Abstract
A single-nucleotide polymorphism-based cluster grouping (SCG) classification system for Mycobacterium tuberculosis was used to examine antibiotic resistance type and resistance mutations in relationship to specific evolutionary lineages. Drug resistance and resistance mutations were seen across all SCGs. SCG-2 had higher proportions of katG codon 315 mutations and resistance to four drugs.
Isoniazid (INH) is an effective agent for treatment of infections with Mycobacterium tuberculosis. Increases in INH-resistant and multidrug-resistant tuberculosis jeopardize drug effectiveness (7, 24), and development of INH resistance is often a first step in multidrug resistance (2, 8). Mutations in specific genes have been linked to INH-related resistance (10, 11), including katG (26), inhA (14), codon 315 of katG (katG315) (13, 16, 19, 23), ahpC (20), the inhA open reading frame and promoter (14, 17, 25), and ndh (22).
Recent studies of drug-resistant M. tuberculosis have found associations among M. tuberculosis strains, drug resistance, and specific gene mutations. M. tuberculosis strains belonging to the Beijing family were associated with drug resistance in Iran, Afghanistan, and Russia (6, 12, 15, 18), although not in Venezuela (5). These associations have also been supported through genetic laboratory studies (1, 21). Thus, it is possible that specific types of drug resistance or drug resistance mutations might occur more commonly in certain evolutionary lineages of M. tuberculosis.
The single-nucleotide polymorphism (SNP) cluster group (SCG) classification system defined in reference 8 gives rise to seven phylogenetically distinct groups and three subgroups that can be used to infer an evolutionary pattern in M. tuberculosis. Here, we analyze 428 M. tuberculosis isolates resistant to at least INH collected across 10 countries and report the prevalence of various INH resistance-associated mutations and prevalences of resistance to two, three, and four drugs according to the major SCG-defined phylogenetic lineages of M. tuberculosis.
M. tuberculosis isolates resistant to at least INH were obtained from laboratories in major medical centers in Australia, Colombia, India, Mexico, New York City, Spain, and Texas (Table 1). The study population and sample selection have been described previously (10, 11), with each collection site performing susceptibility testing to INH, rifampin, streptomycin, and ethambutol.
TABLE 1.
M. bovis and M. tuberculosis SCG types overall and by location
| Location | No. (%) of isolates
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| M. bovis |
M. tuberculosis
|
Total | |||||||||
| SCG-1 | SCG-2 | SCG-3a | SCG-3b | SCG-3c | SCG-4 | SCG-5 | SCG-6a | SCG-6b | |||
| Australia | 0 | 9 (30.0) | 11 (36.7) | 3 (10.0) | 3 (10.0) | 0 | 0 | 3 (10.0) | 0 | 1 (3.3) | 30 (100) |
| Colombia | 0 | 0 | 0 | 1 (0.7) | 33 (23.1) | 1 (0.7) | 6 (4.2) | 84 (58.7) | 17 (11.9) | 1 (0.7) | 143 (100) |
| India | 0 | 2 (6.7) | 1 (3.3) | 23 (76.7) | 1 (3.3) | 0 | 0 | 2 (6.7) | 1 (3.3) | 0 | 30 (100) |
| Mexico | 2 (1.6) | 0 | 1 (0.8) | 1 (0.8) | 44 (35.8) | 11 (8.9) | 14 (11.4) | 31 (25.2) | 15 (12.2) | 4 (3.3) | 123 (100) |
| New York City | 0 | 1 (3.9) | 17 (65.3) | 0 | 2 (7.7) | 1 (3.9) | 0 | 2 (7.7) | 3 (11.5) | 0 | 26 (100) |
| Spain | 0 | 0 | 0 | 0 | 1 (9.1) | 0 | 0 | 8 (72.7) | 2 (18.2) | 0 | 11 (100) |
| Texas | 0 | 5 (7.7) | 8 (12.3) | 0 | 8 (12.3) | 5 (7.7) | 6 (9.2) | 16 (24.6) | 15 (23.1) | 2 (3.1) | 65 (100) |
| Total | 2 (0.5) | 17 (4.0) | 38 (8.9) | 28 (6.5) | 92 (21.5) | 18 (4.2) | 26 (6.1) | 146 (34.1) | 53 (12.4) | 8 (1.8) | 428 (100) |
Isolates were tested for virtually all SNP mutations in the M. tuberculosis katG, kasA, mabA, inhA, oxyR, ahpC, and ndh genes found to be associated with INH resistance in published studies (11). A total of 204 INH resistance-associated alleles were detected (9), with confirmatory testing carried out on alleles, identified mutations, and drug resistance.
Strain types are reported in terms of SCG grouping, based on observed genomic level clustering among identified SNPs (8). SCG assignment was performed by testing each isolate for nine SNPs previously determined to replicate larger SNP-based phylogeny, as described in references 3 and 4.
The genetic, resistance, and SCG data from each set of country-specific isolates were entered into a common database. Prevalence was reported by SCG type, country, selected genes, and resistance type. Chi-square tests of differences in proportions were employed as appropriate. The STATA statistical package was used for all calculations.
The complete sample was tested for 240 alleles previously reported to be associated with INH resistance. Country-specific breakdowns by various types of resistance can be found in references 10 and 11. The overall prevalence of SCG groups is given in Table 1. The Beijing family (strongly associated with SCG-2) is present in 8.9% of the collected isolates. The most prevalent SCG types were SCG-5, SCG-3b, and SCG-6a, and the rarest (excluding Mycobacterium bovis) were SCG-6b, SCG-1, SCG-3c, and SCG-4. SCG prevalence by country shows various distributions. SCG-5 is among the three most prevalent SCG types in Spain, Columbia, Mexico, and Texas. SCG-3b was prevalent in Mexico, Columbia, and Texas; SCG-6a was prevalent in Texas, Spain, and Mexico; SCG-3a was prevalent in India; and SCG-2 was prevalent in Australia and New York City, perhaps reflecting Asian immigrant populations. SCG-3c, SCG-4, and SCG-6b had low prevalence in all countries.
Table 2 reports the prevalence of mutations in a selected set of genes. Only KatG and KatG315 mutations occur across all SCG types (excluding M. bovis). The SCG types with the highest numbers of mutations were SCG-5, SCG-3b, and SCG-1. SCG-1 and SCG-5 had all mutations of interest. kasA and ndh mutations were found in the smallest number of SCG types (three).
TABLE 2.
Prevalence of selected genes within SCG type
| Gene | No. (%) of isolates with gene:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| M. bovis |
M. tuberculosis
|
Total | |||||||||
| SCG-1 | SCG-2 | SCG-3a | SCG-3b | SCG-3c | SCG-4 | SCG-5 | SCG-6a | SCG-6b | |||
| KatG | 0 | 12 (5.0) | 29 (12.1) | 12 (5.0) | 51 (21.3) | 12 (5.0) | 16 (6.7) | 81 (33.9) | 22 (9.2) | 4 (1.8) | 239 (100) |
| KatG315 | 0 | 12 (5.4) | 25 (11.5) | 11 (5.1) | 45 (20.1) | 12 (5.4) | 14 (6.4) | 78 (35.9) | 17 (7.8) | 3 (1.4) | 217 (100) |
| KasA | 0 | 8 (16.7) | 1 (2.1) | 0 | 0 | 0 | 0 | 39 (81.2) | 0 | 0 | 48 (100) |
| InhA | 0 | 3 (7.1) | 4 (9.5) | 2 (4.8) | 10 (23.8) | 1 (2.4) | 2 (4.8) | 12 (28.6) | 8 (19.0) | 0 | 42 (100) |
| Inh promoter | 0 | 3 (7.9) | 4 (10.5) | 2 (5.3) | 9 (23.7) | 1 (2.6) | 2 (5.3) | 10 (26.3) | 7 (18.4) | 0 | 38 (100) |
| Inh open reading frame | 0 | 1 (11.1) | 0 | 1 (11.1) | 2 (22.2) | 0 | 1 (11.1) | 3 (33.3) | 1 (11.1) | 0 | 9 (100) |
| AhpC | 0 | 1 (1.7) | 1 (1.7) | 17 (28.3) | 10 (16.7) | 0 | 11 (18.2) | 13 (21.7) | 6 (10.0) | 1 (1.7) | 60 (100) |
| Ndh | 0 | 1 (7.1) | 0 | 0 | 0 | 1 (7.1) | 0 | 12 (85.7) | 0 | 0 | 14 (100) |
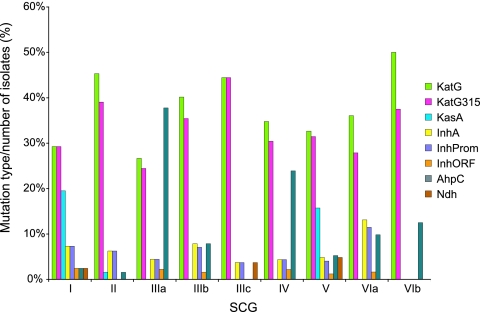
As these measures could be influenced by the proportional representation of each SCG in the sample, we examined the relative proportion of each mutation within each SCG (Fig. 1). The highest proportion of KatG315 mutations occurred in SCG-3c and SCG-2. SCG-6b, SCG-2, and SCG-3c had the highest proportion of mutations in katG. Mutations in inhA and ahpC promoters were found in all SCGs with n > 17.
FIG. 1.
Distribution of mutations by SCG.
To examine antibiotic resistance on a cumulative scale, we examined resistance within each SCG classification type in relation to resistance to one, two, or more antibiotics. All SCG types (excepting M. bovis) had at least 46% of isolates with resistance to one or two of these antibiotics. The range across all SCG types was 46% to 82%. SCG-2 had the highest prevalence (29%) of resistance to all four antibiotics.
This study provides insights into the associations among drug resistance in M. tuberculosis, gene mutation, and genomic SCG-based phylogenetic lineages, utilizing a large number of resistant isolates and examining patterns of relationships across SCG classification type, resistance, gene mutation, and country.
The sample contained 8.9% SCG-2 (Beijing) isolates, similar to Asian samples (18) and comparable to other SCG types, except SCG-3b, SCG-5, and SCG-6a. SCG-2 isolates accounted for more than 10% of the KatG and KatG315 mutations in this study. Most mutations were prevalent in all SCGs.
The prevalence of resistance was high across all SCG types. Table 3 shows that SCG-2 had the highest prevalence of resistance to all antibiotics, but all SCG types display high levels of resistance to one and two antibiotics. SCG-6a displayed a high prevalence of isolates resistant to all four antibiotics. As the SCG classification reflects regions, the presence of antibiotic resistance in so many SCG classifications may reflect several ongoing evolutionary processes and implies a need to maintain a broad perspective on M. tuberculosis antibiotic resistance.
TABLE 3.
Number of isolates within each SCG type with resistance to individual antibiotics
| No. of antibiotics to which isolates show resistance | No. (%) of resistant isolates
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| M. bovis |
M. tuberculosis
|
Total | |||||||||
| SCG-1 | SCG-2 | SCG-3a | SCG-3b | SCG-3c | SCG-4 | SCG-5 | SCG-6a | SCG-6b | |||
| 1 | 2 | 12 (70.59) | 12 (31.58) | 12 (42.86) | 26 (28.26) | 8 (44.44) | 8 (30.77) | 28 (19.18) | 14 (26.42) | 3 (37.5) | 125 (29.21) |
| 2 | 0 | 2 (11.76) | 10 (26.32) | 9 (32.14) | 30 (32.61) | 4 (22.22) | 12 (46.15) | 39 (26.71) | 16 (30.19) | 2 (25.0) | 124 (28.97) |
| 3 | 0 | 2 (11.76) | 5 (13.16) | 6 (21.43) | 21 (22.83) | 5 (27.78) | 4 (15.38) | 53 (36.30) | 10 (18.87) | 3 (37.50) | 109 (25.47) |
| 4 | 0 | 1 (5.88) | 11 (28.95) | 1 (3.57) | 15 (16.3) | 1 (5.56) | 2 (7.69) | 26 (17.81) | 13 (24.53) | 0 | 70 (16.36) |
| Total | 2 | 17 (100) | 38 (100) | 28 (100) | 92 (100) | 18 (100) | 26 (100) | 146 (100) | 53 (100) | 8 (100) | 428 (100) |
Limitations.
This study analyzed M. tuberculosis isolates for mutations associated with INH resistance in previous studies. The results therefore may not be completely representative of mutations relevant to resistance. The overall patterns observed in the SCG classification may have been restricted by the method of sample selection, but the size and diversity of the sample make this unlikely.
Acknowledgments
This work was supported by Public Health Service grants AI-46669 and AI-49352.
Footnotes
Published ahead of print on 10 September 2007.
REFERENCES
- 1.Abebe, F., and G. Bjune. 2006. The emergence of Beijing family genotypes of Mycobacterium tuberculosis and low-level protection by bacille Calmette-Guerin (BCG) vaccines: is there a link? Clin. Exp. Immunol. 145:389-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alland, D., G. E. Kalkut, A. R. Moss, R. A. McAdam, J. A. Hahn, W. Bosworth, E. Drucker, and B. R. Bloom. 1994. Transmission of tuberculosis in New York City. An analysis by DNA fingerprinting and conventional epidemiologic methods. N. Engl. J. Med. 330:1710-1716. [DOI] [PubMed] [Google Scholar]
- 3.Alland, D., D. W. Lacher, M. H. Hazbón, A. S. Motiwala, W. Qi, R. D. Fleischmann, and T. S. Whittam. 2007. Role of large sequence polymorphisms (LSPs) in generating genomic diversity among clinical isolates of Mycobacterium tuberculosis and the utility of LSPs in phylogenetic analysis. J. Clin. Microbiol. 45:39-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alland, D., T. S. Whittam, M. B. Murray, M. D. Cave, M. H. Hazbón, K. Dix, M. Kokoris, A. Duesterhoeft, J. A. Eisen, C. M. Fraser, and R. D. Fleischmann. 2003. Modeling bacterial evolution with comparative-genome-based marker systems: application to Mycobacterium tuberculosis evolution and pathogenesis. J. Bacteriol. 185:3392-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aristimuno L., R. Armengol, A. Cebollada, M. Espana, A. Guilarte, C. Lafoz, M. A. Lezcano, M. J. Revillo, C. Martin, C. Ramirez, N. Rastogi, J. Rojas, A. V. de Salas, C. Sola, and S. Samper. 2006. Molecular characterisation of Mycobacterium tuberculosis isolates in the First National Survey of Anti-tuberculosis Drug Resistance from Venezuela. BMC Microbiol. 6:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox, H. S., T. Kubica, D. Doshetov, Y. Kebede, S. Rusch-Gerdess, and S. Niemann. 2005. The Beijing genotype and drug resistant tuberculosis in the Aral Sea region of Central Asia. Respir. Res. 6:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang, Z., C. Doig, A. Rayner, D. T. Kenna, B. Watt, and K. J. Forbes. 1999. Molecular evidence for heterogeneity of the multiple-drug-resistant Mycobacterium tuberculosis population in Scotland (1990 to 1997). J. Clin. Microbiol. 37:998-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filliol, I., A. S. Motiwala, M. Cavatore, W. Qi, M. H. Hazbón, M. Bobadilla del Valle, J. Fyfe, L. Garcia-Garcia, N. Rastogi, C. Sola, T. Zozio, M. I. Guerrero, C. I. León, J. Crabtree, S. Angiuoli, K. D. Eisenach, R. Durmaz, M. L. Joloba, A. Rendón, J. Sifuentes-Osornio, A. Ponce de Leon, M. D. Cave, R. Fleischmann, T. S. Whittam, and D. Alland. 2006. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J. Bacteriol. 188:759-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hazbón, M. H., and D. Alland. 2004. Hairpin primers for simplified single-nucleotide polymorphism analysis of Mycobacterium tuberculosis and other organisms. J. Clin. Microbiol. 42:1236-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazbón, M. H., M. Bobadilla del Valle, M. I. Guerrero, M. Varma-Basil, I. Filliol, M. Cavatore, R. Colangeli, H. Safi, H. Billman-Jacobe, C. Lavender, J. Fyfe, L. Garcia-Garcia, A. Davidow, M. Brimacombe, C. I. Leon, T. Porras, M. Bose, F. Chaves, K. D. Eisenach, J. Sifuentes-Osornio, A. Ponce de Leon, M. D. Cave, and D. Alland. 2005. Role of embB codon 306 mutations in Mycobacterium tuberculosis revisited: a novel association with broad drug resistance and IS6110 clustering rather than ethambutol resistance. Antimicrob. Agents Chemother. 49:3794-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hazbón, M. H., M. Brimacombe, M. Bobadilla del Valle, M. Cavatore, M. I. Guerrero, M. Varma-Basil, H. Billman-Jacobe, C. Lavender, J. Fyfe, L. García-García, C. Inés León, M. Bose, F. Chaves, M. Murray, K. D. Eisenach, J. Sifuentes-Osornio, M. D. Cave, A. Ponce de León, and D. Alland. 2006. Population genetics study of isoniazid resistance mutations and evolution of multidrug-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 50:2640-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ignatova, A., S. Dubiley, V. Stepanshina, and I. Shemyakin. 2006. Predominance of multi-drug-resistant LAM and Beijing family strains among Mycobacterium tuberculosis isolates recovered from prison inmates in Tula Region, Russia. J. Med. Microbiol. 55:1413-1418. [DOI] [PubMed] [Google Scholar]
- 13.Kapetanaki, S. M., S. Chouchane, S. Yu, X. Zhao, R. S. Magliozzo, and J. P. Schelvis. 2005. Mycobacterium tuberculosis KatG(S315T) catalase-peroxidase retains all active site properties for proper catalytic function. Biochemistry 44:243-252. [DOI] [PubMed] [Google Scholar]
- 14.Musser, J. M., V. Kapur, D. L. Williams, B. N. Kreiswirth, D. van Soolingen, and J. D. van Embden. 1996. Characterization of the catalase-peroxidase gene (katG) and inhA locus in isoniazid-resistant and -susceptible strains of Mycobacterium tuberculosis by automated DNA sequencing: restricted array of mutations associated with drug resistance. J. Infect. Dis. 173:196-202. [DOI] [PubMed] [Google Scholar]
- 15.Pheiffer, C., J. C. Betts, H. R. Flynn, P. T. Lukey, and P. van Helden. 2005. Protein expression by a Beijing strain differs from that of another clinical isolate and Mycobacterium tuberculosis H37Rv. Microbiology 151:1139-1150. [DOI] [PubMed] [Google Scholar]
- 16.Pym, A. S., B. Saint-Joanis, and S. T. Cole. 2002. Effect of katG mutations on the virulence of Mycobacterium tuberculosis and the implication for transmission in humans. Infect. Immun. 70:4955-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramaswamy, S. V., S. J. Dou, A. Rendon, Z. Yang, M. D. Cave, and E. A. Graviss. 2004. Genotypic analysis of multidrug-resistant Mycobacterium tuberculosis isolates from Monterrey, Mexico. J. Med. Microbiol. 53:107-113. [DOI] [PubMed] [Google Scholar]
- 18.Ramazanzadeh, R., P. Farnia, N. N. Amirmozafari, F. Ghazi, Z. Ghadertotonchi, J. Kamran, F. Mohammadi, M. Mirsaedi, and M. Masjedi. 2006. Comparison between molecular epidemiology, geographical regions and drug resistance in Mycobacterium tuberculosis strains isolated from Iranian and Afghan patients. Chemotherapy 52:316-320. [DOI] [PubMed] [Google Scholar]
- 19.Saint-Joanis, B., H. Souchon, M. Wilming, K. Johnsson, P. M. Alzari, and S. T. Cole. 1999. Use of site-directed mutagenesis to probe the structure, function and isoniazid activation of the catalase/peroxidase, KatG, from Mycobacterium tuberculosis. Biochem. J. 338:753-760. [PMC free article] [PubMed] [Google Scholar]
- 20.Sherman, D. R., K. Mdluli, M. J. Hickey, T. M. Arain, S. L. Morris, C. E. Barry III, and C. K. Stover. 1996. Compensatory ahpC gene expression in isoniazid-resistant Mycobacterium tuberculosis. Science 272:1641-1643. [DOI] [PubMed] [Google Scholar]
- 21.Sivkov, A. I., A. N. Boldyrev, M. S. Azaev, S. A. Bodnev, E. V. Medvedeva, O. I. Baranova, A. P. Ivlev-Dantau, L. N. Blinova, A. D. Pasechnikov, and S. I. Tat'kov. 2006. Evaluation of reasons of the MDR M. tuberculosis strains dissemination by analysis of the rifampicin and/or isoniazid resistant isolates. Mol. Gen. Mikrobiol. Virusol. 2006:20-25. [PubMed] [Google Scholar]
- 22.van Soolingen, D., P. E. de Haas, H. R. van Doorn, E. Kuijper, H. Rinder, and M. W. Borgdorff. 2000. Mutations at amino acid position 315 of the katG gene are associated with high-level resistance to isoniazid, other drug resistance, and successful transmission of Mycobacterium tuberculosis in The Netherlands. J. Infect. Dis. 182:1788-1790. [DOI] [PubMed] [Google Scholar]
- 23.Vilchèze, C., T. R. Weisbrod, B. Chen, L. Kremer, M. H. Hazbón, F. Wang, D. Alland, J. C. Sacchettini, and W. R. Jacobs, Jr. 2005. Altered NADH/ NAD+ ratio mediates coresistance to isoniazid and ethionamide in mycobacteria. Antimicrob. Agents Chemother. 49:708-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weis, S. E., J. M. Pogoda, Z. Yang, M. D. Cave, C. Wallace, M. Kelley, and P. F. Barnes. 2002. Transmission dynamics of tuberculosis in Tarrant County, Texas. Am. J. Respir. Crit. Care Med. 166:36-42. [DOI] [PubMed] [Google Scholar]
- 25.Wilson, R. W., Z. Yang, M. Kelley, M. D. Cave, J. M. Pogoda, R. J. Wallace, Jr., J. P. Cegielski, D. F. Dunbar, D. Bergmire-Sweat, L. B. Elliott, and P. F. Barnes. 1999. Evidence from molecular fingerprinting of limited spread of drug-resistant tuberculosis in Texas. J. Clin. Microbiol. 37:3255-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson, T. M., and D. M. Collins. 1996. ahpC, a gene involved in isoniazid resistance of the Mycobacterium tuberculosis complex. Mol. Microbiol. 19:1025-1034. [DOI] [PubMed] [Google Scholar]