Abstract
Seventy-four nonrepetitive uropathogenic fluoroquinolone-resistant or -intermediate extended-spectrum-β-lactamase-producing Klebsiella isolates from Slovenia were screened for the presence of plasmid-mediated quinolone resistance genes. None of the known qnr genes were detected. The aac(6′)-Ib-cr allele was detected on plasmids from 25 transconjugants for which the ciprofloxacin MIC was higher than for the recipient Escherichia coli strain.
Fluoroquinolones are broad-spectrum antimicrobial agents widely used in clinical medicine. Resistance to this class of antibacterials is usually caused by mutations in the chromosomal genes that code for DNA gyrase and/or DNA topoisomerase IV, the target enzymes, and/or mutations resulting in alterations in drug accumulation (14). Recently, three plasmid-mediated quinolone resistance (PMQR) mechanisms have also been described.
The first comprises qnr genes that encode target protection proteins of the pentapeptide repeat family (8, 16). Three qnr genes, qnrA, qnrB, and qnrS, and their allelic variants have been described so far. Although the protein products of qnrA1 through qnrA5, qnrB1, qnrB2, qnrS1, and qnrS2 all provide low-level quinolone resistance, considerable variability can be observed in their amino acid sequences (2, 5, 7, 11).
The second mechanism involves the aac(6′)-Ib-cr gene, which encodes a new variant of the common aminoglycoside acetyltransferase. Two single-nucleotide substitutions at codons 102 and 179 in the wild-type allele aac(6′)-Ib enable the gene product to be capable of acetylating and thus reducing the activity of some fluoroquinolones, including norfloxacin and ciprofloxacin (12).
The prevalence of this PMQR gene has been so far reported from China, the United States, and Nigeria (9, 13, 15). Despite these recent findings, the contributions of the qnr and aac(6′)-Ib-cr genes to the increasing quinolone resistance worldwide and the association between quinolone resistance and extended-spectrum-β-lactamase (ESBL)-producing strains remain largely unknown.
The third mechanism, involving the fluoroquinolone efflux pump protein QepA, has been described very recently (17). The qepA gene was identified on plasmid pHPA of Escherichia coli C316. It resides on a putative transposable element, flanked by two copies of IS26. The similarity of QepA to the MFS-type pumps of the 14 TMS family, a high G+C content, and the codon usage pattern indicate a probable intergeneric transfer of a qepA-like gene from environmental microbes to E. coli (17).
Our study was initiated by the observation that the intermediate resistance phenotype, as determined by the CLSI (Clinical and Laboratory Standards Institute) guidelines (3), to the two fluoroquinolones most frequently used in Slovenia, ciprofloxacin and norfloxacin, almost disappeared in a collection of 74 Klebsiella isolates producing ESBLs, as detected by double-disc synergy tests and interpreted according to CLSI criteria, from the Institute of Public Health of the Republic of Slovenia (IVZ) after 2003, although it was frequently reported before that year. In order to find out whether this could be linked to the emergence of PMQR genes, 74 ESBL-producing Klebsiella isolates, with a ciprofloxacin- or norfloxacin-resistant or -intermediate phenotype were screened for the presence of described PMQR genes. The isolates were from urine culture samples obtained between January 2000 and December 2005 at the IVZ. Only nonrepetitive isolates, including 31 from nursing home residents, 10 from general practice patients, 7 from the urological outpatient clinic at the University Medical Center, Ljubljana, and 26 from Hospital Trbovlje, were included in this study (Table 1).
TABLE 1.
Characteristics of ESBL-producing Klebsiella isolates from the IVZ collection
| Yr of isolation | No. of isolates from:
|
Total no. of nonrepetitive isolates | No. (%) of isolates with different resistance phenotype
|
No. (%) of isolates with detected resistance gene
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nursing home residents (n = 31) | Hospital, Trbovlje (n = 26) | Urological outpatient clinic (n = 7) | General practices (n = 10) | CIPa resistant | CIP intermediate | NORb resistant | NOR intermediate | NOR susceptible | aac(6′)-Ib-crc | aac(6′)-Ib | ||
| 2000 | 5 | 2 | 1 | 0 | 8 | 4 (50) | 4 (50) | 4 (50) | 0 (0) | 4 (50) | 0 (0.0) | 6 (75) |
| 2001 | 2 | 0 | 1 | 0 | 3 | 2 (66) | 1 (33) | 1 (33) | 1 (33) | 1 (33) | 1 (33.3) | 3 (100) |
| 2002 | 3 | 2 | 1 | 0 | 6 | 3 (50) | 3 (50) | 3 (50) | 2 (33) | 1 (17) | 0 (0.0) | 5 (83) |
| 2003 | 7 | 9 | 2 | 2 | 20 | 18 (90) | 2 (10) | 18 (90) | 2 (10) | 0 (0) | 10 (50.0) | 14 (70.0) |
| 2004 | 7 | 6 | 2 | 5 | 20 | 20 (100) | 0 (0) | 19 (95) | 0 (0) | 1 (5) | 9 (45.0) | 11 (55) |
| 2005 | 7 | 7 | 0 | 3 | 17 | 15 (88) | 2 (12) | 15 (88) | 1 (6) | 1 (6) | 5 (29) | 10 (58) |
CIP, ciprofloxacin.
NOR, norfloxacin.
aac(6′)-Ib-cr detected on transconjugants TK1 to TK25.
To find out whether quinolone resistance was transferable, matings with all 74 Klebsiella isolates as donors and E. coli J53 Azr as the recipient were performed on brain heart infusion agar plates as described previously (1). Transconjugants were selected after 24 h of incubation at 37°C on LB agar plates containing sodium azide (150 μg/ml) for counterselection of the donor strains and ampicillin (100 μg/ml), tetracycline (10 μg/ml), trimethoprim (25 μg/ml), and chloramphenicol (30 μg/ml), respectively, for the selection of transconjugants with plasmid-encoded resistance determinants. Grown colonies from all four types of plates were replica plated onto LB agar plates, with and without ciprofloxacin (0.01 μg/ml; Fluka). MICs for the parental isolates and their derived transconjugants in E. coli J53Azr and for E. coli J53Azr were determined by the Etest method (AB Biodisk, Solna, Sweden) according to the manufacturer's recommendations. Twenty-five transconjugants (TK1 to TK25) for which the MIC of ciprofloxacin (0.125 μg/ml) was higher than that for recipient strain J53Azr (0.008 μg/ml) were obtained and further analyzed.
First, transconjugants were screened with a specific PCR for the presence of the qnrA1, qnrB1, and qnrS genes, respectively, with primer pairs QP2/QP1, FQ1/FQ2, and qnrS1/qnrS2 as described previously (6, 7, 10). Since none of the qnr genes could be detected, the resistance determinant was cloned from TK16, sequenced, and identified as the aac(6′)-Ib-cr variant of the aminoglycoside acetyltransferase gene aac(6′)-Ib.
Next, a combined PCR-restriction fragment length polymorphism protocol was developed to check for the presence of the wild-type allele and/or the mutant allele in the transconjugants. A 25-μl reaction mixture containing 5 μl of the bacterial lysate, PCR Master mix (Fermentas UAB, Lithuania), and 10 pg each of primers qac1 (5′-GGCATCACTGCGTGTTCGCTCG-3′) and qac2 (5′-GACTGAGCATGACCTTGCG-3′) was first denatured at 94°C for 5 min and then subjected to 30 PCR cycles of denaturation at 94°C for 40 s, primer annealing at 64°C for 30 s, and elongation at 72°C for 45 s. The 514-bp PCR products were digested with the enzymes NdeI and TaaI (Fermentas UAB, Lithuania) as recommended by the manufacturer. Restriction fragments were analyzed by electrophoresis on 2% agarose gels. Restriction of the wild-type allele aac(6′)-Ib with TaaI yielded three PCR fragments (331, 108, and 75 bp), whereas four fragments (217, 114, 108, and 75 bp) were observed after the restriction of the mutated allele aac(6′)-Ib-cr. Wild-type allele aac(6′)-Ib does not contain the recognition sequence for NdeI, while aac(6′)-Ib-cr is digested by NdeI, yielding two products (451 and 63 bp). According to the sizes of the restriction fragments, the qac1/qac2 PCR product was characterized as part of the aac(6′)-Ib and/or aac(6′)-Ib-cr allele.
The presence of the aac(6′)-Ib-cr allele was demonstrated in all transconjugants for which the ciprofloxacin MIC was higher than 0.008 μg/ml. Although aac(6′)-Ib was not detected in any of the transconjugants, all of them were resistant to kanamycin according to the CLSI guidelines (3). All 25 donor strains carrying the aac(6′)-Ib-cr gene, except the first one detected in 2001, which exhibited an intermediate phenotype, were highly resistant to ciprofloxacin and norfloxacin.
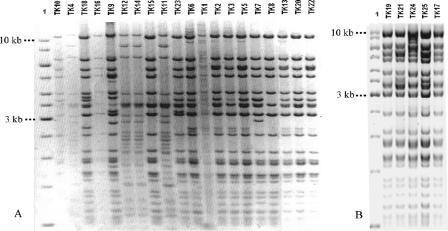
To find out whether the aac(6′)-Ib-cr gene was carried on similar or different plasmids, isolated plasmid DNA from all 25 transconjugants was digested with PstI and subjected to electrophoresis on a 0.7% agarose gel. According to the restriction pattern, the plasmids were classified into two major groups. Group 1 plasmids, observed in four transconjugants, were essentially the same and different from plasmids observed in the rest of the transconjugants. The latter comprised group 2 and appeared to share the same plasmid backbone, although they could be divided into three subgroups based on minor differences in the restriction profiles. All plasmids from subgroups 2a and 2b shared the same restriction pattern within each subgroup, whereas restriction patterns from subgroup 2c plasmids exhibited overall similarity with minor differences (Fig. 1). Regarding the sources of the donor isolates and plasmid groups, identical plasmids could be isolated from different institutions and more than one plasmid type could be isolated from the same institution (Table 2).
FIG. 1.
Plasmid restriction patterns of PstI-digested plasmids isolated from TK1 to TK25. Transconjugant designations are above the lanes; the molecular weight marker in lanes 1 of both panels A and B was the improved Gene Ruler 1-kb DNA ladder (Fermentas UAB, Lithuania).
TABLE 2.
Distribution of plasmid restriction patterns among the IVZ Klebsiella donor isolates collected from different sources between 2000 and 2005
| Yr of isolation, source | Donor | Transconjugant | Membership in restriction pattern group:
|
|||
|---|---|---|---|---|---|---|
| 1 | 2a | 2b | 2c | |||
| 2001, Urologa | U1361a | TK1 | + | |||
| 2003 | ||||||
| DsoKob | D2237a | TK2 | + | |||
| BTc | B2550b | TK3 | + | |||
| BT | B3090a | TK4 | + | |||
| BT | B3117a | TK5 | + | |||
| BT | B3366b | TK6 | + | |||
| BT | B3951c | TK7 | + | |||
| DsoKo | D4319a | TK8 | + | |||
| DsoKo | D4432b | TK9 | + | |||
| BT | B4700 | TK10 | + | |||
| BT | B4743a | TK11 | + | |||
| 2004 | ||||||
| ZDLitd | Z359d | TK12 | + | |||
| BT | B488a | TK13 | + | |||
| ZdLit | Z672a | TK14 | + | |||
| DsoKo | D779 | TK15 | + | |||
| DsoKo | D1449b | TK16 | + | |||
| DsoKo | D2057a | TK17 | + | |||
| BT | B2108a | TK18 | + | |||
| Urolog | U2897a | TK19 | + | |||
| 2005 | ||||||
| ZdSise | Z4703b | TK20 | + | |||
| BT | B788a | TK21 | + | |||
| BT | B1093 | TK22 | + | |||
| BT | B1181a | TK23 | + | |||
| BT | B1981 | TK24 | + | |||
| ZdSis | Z4750a | TK25 | + | |||
Urolog, urological outpatient clinic at the University Medical Center Ljubljana.
DsoKo, nursing home, Kolezija.
BT, hospital, Trbovlje.
ZDLit, general practice, Litija.
ZDSis, general practice, Siska.
According to data from the European surveillance of antimicrobial consumption, the use of norfloxacin decreased in Slovenia from 0.94 defined daily dose (DDD)/1,000 inhabitants per day in 1997 to 0.49 DDD/1,000 inhabitants per day in 2003, whereas the use of ciprofloxacin increased slightly from 0.5 DDD/1,000 inhabitants per day in 1997 to 0.6 DDD/1,000 inhabitants per day in 2003 (4). In the same time period, resistance to both fluoroquinolones constantly increased.
In the collection of isolates studied, the percentage of ciprofloxacin-resistant isolates increased from 50% in 2000 to 88% in 2005 and the percentage of those with an intermediate resistance phenotype decreased from 50% to 12%. The data for norfloxacin are similar. Here the percentage of susceptible isolates decreased from 50% in 2000 to 6% in 2005, and the percentage of resistant isolates increased over the same period from 50% to 88% (Table 1).
Since the increased number of ciprofloxacin-resistant ESBL-positive isolates, the extinction of the intermediate resistance phenotype, and the emergence of the aac(6′)-Ib-cr gene apparently coincide, a role for this low-level PMQR gene in enhancing the selection of chromosomal mutations and resulting in the occurrence and dissemination of clinically relevant resistance levels could be hypothesized.
Acknowledgments
We thank G. A. Jacoby for providing Escherichia coli strains J53Azr and J53Azr/pMG252. We are grateful to Alenka Smole, who, by her precise and dedicated work, collected and identified the strains between 2000 and 2004 at the IVZ.
Footnotes
Published ahead of print on 10 September 2007.
REFERENCES
- 1.Ambrožič, J., H. Ostroveršnik, M. Starčič, I. Kuhar, M. Grabnar, and D. Žgur-Bertok. 1998. Escherichia coli ColV plasmid pRK100: genetic organization, stability and conjugal transfer. Microbiology 144:343-352. [DOI] [PubMed] [Google Scholar]
- 2.Bönemann, G., M. Stiens, A. Puhler, and A. Schluter. 2006. Mobilizable IncQ-related plasmid carrying a new quinolone resistance gene, qnrS2, isolated from the bacterial community of a wastewater treatment plant. Antimicrob. Agents Chemother. 50:3075-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute. 2007. Performance standards for antimicrobial susceptibility testing; seventeenth informational supplement. M100-S17. Clinical and Laboratory Standards Institute, Wayne, PA.
- 4.Ferech, M., S. Coenen, S. Malhotra-Kumar, K. Dvorakova, E. Hendrickx, C. Suetens, and H. Goossens on behalf of the ESAC Project Group. 2006. European surveillance of antimicrobial consumption (ESAC): outpatient quinolone use in Europe. J. Antimicrob. Chemother. 58:423-427. [DOI] [PubMed] [Google Scholar]
- 5.Hata, M., M. Suzuki, M. Matsumoto, M. Takahashi, K. Sato, S. Ibe, and K. Sakae. 2005. Cloning of a novel gene for quinolone resistance from a transferable plasmid in Shigella flexneri 2b. Antimicrob. Agents Chemother. 49:801-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacoby, G. A., N. Chow, and K. B. Waites. 2003. Prevalence of plasmid-mediated quinolone resistance. Antimicrob. Agents Chemother. 47:559-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacoby, G. A., K. E. Walsh, D. M. Mills, V. J. Walker, H. Oh, A. Robicsek, and D. C. Hooper. 2006. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob. Agents Chemother. 50:1178-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martínez-Martínez, L., A. Pascual, and G. A. Jacoby. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797-799. [DOI] [PubMed] [Google Scholar]
- 9.Park, C. H., A. Robicsek, G. A. Jacoby, D. Sahm, and D. C. Hooper. 2006. Prevalence of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme in the United States. Antimicrob. Agents Chemother. 50:3953-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poirel, L., C. Leviandier, and P. Nordmann. 2006. Prevalence and genetic analysis of plasmid-mediated quinolone resistance determinants QnrA and QnrS in Enterobacteriaceae isolates from a French university hospital. Antimicrob. Agents Chemother. 50:3992-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poirel, L., J.-M. Rodriguez-Martinez, H. Mammeri, A. Liard, and P. Nordmann. 2005. Origin of plasmid-mediated quinolone resistance determinant QnrA. Antimicrob. Agents Chemother. 49:3523-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robicsek, A., J. Strahilevitz, G. A. Jacoby, M. Macielag, D. Abbanat, C. H. Park, K. Bush, and D. C. Hooper. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12:83-88. [DOI] [PubMed] [Google Scholar]
- 13.Robicsek, A., G. A. Jacoby, and D. C. Hooper. 2006. The worldwide emergence of plasmid-mediated quinolone resistance. 2006. Lancet Infect. Dis. 6:629-640. [DOI] [PubMed] [Google Scholar]
- 14.Ruiz, J. 2003. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J. Antimicrob. Chemother. 51:1109-1117. [DOI] [PubMed] [Google Scholar]
- 15.Soge, O. O., B. A. Adeniyi, and M. C. Roberts. 2006. New antibiotic resistance genes associated with CTX-M plasmids from uropathogenic Nigerian Klebsiella pneumoniae. J. Antimicrob. Chemother. 58:1048-1053. [DOI] [PubMed] [Google Scholar]
- 16.Tran, J. H., G. A. Jacoby, and D. C. Hooper. 2005. Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob. Agents Chemother. 49:118-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamane, K., J.-I. Wachino, S. Suzuki, K. Kimura, N. Shibata, H. Kato, K. Shibayama, T. Konda, and Y. Arakawa. 2007. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob. Agents Chemother. 51:3354-3360. [DOI] [PMC free article] [PubMed] [Google Scholar]