Abstract
In this study, we analyzed the mechanisms of multiresistance for 22 clinical multiresistant and clonally different Pseudomonas aeruginosa strains from Germany. Twelve and 10 strains originated from cystic fibrosis (CF) and non-CF patients, respectively. Overproduction of the efflux systems MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY-OprM was studied. Furthermore, loss of OprD, alterations in type II topoisomerases, AmpC overproduction, and the presence of 25 acquired resistance determinants were investigated. The presence of a hypermutation phenotype was also taken into account. Besides modifications in GyrA (91%), the most frequent mechanisms of resistance were MexXY-OprM overproduction (82%), OprD loss (82%), and AmpC overproduction (73%). Clear differences between strains from CF and non-CF patients were found: numerous genes coding for aminoglycoside-modifying enzymes and located, partially in combination with β-lactamase genes, in class 1 integrons were found only in strains from non-CF patients. Furthermore, multiple modifications in type II topoisomerases conferring high quinolone resistance levels and overexpression of MexAB-OprM were exclusively detected in multiresistant strains from non-CF patients. Correlations of the detected phenotypes and resistance mechanisms revealed a great impact of efflux pump overproduction on multiresistance in P. aeruginosa. Confirming previous studies, we found that additional, unknown chromosomally mediated resistance mechanisms remain to be determined. In our study, 11 out of 12 strains and 3 out of 10 strains from CF patients and non-CF patients, respectively, were hypermutable. This extremely high proportion of mutator strains should be taken into consideration for the treatment of multiresistant P. aeruginosa.
The ubiquitous environmental bacterium Pseudomonas aeruginosa is one of the most important nosocomial pathogens, especially in intensive care units (ICUs) (39, 42). It is also the pathogen most frequently associated with morbidity and mortality in cystic fibrosis (CF) and other underlying chronic respiratory diseases causing chronic lung infections (6, 8, 22).
Intrinsic and acquired antibiotic resistance makes P. aeruginosa one of the most difficult organisms to treat. The high intrinsic antibiotic resistance of P. aeruginosa is due to several mechanisms: a low outer membrane permeability, the production of an AmpC β-lactamase, and the presence of numerous genes coding for different multidrug resistance efflux pumps (20).
A high number of acquired resistance genes coding for aminoglycoside-modifying enzymes (AME) and β-lactamases have been noted in P. aeruginosa (2). Extended-spectrum β-lactamases have been increasingly reported (3, 7, 33, 41, 44), and metallo-β-lactamases have also started to emerge in P. aeruginosa (43).
Besides horizontal transfer of resistance genes, acquired resistance also occurs via mutations in chromosomal genes. Alterations in the quinolone resistance-determining regions (QRDR) in the genes coding for DNA gyrase and topoisomerase IV play an important role in quinolone resistance in P. aeruginosa (14). Loss of OprD is the most prevalent mechanism of resistance to carbapenems and is associated with resistance to imipenem and reduced susceptibility to meropenem (28). To date, four efflux systems of the RND (resistance nodulation division) family, MexAB-OprM, MexEF-OprN, MexCD-OprJ, and MexXY-OprM, are well characterized and known to contribute significantly to antimicrobial resistance in P. aeruginosa (17, 27, 34, 35). Their overproduction confers cross-resistance or reduced susceptibility to several β-lactams (piperacillin, ticarcillin, ceftazidime, cefepime, and meropenem), quinolones, and aminoglycosides (20). Hyperproduction of the inducible AmpC β-lactamase is mostly due to the inactivation of the amidase AmpD (15) and two additional AmpD homologues (16), leading to an increase of inducer molecules.
Among P. aeruginosa strains from CF patients, a high proportion are hypermutable (30). Hypermutation is characterized by an increased spontaneous-mutation rate and seems to be an advantage for fast adaptation to a heterogeneous and fluctuating environment, like the lung of a chronically infected CF patient (30). Recently, studies found hypermutation to be the key factor in development of mutation-mediated multiresistance in patients with chronic P. aeruginosa lung infections (23). Among strains from patients with acute infections, mutator strains are rare (9).
Accumulation of resistance mechanisms frequently occurs in P. aeruginosa strains from patients with CF, where sequential treatment of the chronic infection with various antibiotics is common. In patients without CF, multiresistant P. aeruginosa strains are less frequent but are associated with severe adverse clinical outcomes (1).
The aim of our study was to investigate the mechanisms of resistance of multiresistant P. aeruginosa strains from CF patients and non-CF patients from Germany. Furthermore, we intended to get insights into the correlation between the genetic resistance determinants detected and the respective resistance phenotypes.
(This study was presented in part at the 45th Interscience Conference on Antimicrobial Agents and Chemotherapy, 16 to 19 December 2005, Washington, DC.)
MATERIALS AND METHODS
Bacterial strains.
Twenty-two multiresistant, clonally different (determined by pulsed-field gel electrophoresis [PFGE]) P. aeruginosa strains (I to XXII) were obtained from three German hospitals participating in the German Network for Antimicrobial Resistance Surveillance (GENARS) from January through March 2004. The strains showed reduced susceptibility (intermediate or resistant according to the guidelines of the German Institute for Standardization) to each of the following class representatives: imipenem, ceftazidime, piperacillin, ciprofloxacin, and gentamicin. A reidentification was performed by verifying growth on cetrimide agar plates and confirming the presence of the P. aeruginosa-specific chromosomal algD gene by PCR (see the supplemental material).
The 22 strains originated from 11 CF and 10 non-CF patients. For three CF patients, two clonal variants (VIa and VIIa) and another clonal isolate of strain XI were also investigated. All CF patients were medicated at ambulant CF wards. Of the non-CF patients, five were inpatients from normal wards (heart vessel surgery, infection, internal medicine, and neurosurgery) and five were from ICU wards (anesthesiology, interdisciplinary ICU, internal medicine, neurology, and neurosurgery). All strains from CF patients were isolated from sputum specimens. The strains isolated from non-CF patients originated from several anatomic sites: respiratory tract (sputum, bronchoalveolar lavage, and tracheal secretion), wound, vagina, and urinary tract.
In vitro susceptibility testing.
Susceptibility testing was performed by broth microdilution with reference to the guidelines of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (5).
PFGE.
Genomic DNA was prepared using GenePath group 3 reagents as recommended by the manufacturer (Bio-Rad, Marnes-la-Coquette, France). Analysis of genomic DNA macrorestriction patterns by PFGE was carried out after digestion with SpeI using the CHEF mapper system (Bio-Rad, Hercules, CA). Clonal relationships were interpreted in accordance with the guidelines proposed by Tenover et al. (40).
Mutation frequency determination.
Mutation frequencies conferring rifampin resistance were determined in triplicate, following a previously described method (29). According to Oliver et al., a strain was considered a mutator strain when the mutation frequency conferring rifampin resistance was at least 20-fold higher than that observed for the wild-type strain PAO1 (30).
Efflux pump inhibition test.
Efflux pump overexpression was detected using an efflux pump inhibitor (EPI) test (21). Levofloxacin MICs (concentrations ranging from 256 mg/liter to 0.06 mg/liter) were tested either with or without 20 μg/ml of the EPI Phe-Arg-β-naphtylamide (Sigma-Aldrich, Taufkirchen, Germany). An at least 16-fold reduction of the levofloxacin MIC in the presence of Phe-Arg-β-naphtylamide indicated the overproduction of efflux pumps.
Cephalosporinase inhibition test.
Overproduction of the chromosomal cephalosporinase AmpC was evaluated by a disk diffusion test with 30-μg ceftazidime discs (Oxoid Limited, Basingstoke, United Kingdom) on Mueller- Hinton agar with or without cloxacillin (Sigma-Aldrich, Taufkirchen, Germany) at 500 mg/liter as described elsewhere (4). The test result was considered positive when the ceftazidime inhibition zone diameter increased by at least 10 mm in the presence of cloxacillin.
EDTA-phenanthroline-imipenem microdilution test.
To detect metallo-β-lactamases, MICs of imipenem (concentrations in the range of 512 mg/liter to 0.25 mg/liter) were tested in the absence or presence of 0.4 mM EDTA and 0.04 mM 1,10-phenanthroline (Sigma-Aldrich, Taufkirchen, Germany) (26).
Molecular analysis techniques.
Extraction of plasmid DNA was performed using a Fast Plasmid mini kit (Eppendorf, Hamburg, Germany). Genomic DNA was purified from P. aeruginosa with a QIAGEN tissue kit (QIAGEN, Hilden, Germany). To analyze chromosomally mutation-mediated resistance mechanisms, the QRDRs of the DNA gyrase and topoisomerase IV genes gyrA and parC, in addition to the gene oprD, encoding the porin OprD, were amplified and sequenced. Furthermore, the efflux regulator-encoding genes mexR, nalC, nfxB, mexZ, mexT, and mexS were sequenced.
The relevance of amino acid changes not previously reported was analyzed. The positions in functional domains were investigated by localizing the changed amino acid either in the tertiary structure if available (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Structure) or in the secondary structure by using LOOPP @ CBSU software, version 3.0 (http://cbsuapps.tc.cornell.edu/loopp.aspx).
Class 1 and 2 integrons were detected using specific primers for the respective integrase genes intI1 and intI2. To detect acquired resistance genes, an elaborate PCR screening for β-lactamase and AME genes and qnrA was carried out.
The genetic context of the detected resistance genes was analyzed by PCR mapping (19). Class 1 integron-specific primers binding to the 5′-conserved segment (intI1) and to the 3′-conserved segment (ΔqacE1) and a set of primers binding to the resistance genes found were used. The identities of PCR screening and mapping products were partially verified by sequencing.
All primers used for PCR techniques and sequencing are listed in the supplemental material (see Table S1 in the supplemental material). Sequencing was performed by SEQLAB Sequence Laboratories Göttingen, Göttingen, Germany.
RESULTS
Hypermutation.
Fourteen of the 22 clonally different multiresistant strains showed at least a 20-fold-higher frequency of mutations conferring rifampin resistance than that observed for the wild-type strain PA01. The mutation frequencies ranged from (5.85 ± 0.68) × 10−7 to (1.43 ± 0.15) × 10−5 (Table 1) .
TABLE 1.
Multiresistant P. aeruginosa strains from CF and non-CF patientsa
| Epidemiology | Phenotypic test result(s)
|
Genotypic test result(s) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | Patient | Material | Ward | Type of ward | Center | Mutation frequencyb | Cephalosporinase inhibition test | MBL test | EPI test | Chromosomal alteration(s) in indicated repressor
|
Acquired resistance and integrase genes | ||||||||
| MexRc | NalCc | NfxBc | MexTc | MexSc | MexZc | OprD | GyrA | ParC | |||||||||||
| From CF patients | |||||||||||||||||||
| I | A | SPU | CF | A | 1 | (3.26 ± 0.65) × 10−9 | Leu138Argd, deletion C595, frameshiftd | Deletion bp 745-748, frameshift | Thr83Ile | − | − | ||||||||
| II | A | SPU | CF | A | 1 | (3.56 ± 1.67) × 10−6 | NR | − | + | − | Gly71Glu | − | Gln134stopd | Trp138stop | − | − | − | ||
| III | B | SPU | CF | A | 1 | (1.09 ± 0.21) × 10−5 | + | NG | − | Tyr49Cysd | Trp277stop, Asp318Asn | Asp87Asn | − | − | |||||
| IV | C | SPU | CF | A | 1 | (2.00 ± 0.19) × 10−6 | NR | − | − | Leu123stopd | Tyr91stop | Asp87Asn | − | − | |||||
| V | D | SPU | CF | N | 1 | (1.27 ± 0.93) × 10−5 | NR | − | + | − | Gly71Glu | Leu62Prod Arg186His | Val58Ala, Gly89Serd, Arg143Glnd | Trp138stop | Thr83Ile | − | − | ||
| VI | E | SPU | CF | A | 1 | (6.57 ± 1.14) × 10−7, | + | NG | − | Functional | Gly68Serd | Arg65His | − | Thr83Ile | − | − | |||
| VIag | SPU | CF | 3.61 × 10−9 | − | − | Val23Glyd | C insertion after A1205, frameshift | − | − | ||||||||||
| VII | F | SPU | CF | A | 1 | (2.50 ± 0.82) × 10−6 | + | − | + | Val126Glu | Gly71Glu, Arg209Ser | − | Functional | Phe273Iled | deletion G142, frameshiftd | − | Thr83Val | − | − |
| VIIag | SPU | CF | Functional Val199Ala | Thr83Ala | − | ||||||||||||||
| VIII | G | SPU | CF | A | 1 | (7.30 ± 4.91) × 10−7 | − | NG | NG | − | Gly71Glu, Arg209Ser | − | Thr32Iled, Ala175Val | Deletion G1017, frameshift | Thr83Ile | − | − | ||
| IX | H | SPU | CF | A | 1 | (1.94 ± 0.24) × 10−6 | + | − | − | Ser44Phed | Trp277stop | Thr83Ile | − | − | |||||
| X | I | SPU | CF | A | 1 | (5.85 ± 0.68) × 10−7 | + | − | − | Asp155Glyd | G insertion after G635, frameshift | Thr83Ile | − | − | |||||
| XI | J | SPU | CF | A | 1 | (1.43 ± 0.15) × 10−5, 1,71 × 10−7 | + | − | − | Arg45Leud | Trp277stop | Asp87Asn | − | − | |||||
| XII | K | SPU | CF | A | 2 | (1.20 ± 0.14) × 10−6 | + | − | + | Val126Glu | Gly71Glu, Arg209Ser | Arg21His Asp56GlydSer167Pro | Leu138Argd, deletion bp 447-630, frameshiftd | Trp277stop | Thr83Ile | − | − | ||
| From non-CF patients | |||||||||||||||||||
| XIII | L | SPU | MED | N | 1 | (8.03 ± 3.68) × 10−9 | + | − | + | − | Gly71Glu, Arg209Ser | − | bp 318-592d,f | Trp277stop | Thr83Ile | − | − | ||
| XIV | M | SPU | INF | N | 1 | (1.47 ± 0.48) × 10−8 | + | − | − | Deletion bp 491-496, frameshiftd | Trp339stop | Thr83Ile | − | − | |||||
| XV | N | BAL | ID | I | 2 | (3.06 ± 1.26) × 10−6 | + | − | − | Leu138Arg, deletion bp 515-532, frameshiftd | Deletion bp 155-167, frameshift | Thr83Ile | − | − | |||||
| XVI | O | BAL | NEU | I | 2 | (2.01 ± 0.66) × 10−8 | + | − | + | Val126Glu | Gly71Glu | − | − | Deletion C393, frameshift | Thr83Ile | Ser80Leu | aac(6′)Ib′, intI1 | ||
| XVII | P | SPU | MED | I | 2 | (3.50 ± 0.66) × 10−8 | + | − | − | Gly89Serd, Asp155Glyd | Trp138stop | Thr83Ile | − | aadA1, aadB, aphA1-IAB, intI1 | |||||
| XVIII | Q | BLO | MED | I | 2 | (3.24 ± 0.55) × 10−8 | + | − | + | Lys44Mete Val126Glu | Gly71Glu Glu153Gln | − | Val48Alae | Deletion A1007, frameshift | Thr83Ile Asp87His | Ser80Leu | aacA7, aacA8, blaOXA-2, intI1 | ||
| XIX | R | URM | HGC | N | 2 | (1.57 ± 0.44) × 10−8 | + | − | − | Functional | − | − | − | Thr83Ile | Ser80Leu | aac(6′)Ib, aadA2, blaPSE-1, intI1 | |||
| XX | S | SPU | INF | N | 3 | (5.86 ± 1.41) × 10−8 | + | − | + | C insertion after G75, frameshiftd | − | Val48Alae | Gln327stop | Thr83Ile | − | aac(6′)Ib′, aadA2, intI1 | |||
| XXI | T | WUP | ANA | I | 3 | (2.26 ± 0.23) × 10−6 | − | − | + | Asn insertion after Leu52d | − | Functional | − | − | − | Thr83Ile Asp87Gly | Ser80Leu | − | |
| XXII | U | TRS | NEC | N | 3 | (5.40 ± 1.65) × 10−6 | − | − | + | Val126Glu | C19 deletion Gly71Glu Ala145Val | Arg21His Asp56Glyd | Val48Alae, Asn186Ser | C insertion after T1002, frameshift | − | − | − | ||
| PAO1 | (3,07 ± 0.58) × 10−8 | − | − | − | |||||||||||||||
MBL, metallo-β-lactamase; BAL, bronchoalveolar lavage sample; BLO, blood culture; SPU, sputum; TRS, tracheal secretion; URM, midstream urine; WUP, wound pus; ANA, anesthesiology; HGC, heart vessel surgery; INF, infection ward; ID, interdisciplinary ICU; MED, internal medicine; NEC, neurosurgery; NEU, neurology; A, ambulant; I, intensive; N, normal; +, positive test result; −, negative test result or no modification or resistance gene found; NR, no result; NG, no growth in the presence of the inhibitors.
Mutation frequencies of mutator strains are indicated in bold. Each strain was tested in triplicate; the additionally tested variant of clone VI and another strain with a PFGE pattern identical to that of clone XI showed divergent results for hypermutability (tested once).
Modifications that are relevant for the function of the regulator are indicated in bold.
Relevance postulated on the basis of the localization and sort of the amino acid change.
Relevance elsewhere experimentally proven.
Repeated sequencing attempts revealed nondifferentiable multisignals for bp 318 to 592.
Clones VIa and VIIa are additionally tested variants of clones VI and VII, respectively, showing differences in mutation frequencies and/or chromosomal alterations.
Eleven out of 12 multiresistant strains from CF patients were hypermutable. Of note, for two hypermutable strains (VI and XI) collected from CF patients, clonally related and identical strains, respectively, that did not show hypermutation phenotypes were found. The clonal variant VIa had a mutation frequency that was 2 orders of magnitude lower than that of strain VI. The same phenomenon was found for an additionally tested strain with a PFGE pattern identical to that of strain XI. All CF patients were infected by multiresistant strains with mutator phenotypes. Interestingly, three mutator strains were also found among the multiresistant strains isolated from non-CF patients. Two of those hypermutable strains originated from the respiratory tract, but one was isolated from wound specimens.
Chromosomally mediated resistance mechanisms. (i) Efflux pump overproduction.
The EPI test indicated efflux pump overexpression for 10 strains (Table 2). For strain VIII, the EPI test yielded no result because the strain showed no growth in the presence of the efflux inhibitor.
TABLE 2.
Correlation of resistance phenotypes and mechanisms relevant for resistance to ureidopenicillins and cephalosporins, carbapenems, aminoglycosides, and quinolones for 22 multiresistant P. aeruginosa strainsa
| Strain | MIC (mg/liter) for:
|
Result for indicated resistance mechanism | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ureidopenicillins and cephalosporins
|
Carbapenems
|
Aminoglycosides
|
Quinolones
|
Efflux
|
OprD status | Type II topoisomerase
|
AmpC overproduction | Acquired resistance gene(s) | ||||||||||||||
| Piperacillin | Piperacillin-tazobactam | Ceftazidime | Cefepime | Imipenem | Meropenem | Gentamicin | Tobramycin | Amikacin | Ciprofloxacin | Levofloxacin | EPI test | MexR, NalC (MexAB-OprM) | NfxB (MexCD-OprJ) | MexT (MexEF-OprN) | MexS (MexEF-OprN) | MexZ (MexXY-OprM) | GyrA | ParC | ||||
| From CF patients | ||||||||||||||||||||||
| I | 128 | 128 | >32 | >32 | 32 | 8 | 16 | 2 | 32 | 4 | 8 | − | + | Loss | Thr83Ile | − | + | − | ||||
| II | >128 | >128 | >32 | >32 | 32 | 8 | 32 | 4 | >32 | 2 | 4 | + | − | − | + | Loss | − | − | NA | − | ||
| III | >128 | >128 | >32 | >32 | 32 | 8 | 32 | 8 | >32 | 4 | 8 | − | + | Loss | Asp87Asn | − | + | − | ||||
| IV | >128 | >128 | >32 | >32 | 8 | 16 | >32 | 32 | >32 | 2 | 4 | − | + | Loss | Asp87Asn | − | NA | − | ||||
| V | >128 | >128 | >32 | >32 | 16 | 16 | >32 | 8 | >32 | 4 | 8 | + | − | + | + | Loss | Thr83Ile | − | NA | − | ||
| VI | 64 | 32 | 32 | 32 | 8 | 4 | >32 | 4 | >32 | 4 | 8 | − | Intact | + | − | NM | Thr83Ile | − | + | − | ||
| VIab | + | Loss | − | |||||||||||||||||||
| VII | >128 | >128 | >32 | >32 | 16 | 16 | 16 | 2 | 32 | 8 | 8 | + | − | − | Intact | + | + | NM | Thr83Val | − | + | − |
| VIIac | Thr83Ala | |||||||||||||||||||||
| VIII | 64 | 32 | 8 | 32 | 8 | 4 | 16 | 2 | >32 | 8 | 16 | NA | − | + | Loss | Thr83Ile | − | − | − | |||
| IX | >128 | >128 | >32 | >32 | >32 | 32 | 16 | 4 | 32 | >8 | 128 | − | + | Loss | Thr83Ile | − | + | − | ||||
| X | >128 | >128 | >32 | 32 | 16 | 4 | 4 | 1 | 8 | 4 | 4 | − | + | Loss | Thr83Ile | − | + | − | ||||
| XI | 128 | 128 | 32 | 32 | 16 | 8 | 16 | 2 | 16 | 2 | 4 | − | + | Loss | Asp87Asn | − | + | − | ||||
| XII | >128 | 128 | >32 | 32 | 16 | 4 | 4 | 4 | 32 | >8 | 16 | + | − | + | + | Loss | Thr83Ile | − | + | − | ||
| From non-CF patients | ||||||||||||||||||||||
| XIII | >128 | >128 | >32 | >32 | 32 | 16 | 32 | 8 | >32 | 4 | 8 | + | − | − | − | Loss | Thr83Ile | − | + | − | ||
| XIV | >128 | >128 | >32 | >32 | >32 | 32 | 16 | 2 | >32 | 4 | 8 | − | + | Loss | Thr83Ile | − | + | − | ||||
| XV | 64 | 32 | 16 | >32 | 32 | 16 | >32 | >32 | >32 | >8 | 32 | − | + | Loss | Thr83Ile | − | + | − | ||||
| XVI | 64 | 32 | 32 | 16 | 16 | 8 | >32 | 16 | 8 | >8 | 32 | + | − | − | − | Loss | Thr83Ile | Ser80Leu | aac(6′)Ib′ | |||
| XVII | 64 | 32 | >32 | >32 | 32 | 16 | >32 | >32 | 16 | >8 | 16 | − | + | Loss | Thr83Ile | − | + | aadA1, aadB, aphA1-IAB | ||||
| XVIII | 32 | 16 | 16 | 16 | 16 | 16 | 32 | 16 | >32 | >8 | 128 | + | + (MexR) | − | + | Loss | Thr83Ile Asp87His | Ser80Leu | + | aacA7, aacA8, blaOXA-2 | ||
| XIX | >128 | >128 | 32 | >32 | 32 | 16 | 16 | 16 | 32 | >8 | 16 | − | Intact | − | − | NM | Thr83Ile | Ser80Leu | + | aac(6′)Ib, aadA2, blaPSE-1 | ||
| XX | >128 | >128 | >32 | >32 | 16 | 8 | >32 | 16 | 16 | >8 | 32 | + | + (MexR) | − | + | Loss | Thr83Ile | − | + | aac(6′)Ib′, aadA2 | ||
| XXI | 32 | 32 | 16 | 8 | 4 | 2 | 8 | 2 | 16 | >8 | 128 | + | + (MexR) | − | Intact | − | − | NM | Thr83Ile Asp87Gly | Ser80Leu | − | − |
| XXII | 32 | 32 | 16 | 16 | 16 | 16 | 2 | 1 | 8 | 2 | 8 | + | + (NalC) | + | + | Loss | − | − | − | − | ||
| PAO1 | 4 | 4 | 2 | 2 | 2 | 0.5 | 2 | 0.5 | 4 | 0.13 | 0.5 | |||||||||||
Variants of strains are listed only for cases in which divergent results for resistance mechanisms were obtained. NA, no analysis possible; NM, no modification; +, positive test result or detection of resistance-relevant modification(s); −, negative test result or no detection of resistance-relevant modification(s) or acquired genes.
Variant of clone VI.
Variant of clone VII.
The phenotypic EPI test is useful only for indicating the collective extent of overproduction of the efflux pumps MexAB-OprM, MexCD-OprJ, and MexEF-OprN, which can all be inhibited by Phe-Arg-β-naphtylamide. To differentiate between those efflux pumps and to investigate the overproduction of MexXY-OprM, we used the following strategy. For strains with positive EPI test results, the efflux regulator-encoding genes mexR (MexAB-OprM) and nfxB (MexCD-OprJ) were sequenced. Strains without modifications or with functionally irrelevant modifications in MexR were additionally investigated for alterations in nalC leading to the overexpression of mexAB-oprM. mexZ (MexXY-OprM) was sequenced for all strains. For strains showing no disruption of oprD, the carbapenem resistance was further investigated by sequencing mexT (MexEF-OprN). As the influence of the oxidoreductase MexS on the expression of MexEF-OprN had been proven previously (38), for these strains the gene mexS was also sequenced.
For 6 (V, XII, XVIII, XX, XXI, and XXII) of the 10 strains with positive EPI test results, functionally relevant modifications in the repressors of MexAB-OprM (MexR/NalC) and/or MexCD-OprJ (NfxB) were detected (Table 2). Functionally relevant modifications in the repressors of both efflux systems were found for only one strain (XXII). For the VI (except the variant VIa), VII, XIX, and XXI strains, whose carbapenem resistance levels were not due to the loss of the porin OprD, MexT was found to be a functional expression activator. For two of those strains, modifications in MexS were detected. A high proportion of strains (18 out of 22) with functionally relevant modifications in MexZ were found.
(ii) Loss of OprD.
Loss of OprD due to premature stop codons or frameshifts was found in 18 out of 22 multiresistant P. aeruginosa strains.
(iii) Type II topoisomerase alterations.
Modifications in the QRDR in the genes coding for the quinolone target enzymes DNA gyrase and topoisomerase IV were found in 20 out of the 22 multiresistant P. aeruginosa strains investigated in this study. Sixteen strains had alterations in GyrA only. Simultaneous mutations in gyrA and parC were found four times, exclusively among strains from non-CF patients. In two strains and one clonal variant (VIa), no target modifications were detected. None of the strains showed mutations in parC only.
(iv) AmpC overproduction.
Up-regulated production of the chromosomal β-lactamase AmpC was shown for 17 of the 22 multiresistant P. aeruginosa strains by using the cephalosporinase inhibition test. For three strains (II, IV, and V), the test revealed no result because, in the presence and absence of the inhibitor cloxacillin, no ceftazidime inhibition zone was detected.
Acquired resistance determinants.
Acquired resistance genes were detected in only five (XVI, XVII, XVIII, XIX, and XX) multiresistant P. aeruginosa strains, all from non-CF patients. intI1, coding for the integrase of class 1 integrons, was detected in these strains. intI2 was not detected. The acquired resistance determinants found were three adenylyltransferase genes (aadA1, aadA2, and aadB), three acetyltransferase genes [aac(6′)-Ib, aac(6′)-Ib′, aacA7, and aacA8], and one phosphotransferase gene (aphA1-IAB). Two β-lactamase genes, blaOXA-2 and blaPSE-1, were detected. The EDTA-phenanthroline-imipenem microdilution test and the PCR screening revealed the absence of metallo-β-lactamases. The qnrA gene lowering quinolone susceptibility was not found.
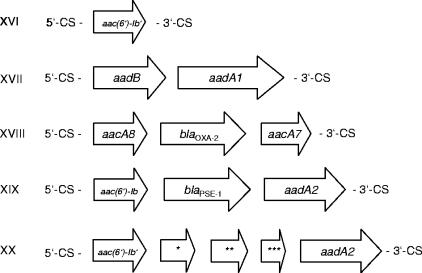
To characterize the genetic context of the acquired resistance genes found, PCR mapping experiments were carried out. As depicted in Fig. 1, all resistance genes detected by PCR but aphA1-IAB, an AME-encoding gene usually not integron associated (36), were located in class 1 integrons.
FIG. 1.
Structures of class 1 integrons detected exclusively in multiresistant P. aeruginosa clones from non-CF patients. Sequence analysis (with the BLASTX program) revealed 79% amino acid sequence homology with QecE (NP_044260) (*), 45% amino acid sequence homology with hypothetical protein MED121_04115 of Marinomonas sp. strain MED121 (ZP_01077982) (**), and 57% amino acid sequence homology with the RecA/RadA recombinase of Yersinia bercovieri (ZP_00823172) (***). 5′-CS, 5′ conserved segment; 3′-CS, 3′ conserved segment.
DISCUSSION
We analyzed the contribution of eight chromosomally mediated resistance mechanisms and 25 acquired resistance determinants to multiresistance for clinical multiresistant P. aeruginosa strains from CF and non-CF patients, taking into account the presence of a hypermutation phenotype.
Ureidopenicillin and cephalosporin resistance.
Regarding the phenotypes resistant to ureidopenicillins (piperacillin) and antipseudomonal cephalosporins (ceftazidime and cefepime), an overall higher resistance level was observed for strains from CF patients (Table 2). Consistent with former studies (20), overproduction of AmpC was, with a proportion of 73%, the predominant and, for the resistance level, the most important mechanism of resistance to ureidopenicillins and cephalosporins among the multiresistant P. aeruginosa strains investigated in this study. Overproduction of AmpC leads usually to MICs of cefepime lower than those of ceftazidime. However, in at least five strains (VIII, XI, XV, XVIII, and XXII) with cefepime MICs as high as or higher than the MICs of ceftazidime, this phenotype may be explained by the (additional) overproduction of MexXY-OprM, as found in a previous study (11). Of note, β-lactam resistance was almost exclusively due to chromosomally mediated resistance mechanisms. The acquired β-lactamase genes blaPSE-1 and blaOXA-2 were detected in only two strains, both from non-CF patients.
Carbapenem resistance.
Metallo-β-lactamases are rare in Germany (10, 43) and were not detected in this study. Confirming former studies (32, 20), loss of OprD was, with a proportion of 82%, the predominant and, for the resistance level, the most important mechanism of imipenem resistance (Table 2). In so-called nfxC mutants, the reduced production of OprD leads to borderline resistance to imipenem (MIC, 4 mg/liter) (18). Accordingly, for two strains (VII and VI) the reduced production of OprD due to modifications in MexT and MexS (nfxC mutants) seems to be the reason for reduced imipenem susceptibility. nfxB mutants overproducing MexCD-OprJ have heterogeneous phenotypes. In this study, the hypersusceptibility to imipenem that was reported for type B nfxB mutants (24, 45) was not found.
For meropenem, several mechanisms reducing susceptibility are known. However, each of these mechanisms alone leads only to a slight decrease in susceptibility, not conferring full resistance. In this study, the most frequent mechanisms lowering meropenem susceptibility were loss of OprD (82%) and overproduction of AmpC (73%), which result in meropenem MICs of 4 mg/liter and up to 2 mg/liter, respectively (16). Meropenem is unlike the imipenem substrates of several efflux pumps: while overproduction of MexCD-OprJ and MexXY-OprM leads to poorly increased MICs of meropenem, overexpression of MexAB-OprM confers meropenem MICs of 4 mg/liter (25). For the two nfxC mutants (strains VII and VI), the interaction of reduced OprD production and AmpC overproduction seems to be the reason for reduced meropenem susceptibility, whereas additional efflux pump overproduction leads to full meropenem resistance in strain VII. For several strains (e.g., IX and XIX) resistant to meropenem and imipenem, mechanisms other than the known mechanisms of carbapenem resistance seem to contribute additionally to resistance. The high levels of resistance seen for these strains are unlikely to be caused solely by the mechanisms detected.
Aminoglycoside resistance.
The analysis of aminoglycoside resistance mechanisms and the associated aminoglycoside susceptibility phenotypes revealed one important difference between multiresistant P. aeruginosa strains from CF and non-CF patients: numerous AME genes were found only in strains from non-CF patients (Table 2). These resistance genes were located in class 1 integrons, partially repeated and found in combination with β-lactamase genes. The finding of acquired resistance genes in half of the multiresistant strains from non-CF patients points to the decisive role of horizontal transfer of resistance mechanisms for P. aeruginosa strains from this group of patients. The absence of any acquired resistance genes in multiresistant strains from CF patients emphasizes the relevance of the mutator phenotype for the autarkic evolution of multiresistant P. aeruginosa strains during chronic infection of the lungs of CF patients. The only chromosomally mediated high-level aminoglycoside resistance mechanism in clinical P. aeruginosa strains known so far, overproduction of MexXY-OprM, was found in 82% of the multiresistant strains. Some studies, including this work, show that the impact of this efflux pump on resistance in P. aeruginosa may have been underestimated so far (11, 12, 37). The overproduction level of MexXY-OprM can vary significantly. Quantitative real-time PCR experiments revealed 3- to 727-fold-increased amounts of MexY mRNA in overproducing strains (12). Variety in mexXY expression levels might explain the broad range of aminoglycoside resistance phenotypes, particularly in multiresistant P. aeruginosa strains from CF patients found in this study. The cause may be the different impacts and numbers of modifications in the repressor MexZ. However, previous studies provided evidence of the existence of additional components involved in mexXY expression regulation besides MexZ, as overproducing strains without modifications in MexZ were detected. Furthermore, the contribution of other, unknown components of aminoglycoside resistance was suggested, as no clear correlation between mexXY expression levels and aminoglycoside MICs was found (12, 37). These components might also be involved in aminoglycoside-resistant strains found in this work that lack modifications in MexZ and acquired AME genes.
Quinolone resistance.
The most noticeable result was the difference in quinolone resistance levels between strains from CF patient and non-CF patient strains: while 7 out of 10 strains from non-CF patients showed ciprofloxacin MICs of >16 mg/liter, only 2 of 12 strains from CF patients had this level of resistance to ciprofloxacin (Table 2). The analysis of surveillance data generated as part of the GENARS project confirmed this phenomenon with a higher number of strains, including the strains investigated in this study: of 207 P. aeruginosa strains from non-CF patients, 32.9% were intermediately resistant and 67.1% were fully resistant to ciprofloxacin. In contrast, of 78 strains from CF patients, 46.2% showed intermediate resistance and 51.3% showed full resistance to ciprofloxacin. Lower concentrations of quinolones in the mucus of the CF lung or biofilm formation may reduce the need for high-level resistance in CF patient strains.
The obvious differences in quinolone resistance between P. aeruginosa strains from CF and non-CF patients were also found on a molecular level. High-level quinolone resistance conferred by modifications in ParC in combination with (multiple) modifications in GyrA was exclusively detected in P. aeruginosa strains from non-CF patients. Besides target modifications, the overproduction of MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY-OprM can also contribute to quinolone resistance. A previous study of quinolone resistance in P. aeruginosa strains from CF patients revealed no modifications in mexR or parC, but overproduction of MexCD-OprJ was most prevalent (13). In a comparative study with quinolone-resistant P. aeruginosa isolates from urinary tract and wound infections, numerous modifications in gyrA, parC, and mexR were found, while overproduction of MexCD-OprJ played a minor role (14). This study confirms the results of both studies. However, several strains with different resistance levels showing modifications in GyrA and MexZ only were found in this study. Their quinolone resistance levels may be due to different high expression levels of mexXY comparable to those found for aminoglycoside resistance due to MexXY-OprM overproduction. These findings emphasize the need for further investigations about the variable contribution of MexXY-OprM to multiresistance in P. aeruginosa.
Mutator strains.
A high proportion of resistance mechanisms detected in the multiresistant P. aeruginosa strains were mutation mediated; therefore, we were interested in the mutator phenotypes of the strains.
In our study, all but one multiresistant P. aeruginosa strain from CF patients were hypermutable. This extremely high proportion of mutator phenotypes is in concordance with recent studies that found hypermutation to be the key factor in development of mutation-mediated multiresistance in patients with chronic P. aeruginosa lung infections (23, 31). The extremely high proportion of mutator strains found in this study is expected to have important consequences for the treatment of multiresistant P. aeruginosa strains from CF patients. A multiresistant P. aeruginosa strain from a CF patient is most probably also hypermutable and very likely to acquire additional resistance properties during therapy. Three mutator strains were also detected among the multiresistant strains from non-CF patients. Two of those originated from respiratory tract specimens and might be assigned to chronic lung infections other than CF that seem to favor in the same way the selection of hypermutability (23). Interestingly, one hypermutable P. aeruginosa strain was isolated twice from wound specimens of a patient who was medicated in an ICU anesthesiology ward and subsequently in a normal abdominal surgery ward. To our knowledge, this is the first report of a P. aeruginosa mutator strain that was not isolated in the context of a chronic lung infection.
Supplementary Material
Acknowledgments
We thank Reinhard Marre, Heike von Baum, Sebastian Suerbaum, and Stefan Ziesing for providing us with clinical P. aeruginosa strains. We are grateful to Robert E. W. Hancock for providing us with P. aeruginosa PAO1. We also thank Aida Duarte, Angelika Miko, Ronald Jones, and Mark Toleman for providing us with positive PCR controls. We are grateful to Ines Noll for compiling statistics on quinolone resistance data from GENARS.
Beate Henrichfreise received a fellowship from the German national academic foundation (Studienstiftung des deutschen Volkes). Financial support was also obtained from AstraZeneca.
Footnotes
Published ahead of print on 17 September 2007.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Aloush, V., S. Navon-Venezia, Y. Seigman-Igra, S. Cabili, and Y. Carmeli. 2006. Multidrug-resistant Pseudomonas aeruginosa: risk factors and clinical impact. Antimicrob. Agents Chemother. 50:43-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonomo, R. A., and D. Szabo. 2006. Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin. Infect. Dis. 43:S49-S56. [DOI] [PubMed] [Google Scholar]
- 3.Castanheira, M., R. E. Mendes, T. R. Walsh, A. C. Gales, and R. N. Jones. 2004. Emergence of the extended-spectrum β-lactamase GES-1 in a Pseudomonas aeruginosa strain from Brazil: report from the SENTRY antimicrobial surveillance program. Antimicrob. Agents Chemother. 48:2344-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Champs, C., L. Poirel, R. Bonnet, D. Sirot, C. Chanal, J. Sirot, and P. Nordmann. 2002. Prospective survey of β-lactamases produced by ceftazidime-resistant Pseudomonas aeruginosa isolated in a French hospital in 2000. Antimicrob. Agents Chemother. 46:3031-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.European Committee for Antimicrobial Susceptibility Testing (EUCAST). 2003. Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin. Microbiol. Infect. Dis. 9:1-7. [Google Scholar]
- 6.Gibson, R. L., J. L. Burns, and B. W. Ramsey. 2003. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am. J. Respir. Crit. Care Med. 168:918-951. [DOI] [PubMed] [Google Scholar]
- 7.Girlich, D., T. Naas, A. Leelaporn, L. Poirel, M. Fennewald, and P. Nordmann. 2002. Nosocomial spread of the integron-located veb-1-like cassette encoding an extended-spectrum β-lactamase in Pseudomonas aeruginosa in Thailand. Clin. Infect. Dis. 34:603-611. [DOI] [PubMed] [Google Scholar]
- 8.Govan, J. R. W., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutierrez, O., C. Juan, J. L. Perez, and A. Oliver. 2004. Lack of association between hypermutation and antibiotic resistance development in Pseudomonas aeruginosa isolates from intensive care unit patients. Antimicrob. Agents Chemother. 48:3573-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henrichfreise, B., I. Wiegand, K. J. Sherwood, and B. Wiedemann. 2005. Detection of VIM-2 metallo-β-lactamase in Pseudomonas aeruginosa from Germany. Antimicrob. Agents Chemother. 49:1668-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hocquet, D., P. Nordmann, F. El Garch, L. Cabanne, and P. Plesiat. 2006. Involvement of the MexXY-OprM efflux system in emergence of cefepime resistance in clinical strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 50:1347-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Islam, S., S. Jalal, and B. Wretlind. 2004. Expression of the MexXY efflux pump in amikacin-resistant isolates of Pseudomonas aeruginosa. Clin. Microbiol. Infect. 10:877-883. [DOI] [PubMed] [Google Scholar]
- 13.Jalal, S., O. Ciofu, N. Hoiby, N. Gotoh, and B. Wretlind. 2000. Molecular mechanisms of fluoroquinolone resistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob. Agents Chemother. 44:710-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jalal, S., and B. Wretlind. 1998. Mechanisms of quinolone resistance in clinical strains of Pseudomonas aeruginosa. Microb. Drug Resist. 4:257-261. [DOI] [PubMed] [Google Scholar]
- 15.Juan, C., M. D. Macia, O. Gutierrez, C. Vidal, J. L. Perez, and A. Oliver. 2005. Molecular mechanisms of β-lactam resistance mediated by AmpC hyperproduction in Pseudomonas aeruginosa clinical strains. Antimicrob. Agents Chemother. 49:4733-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Juan, C., B. Moya, J. L. Perez, and A. Oliver. 2006. Stepwise upregulation of the Pseudomonas aeruginosa chromosomal cephalosporinase conferring high-level β-lactam resistance involves three AmpD homologues. Antimicrob. Agents Chemother. 50:1780-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koehler, T., M. Michea-Hamzehpour, U. Henze, N. Gotoh, L. K. Curty, and J. C. Pechere. 1997. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 23:345-354. [DOI] [PubMed] [Google Scholar]
- 18.Koehler, T., C. van Delden, L. K. Curty, M. M. Hamzehpour, and J. C. Pechere. 2001. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J. Bacteriol. 183:5213-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levesque, C., L. Piche, C. Larose, and P. H. Roy. 1995. Pcr mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livermore, D. M. 2002. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 34:634-640. [DOI] [PubMed] [Google Scholar]
- 21.Lomovskaya, O., M. S. Warren, A. Lee, J. Galazzo, R. Fronko, M. Lee, J. Blais, D. Cho, S. Chamberland, T. Renau, R. Leger, S. Hecker, W. Watkins, K. Hoshino, H. Ishida, and V. J. Lee. 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 45:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macia, M. D., D. Blanquer, B. Togores, J. Sauleda, J. L. Perez, and A. Oliver. 2005. Hypermutation is a key factor in development of multiple-antimicrobial resistance in Pseudomonas aeruginosa strains causing chronic lung infections. Antimicrob. Agents Chemother. 49:3382-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masuda, N., N. Gotoh, S. Ohya, and T. Nishino. 1996. Quantitative correlation between susceptibility and OprJ production in NfxB mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 40:909-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-oprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:3322-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Migliavacca, R., J. D. Docquier, C. Mugnaioli, G. Amicosante, R. Daturi, K. W. Lee, G. M. Rossolini, and L. Pagani. 2002. Simple microdilution test for detection of metallo-β-lactamase production in Pseudomonas aeruginosa. J. Clin. Microbiol. 40:4388-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mine, T., Y. Morita, A. Kataoka, T. Mizushima, and T. Tsuchiya. 1999. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:415-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ochs, M. M., M. P. McCusker, M. Bains, and R. E. Hancock. 1999. Negative regulation of the Pseudomonas aeruginosa outer membrane porin OprD selective for imipenem and basic amino acids. Antimicrob. Agents Chemother. 43:1085-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oliver, A., F. Baquero, and J. Blazquez. 2002. The mismatch repair system (mutS, mutL and uvrD genes) in Pseudomonas aeruginosa: molecular characterization of naturally occurring mutants. Mol. Microbiol. 43:1641-1650. [DOI] [PubMed] [Google Scholar]
- 30.Oliver, A., R. Canton, P. Campo, F. Baquero, and J. Blazquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251-1254. [DOI] [PubMed] [Google Scholar]
- 31.Oliver, A., B. R. Levin, C. Juan, F. Baquero, and J. Blazquez. 2004. Hypermutation and the preexistence of antibiotic-resistant Pseudomonas aeruginosa mutants: implications for susceptibility testing and treatment of chronic infections. Antimicrob. Agents Chemother. 48:4226-4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pai, H., J. W. Kim, J. Kim, J. H. Lee, K. W. Choe, and N. Gotoh. 2001. Carbapenem resistance mechanisms in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents and Chemother. 45:480-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poirel, L., E. Lebessi, M. Castro, C. Fevre, M. Foustoukou, and P. Nordmann. 2004. Nosocomial outbreak of extended-spectrum β-lactamase SHV-5-producing isolates of Pseudomonas aeruginosa in Athens, Greece. Antimicrob. Agents Chemother. 48:2277-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poole, K., N. Gotoh, H. Tsujimoto, Q. Zhao, A. Wada, T. Yamasaki, S. Neshat, J. Yamagishi, X. Z. Li, and T. Nishino. 1996. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol. Microbiol. 21:713-724. [DOI] [PubMed] [Google Scholar]
- 35.Poole, K., K. Krebes, C. Mcnally, and S. Neshat. 1993. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 175:7363-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaw, K. J., P. N. Rather, R. S. Hare, and G. H. Miller. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57:138-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sobel, M. L., G. A. McKay, and K. Poole. 2003. Contribution of the MexXY multidrug transporter to aminoglycoside resistance in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 47:3202-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sobel, M. L., S. Neshat, and K. Poole. 2005. Mutations in PA2491 (mexS) promote MexT-dependent mexEF-oprN expression and multidrug resistance in a clinical strain of Pseudomonas aeruginosa. J. Bacteriol. 187:1246-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 40.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vahaboglu, H., R. Ozturk, G. Aygun, F. Coskunkan, A. Yaman, A. Kaygusuz, H. Leblebicioglu, I. Balik, K. Aydin, and M. Otkun. 1997. Widespread detection of PER-1-type extended-spectrum β-lactamases among nosocomial Acinetobacter and Pseudomonas aeruginosa isolates in Turkey: a nationwide multicenter study. Antimicrob. Agents Chemother. 41:2265-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vincent, J. L., D. J. Bihari, P. M. Suter, H. A. Bruining, J. White, M. H. Nicolaschanoin, M. Wolff, R. C. Spencer, and M. Hemmer. 1995. The prevalence of nosocomial infection in intensive care units in Europe. Results of the European prevalence of infection in intensive care (EPIC) study. JAMA 274:639-644. [PubMed] [Google Scholar]
- 43.Walsh, T. R., M. A. Toleman, L. Poirel, and P. Nordmann. 2005. Metallo-β-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18:306-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weldhagen, G. F., L. Poirel, and P. Nordmann. 2003. Ambler class A extended-spectrum β-lactamases in Pseudomonas aeruginosa: novel developments and clinical impact. Antimicrob. Agents Chemother. 47:2385-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolter, D. J., N. D. Hanson, and P. D. Lister. 2005. AmpC and OprD are not involved in the mechanism of imipenem hypersusceptibility among Pseudomonas aeruginosa isolates overexpressing the mexCD-oprJ efflux pump. Antimicrob. Agents Chemother. 49:4763-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.