Abstract
The genetic elements carrying macrolide resistance genes in Streptococcus pneumoniae isolates belonging to CC271 were investigated. The international clone Taiwan19F-14 was found to carry Tn2009, a Tn916-like transposon containing tet(M) and mef(E). The dual erm(B) mef(E) isolates carried Tn2010, which is similar to Tn2009 with the addition of a putative new transposon, the erm(B) genetic element.
Macrolide resistance in Streptococcus pneumoniae is mediated by one of two main mechanisms, (i) target modification due to a ribosomal methylase encoded by erm(B), which confers high-level resistance to macrolides, lincosamides, and streptogramin B (MLSB phenotype), or (ii) an efflux transport system associated with the mef gene, conferring resistance to 14- and 15-membered macrolides only (M phenotype) (10). erm(B)-mediated erythromycin resistance is the most common mechanism in many areas of the world, including some European and Asian countries (14, 15, 17), whereas mef, with its most common variants mef(A) and mef(E), is predominant in the United States (5), Canada (8), and the United Kingdom (1). In recent years, pneumococcal isolates carrying both macrolide resistance genes erm(B) and mef were observed with increasing frequency, particularly in some Asian countries, in South Africa, and in the United States (4, 9, 12). In the United States, an increase in the proportion of pediatric isolates carrying dual macrolide resistance genes was noted following the introduction of the conjugate pneumococcal vaccine (5). The dual-gene isolates were mainly of serotype 19F or 19A and were multidrug resistant, being resistant to penicillin and tetracycline besides erythromycin and clindamycin (6). By multilocus sequence typing, most of these isolates were found to be genetically related, belonging mainly to three sequence types (STs), ST236, ST271, and ST320, that are included in clonal complex CC271 (4). CC271 also includes one of the internationally spread multidrug-resistant pneumococcal clones, designated Taiwan19F-14, which is resistant to penicillin, erythromycin, and tetracycline and carries the mef gene (13). This clone emerged at the beginning of the 1990s in Taiwan and spread to other Asian countries (9).
The aim of this study was to investigate the genetic elements present in the multidrug-resistant dual-gene isolates belonging to CC271. We have recently described two composite transposons of the Tn916 family, Tn2009 and Tn2010, both carrying mef(E). Tn2010 includes an additional erm(B) element and therefore carries dual macrolide resistance genes (3).
Thirteen multidrug-resistant S. pneumoniae isolates belonging to CC271 (ST236, ST271, or ST320) were examined (Table 1). These isolates included the Taiwan19F-14 type strain carrying mef (13) and 12 isolates carrying both erm(B) and mef, selected among a large collection of strains obtained from different geographical areas in 1999 to 2003, mainly from community-acquired lower respiratory tract infections (4, 5). All of these isolates were multidrug resistant, being nonsusceptible to penicillin (MIC range, 2 to 8 μg/ml), resistant to high levels of erythromycin and clindamycin, and also resistant to tetracycline in all cases but one.
TABLE 1.
Characteristics of the S. pneumoniae isolates used in this study
| Strain | Country of origin | Year(s) isolated | Serotype | Resistance patterna | Presence of resistance gene:
|
ST | ||
|---|---|---|---|---|---|---|---|---|
| mef(E) | erm(B) | tet(M) | ||||||
| Taiwan19F-14 | Taiwan | 1997 | 19F | Penr Eryr Tetr | + | − | + | 236 |
| GRMS01 | United States | 1999-2000d | 19F | Penr Eryr Clir Tetr | + | + | + | 271 |
| GRMS03 | United States | 1999-2000d | 19A | Penr Eryr Clir Tetr | + | + | + | 271 |
| GRMS05 | South Korea | 1999-2000d | 19F | Penr Eryr Clir Tetr | + | + | + | 271 |
| GRMS06 | United States | 2002-2003d | 19F | Penr Eryr Clir | + | + | +b | 271 |
| GRMS09 | South Africa | 2001-2002d | 19F | Penr Eryr Clir Tetr | + | + | + | 271 |
| GRMS13 | China | 2001-2002d | 19F | Penr Eryr Clir Tetr | + | + | + | 271 |
| GRMS17 | Brazil | 2001-2002d | 19F | Penr Eryr Clir Tetr | + | + | + | 271 |
| GRMS19 | United States | 1999-2000d | 19A | Penr Eryr Clir Tetr | + | + | + | 320 |
| GRMS20 | United States | 1999-2000d | 19F | Penr Eryr Clir Tetr | + | + | + | 236 |
| GRMS22 | South Korea | 1999-2000d | 19F | Penr Eryr Clir Tetr | + | + | + | 236 |
| GRMS23 | South Korea | 1999-2000d | 19A | Penr Eryr Clir Tetr | + | + | + | 320 |
| GRMS25 | South Korea | 1999-2000d | 19F | Penr Eryr Clir Tetr | + | + | + | 320 |
| PGX1416c | Unknown | Unknown | 19F | Penr Eryr Clir Tetr | + | + | + | 271 |
Penr, penicillin resistance; Eryr, erythromycin resistance; Clir, clindamycin resistance; Tetr, tetracycline resistance.
The gene sequence contains a frameshift mutation (see text).
Control strain.
Winter season.
PN150 and PGX1416, carrying Tn2009 and Tn2010, respectively, were used as control strains in PCR assays (2). The two mef variants, mef(A) and mef(E), erm(B), and tet(M) were detected by PCR (3, 14).
Analysis of the genetic elements in the isolates under study was performed by PCR mapping (2, 3). The sizes of the amplicons obtained were compared with those obtained with the control strains.
On the basis of the location of Tn2010 in PGX1416 (2), the insertions of the genetic elements were explored by two PCR assays. The left (L) junction was detected by primer SQ8 (5′-CAAAGCTASTTTTTTACCATAG-3′), which anneals to the pneumococcal chromosome, and primer TN4 (3), which anneals to Tn916. The right (R) junction was detected by primer TN6, which anneals to Tn916 (3), and primer SQ7 (5′-GTAATACAATTCCTTACCAACAG-3′), which anneals to the pneumococcal chromosome. In selected isolates, sequencing of both the L and R junctions was performed.
In the Taiwan19F-14 strain, erythromycin resistance was conferred by the mef(E) variant of mef and tetracycline resistance was conferred by tet(M). By PCR mapping, the fragments obtained were of the same size as those obtained from control strain PN150, indicating that Taiwan19F-14 carries Tn2009, which encompasses mef(E) and tet(M).
In all of the 12 dual-gene S. pneumoniae isolates, erm(B), mef(E), and tet(M) were detected. The tetracycline susceptibility of strain GMRS06 (MIC, 0.25 μg/ml) is likely explained by the one-nucleotide insertion found in the tet(M) coding sequence, causing an early stop codon. By PCR mapping, the fragments obtained with these isolates were of the same size as those obtained with control strain PGX1416. This indicates a genetic organization corresponding to Tn2010, a composite transposon similar to Tn2009, with the addition of an erm(B) genetic element integrated into an open reading frame (ORF) corresponding to orf20 of Tn916 (2). The erm(B) genetic element and the adjacent Tn916 regions are identical to sequences of S. pneumoniae strains 670-6B and G54 and of Tn916Erm (2), recently designated CTn6002, a transposon found in Streptococcus cristatus.
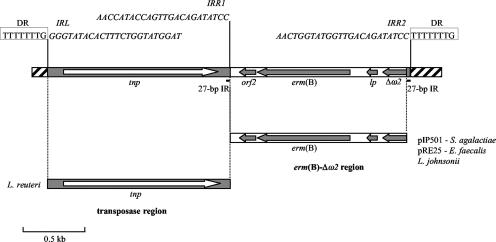
Detailed sequence analysis of the erm(B) genetic element revealed two different regions. A left (L) region (transposase region) of 1,438 bp was 96% identical to a sequence present in the Lactobacillus reuteri genome (accession no. NZ_AAOV01000043) that includes a putative transposase (tnp) homologous to those encoded by ISL3 family insertion sequences (http://www-is.biotoul.fr/is.html) (11).
A right (R) region [erm(B)-Δω2 region] of 1,367 bp, comprising the MLS leader peptide (lp), the erm(B) methylase, and two additional ORFs (Fig. 1), shows 100% identity to sequences present in different multiresistant plasmids belonging to the inc18 incompatibility group, such as pIP501 of Streptococcus agalactiae (18) and pRE25 of Enterococcus faecalis (16), and also to a region found in the Lactobacillus johnsonii chromosome that is designated the erm(B) locus (accession no. DQ518904) (7). The last ORF of this region, designated Δω2, is 100% identical to the C terminus of the ω2 protein (accession no. AJ549242), an additional copy of the ω protein, involved in copy number control of inc18 plasmids (19). The erm(B)-Δω2 region is flanked by perfect 27-bp inverted repeats (IRs). Putative imperfect IRs can be identified, two at both ends of the transposase region (IRL and IRR1) and an additional one at the R end of the erm(B)-Δω2 region (IRR2). The sequences of IRR1and IRR2 partly overlap the sequences of the 27-bp IRs (Fig. 1). On the basis of these observations, it could be hypothesized that the erm(B) genetic element is a putative transposon derived from the assembly of a transposase region from L. reuteri, with an erythromycin resistance region of likely plasmid origin. Consistent with the erm(B) genetic element arriving by transposition was the identification of 8-bp direct repeats flanking the element, likely representing target duplication (Fig. 1).
FIG. 1.
Organization of the erm(B) genetic element of Tn2010. The two distinct regions composing the element [the transposase region and the erm(B)-Δω2 region] are shown below the element, with the respective indications of the origin of identical sequences (microorganism name or plasmid designation). Arrows correspond to ORFs, with the direction of transcription indicated by the arrowheads. The striped bar corresponds to Tn916 sequences at the insertion of the erm(B) element (orf20). The 8-bp direct repeats (DR) flanking the element are boxed. The positions of the putative imperfect IRs (IRL, IRR1, and IRR2) and their corresponding sequences (in italics) are indicated. The positions of the 27-bp perfect IRs are indicated by the black bars.
The analysis of the chromosomal location showed that both Tn2009 in Taiwan19F-14 and Tn2010 in the dual-gene isolates were inserted at the same site, previously identified in strain PGX1416, inside an ORF corresponding to spr1764 of R6 (2). This finding supports the hypothesis that erm(B) mef(E) dual-gene isolates belonging to CC271 have evolved from multidrug-resistant Taiwan19F-14 by acquisition of the erm(B) genetic element (4, 9). Although the erm(B) genetic element does not appear to be transferable by conjugation among pneumococci, it cannot be excluded that CC271 isolates have acquired this element from other species of streptococci or other gram-positive bacteria by conjugation.
The selective advantage of acquiring erm(B) on top of mef(E) consists of gaining not only high-level erythromycin resistance but also an increased number of antimicrobial classes to which the bacteria are resistant, including lincosamides and streptogramins.
Nucleotide sequence accession number.
The sequence of the erm(B) element in PGX1416 has been assigned GenBank accession no. EF592165.
Acknowledgments
This study was supported in part by the DRESP2 contract with the European Commission (6th Framework Program) and by the FIRB project from the Italian MIUR.
Footnotes
Published ahead of print on 20 August 2007.
REFERENCES
- 1.Amezaga, M. R., P. E. Carter, P. Cash, and H. McKenzie. 2002. Molecular epidemiology of erythromycin resistance in Streptococcus pneumoniae isolates from blood and noninvasive sites. J. Clin. Microbiol. 40:3313-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Del Grosso, M., R. Camilli, F. Iannelli, G. Pozzi, and A. Pantosti. 2006. The mef(E)-carrying genetic element (mega) of Streptococcus pneumoniae: insertion sites and association with other genetic elements. Antimicrob. Agents Chemother. 50:3361-3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Del Grosso, M., A. Scotto d'Abusco, F. Iannelli, G. Pozzi, and A. Pantosti. 2004. Tn2009, a Tn916-like element containing mef(E) in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 48:2037-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farrell, D. J., S. G. Jenkins, S. D. Brown, M. Patel, B. S. Lavin, and K. P. Klugman. 2005. Emergence and spread of Streptococcus pneumoniae with erm(B) and mef(A) resistance. Emerg. Infect. Dis. 11:851-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farrell, D. J., K. P. Klugman, and M. Pichichero. 2007. Increased antimicrobial resistance among nonvaccine serotypes of Streptococcus pneumoniae in the pediatric population after the introduction of 7-valent pneumococcal vaccine in the United States. Pediatr. Infect. Dis. J. 26:123-128. [DOI] [PubMed] [Google Scholar]
- 6.Farrell, D. J., I. Morrissey, S. Bakker, L. Morris, S. Buckridge, and D. Felmingham. 2004. Molecular epidemiology of multiresistant Streptococcus pneumoniae with both erm(B)- and mef(A)-mediated macrolide resistance. J. Clin. Microbiol. 42:764-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flórez, A. B., M. S. Ammor, S. Delgado, and B. Mayo. 2006. Molecular analysis of a chromosome-carried erm(B) gene and its flanking insertion points in Lactobacillus johnsonii G41. Antimicrob. Agents Chemother. 50:4189-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoban, D. J., A. K. Wierzbowski, K. Nichol, and G. G. Zhanel. 2001. Macrolide-resistant Streptococcus pneumoniae in Canada during 1998-1999: prevalence of mef(A) and erm(B) and susceptibilities to ketolides. Antimicrob. Agents Chemother. 45:2147-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ko, K. S., and J. H. Song. 2004. Evolution of erythromycin-resistant Streptococcus pneumoniae from Asian countries that contains erm(B) and mef(A) genes. J. Infect. Dis. 190:739-747. [DOI] [PubMed] [Google Scholar]
- 10.Leclercq, R., and P. Courvalin. 2002. Resistance to macrolides and related antibiotic in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:2727-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGee, L., K. P. Klugman, A. Wasas, T. Capper, A. Brink, and the Antibiotics Surveillance Forum of South Africa. 2001. Serotype 19F multiresistant pneumococcal clone harboring two erythromycin resistance determinants [erm(B) and mef(A)] in South Africa. Antimicrob. Agents Chemother. 45:1595-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hakenbeck, W. Hryniewicz, J. C. Lefevre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J. Clin. Microbiol. 39:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monaco, M., R. Camilli, F. D'Ambrosio, M. Del Grosso, and A. Pantosti. 2005. Evolution of erythromycin resistance in Streptococcus pneumoniae in Italy. J. Antimicrob. Chemother. 55:256-259. [DOI] [PubMed] [Google Scholar]
- 15.Reinert, R. R., A. Ringelstein, M. van der Linden, M. Y. Cil, A. Al-Lahham, and F. J. Schmitz. 2005. Molecular epidemiology of macrolide-resistant Streptococcus pneumoniae isolates in Europe. J. Clin. Microbiol. 43:1294-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwarz, F. V., V. Perreten, and M. Teuber. 2001. Sequence of the 50-kb conjugative multiresistance plasmid pRE25 from Enterococcus faecalis RE25. Plasmid 46:170-187. [DOI] [PubMed] [Google Scholar]
- 17.Song, J. H., H. H. Chang, J. Y. Suh, K. S. Ko, S. I. Jung, W. S. Oh, K. R. Peck, N. Y. Lee, Y. Yang, A. Chongthaleong, N. Aswapokee, C. H. Chiu, M. K. Lalitha, J. Perera, T. T. Yee, G. Kumararasinghe, F. Jamal, A. Kamarulazaman, N. Parasakthi, P. H. Van, T. So, T. K. Ng, and the ANSORP Study Group. 2004. Macrolide resistance and genotypic characterization of Streptococcus pneumoniae in Asian countries: a study of the Asian Network for Surveillance of Resistant Pathogens (ANSORP). J. Antimicrob. Chemother. 53:457-463. [DOI] [PubMed] [Google Scholar]
- 18.Thompson, J. K., and M. A. Collins. 2003. Completed sequence of plasmid pIP501 and origin of spontaneous deletion derivatives. Plasmid 50:28-35. [DOI] [PubMed] [Google Scholar]
- 19.Zúñiga, M., I. Pardo, and S. Ferrer. 2003. Conjugative plasmid pIP501 undergoes specific deletions after transfer from Lactococcus lactis to Oenococcus oeni. Arch. Microbiol. 180:367-373. [DOI] [PubMed] [Google Scholar]