Abstract
Five highly amikacin-resistant Acinetobacter baumannii isolates were collected at a medical center in Pennsylvania. The aminoglycoside resistance was due to the production of the 16S rRNA methylase ArmA. Two of the isolates coproduced OXA-23 β-lactamase and were highly resistant to carbapenems as well. The isolates were genetically closely related by pulsed-field gel electrophoresis.
Acinetobacter baumannii is increasingly becoming a major nosocomial pathogen worldwide, particularly in the setting of ventilator-associated pneumonia or bloodstream infections in intensive care units (6). The treatment is complicated by its tendency to acquire resistance to multiple classes of antimicrobials (2). When these strains are encountered, empirical salvage regimens may include such agents as colistin, tigecycline, and amikacin (1, 13). Amikacin is an aminoglycoside that generally continues to retain good activity against A. baumannii (3, 11). Resistance to amikacin in A. baumannii is primarily mediated by structural modification of the agent through the actions of aminoglycoside-modifying enzymes that are produced by resistant strains (14). In recent years, the production of 16S rRNA methylases has been implicated in aminoglycoside resistance among gram-negative pathogens (4). Five such enzymes have been identified, ArmA and RmtA through -D. They confer high-level resistance to all parenterally formulated aminoglycosides, effectively eliminating the entire class, including amikacin, as a therapeutic option. The presence of these 16S rRNA methylases has already been reported worldwide, but no strains with this resistance mechanism have been reported in North America so far.
Between December 2006 and March 2007, five nonrepetitive A. baumannii isolates (isolates A through E) with high-level resistance to amikacin, tobramycin, and gentamicin (defined by MICs of >512 μg/ml) were recovered from inpatients at the University of Pittsburgh Medical Center. Multiplex PCR for the five known 16S rRNA methylase genes (4) yielded amplicons consistent with armA for all five isolates, which was verified by sequencing. No amplicons were obtained for the other 16S rRNA methylase genes. In addition, isolates A and B, both of which were highly resistant to carbapenems, were positive for blaOXA-23 by PCR and sequencing (5). OXA-23 is a carbapenem-hydrolyzing β-lactamase that is known to cause clinically relevant carbapenem resistance (10).
The genomic DNA of isolate B was then prepared, digested with HindIII (New England BioLabs, Beverly, MA), and ligated with cloning vector pUC19. Electrocompetent Escherichia coli DH10B (Invitrogen Corporation, Carlsbad, CA) was transformed with the resultant recombinant plasmids. As a result, pUCarmA, a pUC19 derivative with a 4-kb insert containing armA, was obtained. E. coli DH10B(pUCarmA) exhibited high-level resistance (MICs, ≥256 μg/ml) to amikacin, tobramycin, and gentamicin. The genetic environment surrounding armA was identical to those reported earlier as Tn1548 for strains belonging to the family Enterobacteriaceae (7, 8). The 3′ end of orf513, the gene that encodes a putative transposase that characterizes ISCR1 and that is commonly associated with class 1 integrons (16), and tnpU, another putative transposase gene, were located upstream of armA. It was followed downstream by the 5′ end of yet another putative transposase gene, tnpD. These findings suggest that Tn1548 is serving as an efficient vehicle to mobilize armA across phylogenetically distant gram-negative species.
A reverse transcription assay was conducted to confirm the expression of armA in A. baumannii. Total RNAs of isolate B (armA positive) and isolate F (armA negative) were prepared with the RNeasy Maxi kit (QIAGEN Inc., Valencia, CA). Reverse transcription was performed by the use of a high-capacity cDNA reverse transcription kit (Applied Biosystems). The resultant cDNA was then used as the template for a PCR (4). The presence of mRNA transcripts for armA was observed in isolate B (armA positive) but not isolate F (armA negative). No amplicon was obtained for either isolate when reverse transcriptase was absent. These results confirmed the expression of armA in A. baumannii.
Curing of armA was attempted with A. baumannii isolate B by serial passage of the strain in Luria-Bertani broth containing ethidium bromide at a subinhibitory concentration. One of the strains obtained by this procedure, A. baumannii 231, was found to be susceptible to amikacin. Loss of armA was confirmed by PCR. Conjugation experiments were conducted with the five study isolates as the donors and azide-resistant E. coli J53 or colistin-resistant A. baumannii 213 as the recipient by using standard broth mating techniques. No amikacin-resistant transconjugants were obtained for any of the study isolates producing ArmA with either recipient.
To further localize the armA gene, both plasmid and genomic DNAs from isolate A were subjected to DNA hybridization. Plasmids from isolate A were extracted and subjected to electrophoresis. Subsequently, they were transferred to a nylon membrane by the method of Southern (15) and hybridized with digoxigenin-labeled armA gene fragments by use of the PCR DIG detection system (Roche Diagnostics, Indianapolis, IN). The genomic DNA of isolate A was digested with restriction enzyme CeuI (New England BioLabs) and subjected to pulsed-field gel electrophoresis (PFGE) according to the method of Liu et al. (12). Hybridization was carried out in the same fashion by using probes specific for either armA or the 16S and 23S rRNA genes (9). A plasmidic band from isolate A hybridized with the armA probe, whereas none of the chromosomal bands obtained by CeuI digestion hybridized with it. These results, along with the curability of armA, suggested a plasmidic location of armA.
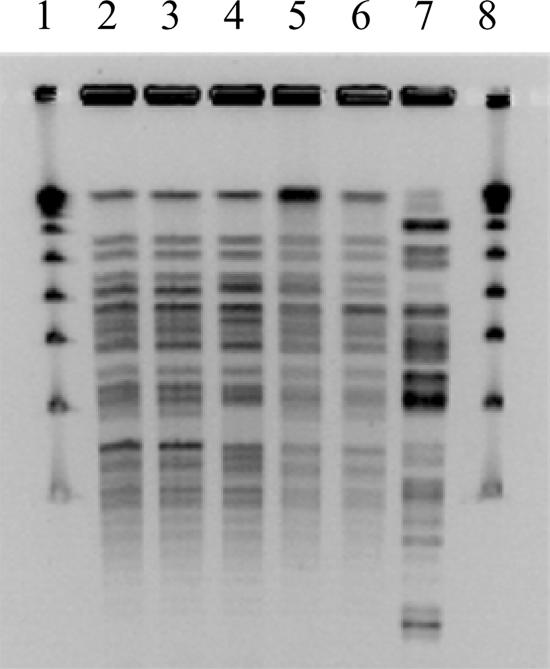
To assess the genetic relatedness of the study isolates, genomic DNA was isolated and digested with ApaI (New England BioLabs). PFGE was performed with the CHEF III system (Bio-Rad, Hercules, CA) with the following run parameters: block I, switch time of 3 to 8 s and run time of 10 h; block II, switch time of 12 to 20 s and run time of 10 h. The five isolates that carry armA were either identical or closely related to each other, differing by up to three bands (Fig. 1). Thus, the spread of armA in our study population appeared to be due to the dissemination of closely related clones of A. baumannii that carry armA on a nonconjugative plasmid.
FIG. 1.
Result of PFGE. Lanes 1 and 8, molecular weight marker; lanes 2 to 6, isolates A to E; lane 7, armA-negative, amikacin-resistant clinical isolate from the study period (control).
In summary, we report the first identification of 16S rRNA methylase as a mechanism of high-level resistance to aminoglycosides in North America. Some of the A. baumannii strains were simultaneously resistant to other classes of antimicrobials, including carbapenems. More research is required to add to the understanding of the increasingly complex nature of the multidrug resistance in this troublesome organism.
Nucleotide sequence accession number.
The nucleotide sequence determined in this study appears in the EMBL/GenBank/DDBJ databases under accession number EU014811.
Acknowledgments
We thank the clinical microbiology laboratory staff at University of Pittsburgh Medical Center for the identification and provision of the study isolates.
Y.D. is supported by NIH training grant T32 AI007333. D.L.P. is supported in part by NIH research grant R01AI070896-01A1.
Footnotes
Published ahead of print on 4 September 2007.
REFERENCES
- 1.American Thoracic Society and Infectious Diseases Society of America. 2005. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am. J. Respir. Crit. Care Med. 171:388-416. [DOI] [PubMed] [Google Scholar]
- 2.Bonomo, R. A., and D. Szabo. 2006. Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin. Infect. Dis. 43(Suppl. 2):S49-S56. [DOI] [PubMed] [Google Scholar]
- 3.Bouchillon, S. K., D. J. Hoban, B. M. Johnson, J. L. Johnson, A. Hsiung, and M. J. Dowzicky. 2005. In vitro activity of tigecycline against 3989 gram-negative and gram-positive clinical isolates from the United States Tigecycline Evaluation and Surveillance Trial (TEST Program; 2004). Diagn. Microbiol. Infect. Dis. 52:173-179. [DOI] [PubMed] [Google Scholar]
- 4.Doi, Y., and Y. Arakawa. 2007. 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin. Infect. Dis. 45:88-94. [DOI] [PubMed] [Google Scholar]
- 5.Donald, H. M., W. Scaife, S. G. Amyes, and H. K. Young. 2000. Sequence analysis of ARI-1, a novel OXA β-lactamase, responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob. Agents Chemother. 44:196-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fournier, P. E., and H. Richet. 2006. The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin. Infect. Dis. 42:692-699. [DOI] [PubMed] [Google Scholar]
- 7.Galimand, M., S. Sabtcheva, P. Courvalin, and T. Lambert. 2005. Worldwide disseminated armA aminoglycoside resistance methylase gene is borne by composite transposon Tn1548. Antimicrob. Agents Chemother. 49:2949-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.González-Zorn, B., A. Catalan, J. A. Escudero, L. Dominguez, T. Teshager, C. Porrero, and M. A. Moreno. 2005. Genetic basis for dissemination of armA. J. Antimicrob. Chemother. 56:583-585. [DOI] [PubMed] [Google Scholar]
- 9.Héritier, C., L. Poirel, D. Aubert, and P. Nordmann. 2003. Genetic and functional analysis of the chromosome-encoded carbapenem-hydrolyzing oxacillinase OXA-40 of Acinetobacter baumannii. Antimicrob. Agents Chemother. 47:268-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Héritier, C., L. Poirel, T. Lambert, and P. Nordmann. 2005. Contribution of acquired carbapenem-hydrolyzing oxacillinases to carbapenem resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:3198-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoban, D. J., S. K. Bouchillon, B. M. Johnson, J. L. Johnson, and M. J. Dowzicky. 2005. In vitro activity of tigecycline against 6792 gram-negative and gram-positive clinical isolates from the global Tigecycline Evaluation and Surveillance Trial (TEST Program, 2004). Diagn. Microbiol. Infect. Dis. 52:215-227. [DOI] [PubMed] [Google Scholar]
- 12.Liu, S. L., A. Hessel, and K. E. Sanderson. 1993. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. USA 90:6874-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rice, L. B. 2006. Challenges in identifying new antimicrobial agents effective for treating infections with Acinetobacter baumannii and Pseudomonas aeruginosa. Clin. Infect. Dis. 43(Suppl. 2):S100-S105. [DOI] [PubMed] [Google Scholar]
- 14.Shaw, K. J., P. N. Rather, R. S. Hare, and G. H. Miller. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57:138-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 16.Toleman, M. A., P. M. Bennett, and T. R. Walsh. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 70:296-316. [DOI] [PMC free article] [PubMed] [Google Scholar]