, E. Machado, J.-C. Galán, L. Peixe, A. Carattoli, F. Baquero, and T. M. Coque. 2006. Dissemination and persistence of blaCTX-M-9 are linked to class 1 integrons containing CR1 associated with defective transposon derivatives from Tn402 located in early antibiotic resistance plasmids of IncHI2, IncP1-alpha, and IncFI groups. Antimicrob. Agents Chemother. 50:2741-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Novais, A., R. Cantón, R. Moreira, L. Peixe, F. Baquero, and T. M. Coque. 2007. Emergence and dissemination of CTX-M-1-like-producing Enterobacteriaceae in Spain is caused by IncFII (CTX-M-15) and broad host range (CTX-M-1, -3, -32) plasmids. Antimicrob. Agents Chemother. 51:796-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olson, A. B., M. Silverman, D. A. Boyd, A. McGeer, B. M. Willey, V. Pong-Porter, N. Daneman, and M. R. Mulvey. 2005. Identification of a progenitor of the CTX-M-9 group of extended-spectrum β-lactamases from Kluyvera georgiana isolated in Guyana. Antimicrob. Agents Chemother. 49:2112-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palucha, A., B. Mikiewicz, W. Hryniewicz, and M. Gniadkowski. 1999. Concurrent outbreaks of extended-spectrum β-lactamase-producing organisms of the family Enterobacteriaceae in a Warsaw hospital J. Antimicrob. Chemother. 44:489-499. [DOI] [PubMed] [Google Scholar]
- 45.Pitout, J. D. D., K. B. Laupland, D. L. Church, M. L. Menard, and J. R. Johnson. 2005. Virulence factors of Escherichia coli isolates that produce CTX-M-type extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 49:4667-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poirel, L., P. Kämpfer, and P. Nordmann. 2002. Chromosome-encoded Ambler class A β-lactamase of Kluyvera georgiana, a probable progenitor of a subgroup of CTX-M extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 46:4038-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poirel, L., J.-W. Decousser, and P. Nordmann. 2003. Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob. Agents Chemother. 47:2938-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poirel, L., M.-F. Lartigue, J.-W. Decousser, and P. Nordmann. 2005. ISEcp1B-mediated transposition of blaCTX-M in Escherichia coli. Antimicrob. Agents Chemother. 49:447-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodríguez, M. M., P. Power, M. Radice, C. Vay, A. Famiglietti, M. Galleni, J. A. Ayala, and G. Gutkind. 2004. Chromosome-encoded CTX-M-3 from Kluyvera ascorbata: a possible origin of plasmid-borne CTX-M-1-derived cefotaximases. Antimicrob. Agents Chemother. 48:4895-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sabaté, M., F. Navarro, E. Miró, S. Campoy, B. Mirelis, J. Barbé, and G. Prats. 2002. Novel complex sul1-type integron in Escherichia coli carrying blaCTX-M-9. Antimicrob. Agents Chemother. 46:2656-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sagulenko, E., V. Sagulenko, J. Chen, and P. J. Christie. 2001. Role of Agrobacterium tumefaciens VirB11 ATPase in T-pilus assembly and substrate selection J. Bacteriol. 183:5813-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salzberg, S. L., A. Delcher, S. Kasif, and O. White. 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 16:544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sambrook, E. F., J. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 54.Stokes, H. W., C. Tomaras, Y. Parsons, and R. M. Hall. 1993. The partial 3′-conserved segment duplications in the integrons In6 from pSa and In7 from pDGO100 have a common origin. Plasmid 30:39-50. [DOI] [PubMed] [Google Scholar]
- 55.Takami, H., K. Nakasone, Y. Takaki, G. Maeno, R. Sasaki, N. Masui, F. Fuji, C. Hirama, Y. Nakamura, N. Ogasawara, S. Kuhara, and K. Horikoshi. 2000. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 28:4317-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thomas, C. M., et al. 2000. The horizontal gene pool. Bacterial plasmids and gene spread. Harwood Academic Publishers, Newark, NJ.
- 57.Thompson, J., D. Higgins, and T. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Toleman, M. A., P. M. Bennett, and T. R. Walsh. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 70:296-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsuchimoto, S., H. Ohtsubo, and E. Ohtsubo. 1988. Two genes, pemI and pemK, responsible for stable maintenance of resistance plasmid R100. J. Bacteriol. 170:1461-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams, L. E., C. Detter, K. Barry, A. Lapidus, and A. O. Summers. 2006. Facile recovery of individual high-molecular-weight, low-copy-number natural plasmids for genomic sequencing. Appl. Environ. Microbiol. 72:4899-4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wright, G. D. 2007. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 5:175-186. [DOI] [PubMed] [Google Scholar]