Abstract
Antimicrobial peptides (AMPs) are naturally occurring, broad-spectrum antimicrobial agents that have recently been examined for their utility as therapeutic antibiotics. Unfortunately, they are expensive to produce and are often sensitive to protease digestion. To address this problem, we have examined the activity of a peptide mimetic whose design was based on the structure of magainin, exhibiting its amphiphilic structure. We demonstrate that this compound, meta-phenylene ethynylene (mPE), exhibits antimicrobial activity at nanomolar concentrations against a variety of bacterial and Candida species found in oral infections. Since Streptococcus mutans, an etiological agent of dental caries, colonizes the tooth surface and forms a biofilm, we quantified the activity of this compound against S. mutans growing under conditions that favor biofilm formation. Our results indicate that mPE can prevent the formation of a biofilm at nanomolar concentrations. Incubation with 5 nM mPE prevents further growth of the biofilm, and 100 nM mPE reduces viable bacteria in the biofilm by 3 logs. Structure-function analyses suggest that mPE inhibits the bioactivity of lipopolysaccharide and binds DNA at equimolar ratios, suggesting that it may act both as a membrane-active molecule, similar to magainin, and as an intracellular antibiotic, similar to other AMPs. We conclude that mPE and similar molecules display great potential for development as therapeutic antimicrobials.
Innate immunity, the first line of defense against pathogen invasion, utilizes broad-spectrum antimicrobial peptides. Characteristic physical hallmarks of these peptides include amphipathic mixtures of α-helical and β-sheet structures and an overall cationic charge (24). Their mode of action often involves binding to the negatively charged lipopolysaccharide (LPS) moieties on the microbial membrane. Once a sufficient aggregate has formed, these peptides destabilize the lipid head groups, form toroidal pores, and disrupt the cellular membrane (24). Due to their attraction to negatively charged structural molecules on the bacterial membrane, development of resistance to these peptides is rare (35), making them potentially useful as antibiotics. These peptides are also cell specific and are able to discriminate host and nonhost cells based on charge (30). Unfortunately, they are difficult and expensive to produce in large quantities and are often sensitive to protease digestion (24). The quest for new and improved antimicrobial peptides has led to the study of peptide mimetics (30). Investigators in this field have developed a series of inexpensive nonpeptidic oligomers and polymers, modeled after compounds found in nature, that adopt amphiphilic secondary structures and exhibit potent and selective antimicrobial activity (25). Modifications of these molecules have resulted in the identification of small-molecule oligomers that have molecular masses ranging from 690 to 1,000 Da, that potently inhibit the growth of both gram-negative and gram-positive bacteria, and that exhibit low hemolytic activity (29). When these small-molecule oligomers were tested against multiple strains of a variety of human-pathogenic bacteria, little variability in sensitivity was observed, and no resistance was developed after 16 passages at sub-MIC levels (29).
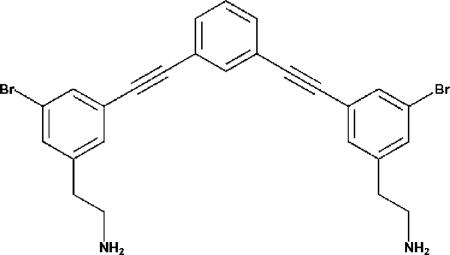
New antimicrobial compounds for use against oral pathogens are in high demand. In this study, we present a novel peptide mimetic, meta-phenylene ethynylene (mPE), modeled after magainin, an antimicrobial peptide from the skin of the African frog Xenopus laevis (Fig. 1) (36). Through the use of sum frequency generation vibrational spectroscopy, Chen et al. showed that similar mimetic compounds orient themselves perpendicularly into the cellular membrane of bacterial cell walls at low physiological concentrations (3, 29). Similar compounds have previously shown potent activity against a number of clinical pathogens (29). Here, we explore the efficacy of mPE against pathogens found in the oral cavity. We also investigated the structure-function relationship of mPE to other antimicrobial peptides.
FIG. 1.
Chemical structure of mPE.
MATERIALS AND METHODS
Bacterial strains and growth.
Strains were maintained as follows. Streptococcus mutans (strain 33402 and clinical isolates) in brain heart infusion (BHI; BD BBL) medium, Staphylococcus aureus (ATCC 27660) and Escherichia coli (D31) in LB, and Actinomyces viscosus in tryptic soy broth (TSB) were maintained at 37°C aerobically. Porphyromonas gingivalis (ATCC 53978) was grown in TSB with yeast extract in an anaerobic chamber at 37°C. Actinobacillus actinomycetemcomitans (CU1000) was grown in TSB supplemented with 6 g/liter yeast extract, 0.8 g/liter dextrose, 0.4 g/liter sodium bicarbonate, 75 mg/liter bacitracin, and 5 mg/liter vancomycin in a 100-mm polystyrene tissue culture dish (TPP, Switzerland) at 37°C-10% CO2. Clinical isolates of S. mutans were obtained from the Department of Oral Biology, New Jersey Dental School, and were identified by growth on selective medium and microscopic evaluation.
Candida strains and growth.
Strains of Candida are shown in Table 1. All strains were grown under standard conditions, specifically at 35°C in YPD medium (1% yeast extract, 2% peptone, 2% dextrose [pH 5.7]).
TABLE 1.
Strains of Candida used in the study
| Species | Strain | Strain type | Drug susceptibility |
|---|---|---|---|
| C. albicans | ATCC 90028 | Reference | Azole susceptible |
| C. albicans | 44 | Laboratory | Azole and echinocandin resistant |
| C. albicans | M70 | Clinical | Azole resistant |
| C. dubliniensis | NCPF3949 | Reference | Azole susceptible |
| C. glabrata | ATCC 90030 | Reference | Azole susceptible |
| C. krusei | ATCC 6258 | Reference | Inherently azole resistant |
| C. parapsilosis | ATCC 22019 | Reference | Azole susceptible |
| C. tropicalis | ATCC 750 | Reference | Azole susceptible |
Antimicrobial assays.
MICs were determined for aerobic bacteria as described by Cole et al. (5), with modifications for each organism as necessary. In brief, the appropriate growth medium for each organism was used to prepare 5-ml overnight cultures to an exponential phase. Bacteria were adjusted to a concentration of 4.5 × 105 CFU/ml, added to various concentrations of antibiotic in 96-well plates, and incubated at 37°C for a period of 18 to 24 h in an incubator in a humidified container. The MIC was defined as the lowest concentration that prevented visible turbidity. Minimum bactericidal concentrations (MBCs) were determined by plating the wells with the MIC as well as one dilution above and one below. After 24 to 48 h of growth, the MBC was determined as the lowest concentration that did not permit visible growth on the surface of the agar. All MIC assays were performed in triplicate.
P. gingivalis was maintained in cooked meat medium with glucose, hemin, and vitamin K (BBL prepared media; BD Biosciences) at 37°C in an anaerobic chamber using CO2 packets (gas packs; BBL). All MICs were carried out in both cooked meat medium and TSB following the same protocol as that used for the aerobic bacteria.
Susceptibility testing for Candida was performed using the broth microdilution method of CLSI (formerly NCCLS) document M27-A2 (23). Briefly, colonies from a 24-h culture grown in YPD agar were resuspended in 0.85% sterile saline. A spectrophotometer was used to standardize the yeast inoculum, using a 0.5 McFarland turbidity standard read at an optical density at 530 nm. Cells were further diluted to a concentration of 1.0 × 103 to 5.0 × 103 CFU/ml in RPMI 1640 medium supplemented with 0.165 M MOPS (morpholinepropanesulfonic acid), pH 7.0. Stock drug solutions of mPE (dissolved in dimethyl sulfoxide) were diluted in RPMI 1640 medium plus 0.165 M MOPS to two times the final concentration (0.016 to 16 μg/ml) in 100 μl and were individually added to a 96-well microdilution tray. Cells (final concentration of 0.5 × 103 to 2.5 × 103 CFU/ml in 100 μl) were added to the drug-containing microtiter plate, which was placed in a 35°C incubator. MICs were visually read at 24 h. All strains were tested in duplicate. The lowest concentration at which there was no visible growth observed compared to growth in the drug-free well was considered the MIC. Cultures were grown for a further 24 h, and the lowest concentration at which no blastoconidia were observed was considered the minimal fungicidal concentration.
Bactericidal kinetics.
Fresh cultures of bacteria were diluted to 1 × 105 CFU/ml and resuspended in 1× phosphate-buffered saline (PBS). After the addition of mPE, aliquots were taken at several time points and plated on BHI plates for colony counts. All experiments were performed in triplicate for statistical analysis.
Quantification of activity against biofilms.
mPE activity against biofilm formation during the initiation stage was measured using a simple biofilm model. Similar to the MIC protocol, mPE was diluted to various concentrations in 96-well plates and added to each well with S. mutans (ATCC 33402) in BHI medium supplemented with 1% sucrose. The plates were incubated overnight at 37°C. The MIC was defined as the lowest concentration of the compound that prevented the formation of a visible biofilm. The biofilms were stained with 0.1% crystal violet for visualization.
To measure activity against mature biofilms, S. mutans was seeded into 96-well flat-bottom plates and allowed to grow for 1 or 2 days in BHI medium supplemented with 1% sucrose before being treated with various concentrations of mPE for three consecutive days. Biofilm growth and susceptibility were determined as described by Wei et al. (33). Planktonic cells were removed by washing the biofilms with PBS. Biofilms were fixed with 1% methanol and stained with crystal violet to determine the extent of biofilm formation. The viability of bacteria in the biofilm was measured using the protocol of Merritt et al. (19). Briefly, biofilms were washed using 1× PBS and individual wells were placed in a 5-ml tube containing 2 ml of PBS. Afterward, they were sonicated at an output level of 6 in a duty cycle of 60% for 30 seconds (VWR Branson Sonifier 450; MicroTip). Each sample was serially diluted and plated on mitis agar to count colonies. The wells were removed from the tubes and stained with 0.4% crystal violet to observe any residual biofilms.
To further confirm the effect, biofilms were subjected to treatment with mPE, washed three times with PBS, and subjected to staining using a LIVE/DEAD kit (Molecular Probes, Eugene, OR). One microliter of dye was diluted 1:1,000, and cultures were stained for 15 min in the dark, followed by one wash with PBS. A Zeiss confocal laser scanning microscope (CLSM 510) mounted to a Zeiss Axiovert 100M base was used to visualize the biofilms. Images were obtained by using the stack 8-bit scan mode with a water-immersible C-Apochromat lens. An argon laser was used for the 488-nm wavelength, and a helium-neon laser was used for the 543-nm wavelength in order to simultaneously collect the two different fluorescent signals. Images were acquired at ×400 magnification. Image analysis was performed using Zeiss LSM Image Browser software. The entire three-dimensional structure (xyz direction) of the biofilm was reconstructed by summing up each of the sectional analyses (xy direction) of 0.45-μm thickness (xz direction).
Synergistic bactericidal activity.
A standard checkerboard assay according to the protocol of Cole et al. (4) was performed to test the synergistic effects of other antimicrobial compounds in conjunction with mPE. Briefly, concentrations of multiple compounds were combined in standard MIC format along with 4.5 × 105 CFU/ml of S. mutans and grown overnight. A fractional inhibitory concentration (FIC) index was then determined by the following equation (14): FIC index = FIC(A) + FIC(B) = [A]/MIC(A) + [B]/MIC(B), where [A] is the lowest inhibitory concentration of compound A in the presence of compound B, MIC(A) is the MIC of compound A alone, and FIC(A) is the FIC of compound A; [B], MIC(B), and FIC(B) are the corresponding values for compound B. Synergy is defined as an FIC index of <1, antagonism as an FIC index of >1, and additivity as an FIC index of 1.
Inhibition of LPS bioactivity.
The ability to inhibit the activity of LPS in vivo was tested through the measurement of tumor necrosis factor alpha (TNF-α) secretion from RAW 264.7 macrophage cells as described by Rosenfeld et al. (26). Cells were cultured overnight in six-well plates (1 × 106 cells/well). The following day, the cells were treated with 100 ng/ml Pseudomonas aeruginosa LPS in the presence of various concentrations of mPE or polymyxin B (10 ng/ml, 7 μM) for 6 h at 37°C. Afterward, the medium for each sample was collected and TNF-α concentrations were evaluated using a mouse TNF-α enzyme-linked immunosorbent assay (ELISA) kit according to the manufacturer's protocol (BD Biosciences).
In silico LPS docking.
A computational approach was used to test LPS binding in silico using the structural file 1QFG (E. coli ferric hydroxamate receptor [FhuA]) (8) and the structural file for mPE. The LPS was removed from the FhuA receptor file and opened in Swiss-PdbViewer v3.7 (10) to rebuild missing side chains. Afterward, both structures, minimized in Sybyl 7.2 (Tripos), were prepared with the addition of polar hydrogens and charges by using the Gasteiger-Marsili method (9). The resulting files were then prepared with the AutoTors utility in AutoDock Tools (13). Docking was performed using the Lamarckian genetic algorithm in AutoDock 3.0 (21). The experiment was performed three times, with a total of 10 docked conformations produced each time. The best fit was chosen based on the lowest energetic conformation of the structure.
DNA binding.
The ability of mPE to bind DNA was investigated both in silico and in vitro. In vitro studies were carried out according to the protocol of Hsu et al. (12). Briefly, both single- and double-stranded oligonucleotides were added at various molar ratios with mPE. After a 15-min incubation at 37°C, they were separated by agarose (1%) gel electrophoresis and poststained with ethidium bromide. The sequences of the oligonucleotides were as follows: GC-rich oligonucleotide, 5′-CGC GCG CGT TTT CGC GCG CG-3′; and AT-rich oligonucleotide, 5′-CAT ATA TAT CCCC CAT ATA TAT G-3′.
AutoDock protocol.
To predict the binding ability of mPE to DNA, a computational approach was used. A structural file for mPE was created using ChemDraw software (CambridgeSoft). The protein database file 1AGL, a single-stranded DNA previously bound to an inhibitor, was used as the docking template. Both structures were prepared with the addition of polar hydrogens and charges by using the Gasteiger-Marsili method (9) and minimized in Sybyl 7.2 (Tripos). The resulting files were then prepared with the AutoTors utility in AutoDock Tools. Docking was performed using the Lamarckian genetic algorithm in AutoDock 3.0 (21). The experiment was performed three times, with a total of 10 docked conformations produced each time. The best fit was chosen based on the lowest binding energy.
RESULTS
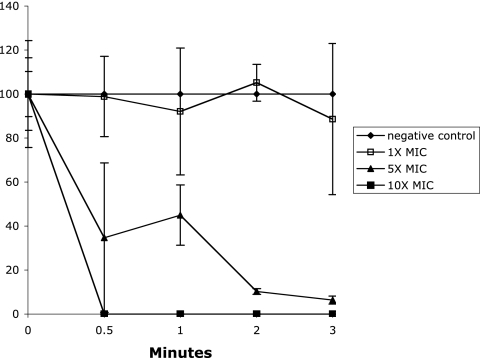
In order to determine the efficacy of the compound mPE, the antimicrobial activity was measured against several common and oral bacterial pathogens, including S. aureus, P. gingivalis, and S. mutans (both laboratory and clinical isolates). The results shown in Table 2 demonstrate that mPE exhibits activity against these pathogens within a range of 0.4 to 2.5 μg/ml (0.75 to 4.7 nM). Identical results were obtained with the standard growth medium for MIC assays (Mueller-Hinton broth) (28) and with the appropriate growth medium for each organism. The rate of bactericidal activity against S. mutans was determined by time course analysis. As shown in Fig. 2, a >2-log killing was observed within 1 minute at 10× MIC. At 1× MIC, no viable bacteria remained by 1 hour (data not shown).
TABLE 2.
MICs and MBCs of mPE against bacterial pathogens
| Bacterium (strain) | MIC (μg/ml) | MBC (μg/ml) |
|---|---|---|
| S. mutans(ATCC 33402) | 0.5-1 | 1.25 |
| S. mutans (isolate N32) | 0.5 | 1.25 |
| S. mutans (isolate N43) | 0.5 | 1.25 |
| S. aureus (ATCC 27660) | 0.25-0.5 | 0.8 |
| P. gingivalis (ATCC 53978) | 2.5 | 2.5 |
| A. actinomycetemcomitans (CU1000) | 0.4 | 1.5 |
| A. viscosus | 0.8 | 1.5 |
FIG. 2.
Kinetics of bactericidal activity of mPE against S. mutans. A total of 2.5 × 103 CFU was incubated at 37°C with increasing concentrations of mPE. Aliquots were removed and plated at the times indicated. The reactions were terminated by dilution in PBS. Samples were plated on BHI agar and incubated overnight. Results are presented as the percentage of viable colonies remaining after treatment.
To determine whether this activity extends to fungal pathogens, we determined the MIC against Candida albicans as well as several non-albicans Candida species which have been identified in oral candidal infections. As shown in Table 3, mPE exhibits growth inhibitory activity against all Candida species tested between 0.5 and 1 μg/ml.
TABLE 3.
Activity of mPE against Candida species
| Strain | MIC (μg/ml) | MFC (μg/ml)a |
|---|---|---|
| C. albicans ATCC 90028 | 1.0 | 2.0 |
| C. albicans 44 | 0.5 | 1.0 |
| C. albicans M70 | 0.5 | 1.0 |
| C. dubliniensis NCPF3949 | 1.0 | 2.0 |
| C. glabrata ATCC 90030 | 1.0 | 2.0 |
| C. krusei ATCC 6258 | 1.0 | 2.0 |
| C. parapsilosis ATCC 22019 | 1.0 | 2.0 |
| C. tropicalis ATCC 750 | 1.0 | 2.0 |
MFC, minimal fungicidal concentration.
Since S. mutans is generally found in a biofilm on the tooth surface, we tested the ability of mPE to prevent the formation of a biofilm. Free-growing (planktonic) S. mutans bacteria were seeded in plastic wells in the presence of 1% sucrose to promote the formation of a biofilm, in the presence or absence of increasing concentrations of mPE. The presence of a biofilm was determined by crystal violet staining. We did not observe the formation of adherent cultures at any concentration above 1 μg/ml.
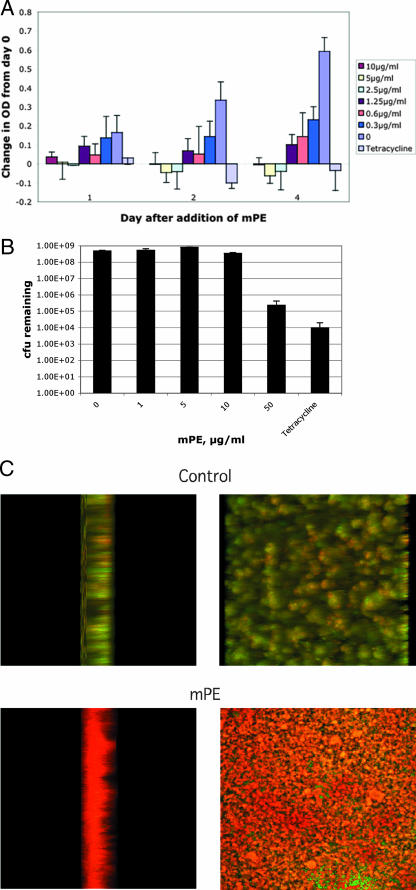
We next tested the activity of mPE against the bacteria growing in a simple biofilm on plastic in the presence of sucrose. Biofilm formation was demonstrated by repeated washing with buffer followed by staining with crystal violet. After 1 day, a significant adherent culture was obtained (not shown). When a 1-day biofilm of S. mutans was further cultured in the presence of the compound, the continued growth of the adherent biomass was inhibited at concentrations higher than 2.5 μg/ml (Fig. 3A). To determine the bactericidal activity against S. mutans in the growing biofilm, we disrupted the biofilm after treatment, followed by plating to determine the number of viable bacteria remaining. A 3-log reduction in viable bacteria was obtained at a concentration of 50 μg/ml (Fig. 3B), comparable to that observed with tetracycline (500 μg/ml). To visually confirm this effect, the biofilms were subjected to treatment with 0 or 50 μg/ml mPE for 24 h and treated with LIVE/DEAD staining. The viability of the cultures was then observed using fluorescence confocal microscopy. As can be seen in Fig. 3C, while the control culture appears predominantly green (demonstrating live organisms), the treated culture is predominantly red (indicating dead organisms). The dead cells are observed throughout the thickness of the culture (left panel), indicating that the compound was able to exert its antimicrobial activity in the biofilm.
FIG. 3.
Activity of mPE against S. mutans in a biofilm. Bacteria (1 × 105 CFU) were seeded in wells of a 96-well plate in the presence of 1% sucrose. (A) Activity against growing biofilm. After formation of a biofilm, mPE was added at increasing concentrations. Growth in the biofilm was quantified by reading the absorbance at 595 nm, and results are presented as the change in absorbance from day 2 cultures prior to the addition of compound. Controls included no mPE (0) and 500 μg/ml tetracycline. The experiment was performed in triplicate. Error bars equal 1 standard deviation. (B) Bactericidal activity against bacteria in a biofilm. Bacteria were plated in sucrose for 2 days, followed by the addition of mPE. After 24 h, biofilms were disrupted by sonication and plated to measure the remaining viable bacteria. Error bars equal 1 standard deviation. (C) Confocal microscopy analysis of biofilm viability. Biofilms were treated for 24 h with 0 or 50 μg/ml mPE and stained using the LIVE/DEAD kit. Cultures were observed using a confocal laser scanning microscope at 488 nm and 543 nm. Merged images were produced using Zeiss LSM Image Browser software. Cultures are visualized from the side (left panel) and the top (right panel).
To determine whether mPE would remain active in the presence of saliva, MIC assays were performed in the presence of increasing concentrations of saliva. Unstimulated, pooled saliva from three individuals was clarified by centrifugation. The addition of clarified human saliva at concentrations of 10% and 25% showed no effect (MIC, 1 μg/ml), while the presence of 50% saliva reduced the MIC twofold (0.5 μg/ml), demonstrating no inhibitory effect. To determine whether this may be due to the synergistic activity of antibacterial salivary proteins, such as salivary histatin (6), lysozyme, and lactoferrin (31), we carried out standard checkerboard assays with S. mutans. None of the proteins demonstrated any synergistic effect on the MIC (data not shown).
To support our hypothesis that mPE would be useful as an antimicrobial, we investigated the potential synergy with a commonly used oral therapeutic compound, chlorhexidine. When examined using a standard checkerboard assay, chlorhexidine exhibited dramatic synergy with mPE, with an FIC index of 0.14.
The observation of synergy with other compounds suggested that mPE may be facilitating the introduction of these microbicides into the organism. To better understand the mechanism of activity, a structural comparison with mPE was performed by searching all free, publicly available biochemical compound databases for similar matches. The pharmacophoric criteria to be used for searching include the SMILES (simplified molecular input line entry system) formula, percent similarity, biological activity, and super- and substructures (20, 27). The results were manually curated and reduced to less than five. These molecules were compared with mPE based on the proximity of the functional groups in relation to the backbone. The molecules found in the database matched either the functional groups or the backbone but not both (not shown), demonstrating the unique structure of mPE.
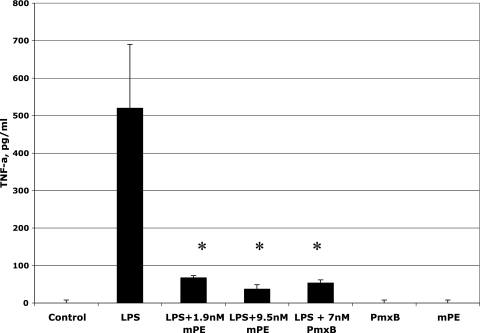
mPE was built using the structural characteristics of the cationic peptide magainin (29). It has been hypothesized that the rapid bactericidal activity of such peptides is based on an initial binding to LPS from gram-negative bacteria (32). To determine whether this functional component is conserved in the mimetic, we examined the interaction of mPE with LPS by using a bioassay. Mouse RAW 264.7 macrophage cells were stimulated simultaneously with 100 ng of Pseudomonas aeruginosa LPS in the presence of mPE, and secretion of TNF-α was measured by ELISA. The results demonstrate a significant reduction in LPS-mediated TNF-α secretion in the presence of mPE at concentrations comparable to that of the LPS-inhibiting peptide polymyxin B (Fig. 4). No effect was seen with mPE or polymyxin B alone. To examine this mPE-LPS interaction on a structural level, an in silico analysis was performed. From the docking model, we predict mPE binding between the inner and outer cores near the O antigen of LPS, with the carbon backbone of mPE closest to the sugar backbone of LPS. This bonding is strengthened by the amine groups' forming hydrogen bonds with the surrounding moieties found on the LPS side chains (see Fig. S1 in the supplemental material).
FIG. 4.
Inhibition of LPS bioactivity by mPE. RAW 264.7 cells were treated with 100 ng/ml E. coli LPS in the presence of 0, 1.9, or 9.5 mM mPE or 7 nM polymyxin B (PmxB) for 6 h at 37°C. Controls were 9.5 nM mPE or 7 nM polymyxin B alone (no LPS). TNF-α (TNF-a) levels were quantified by ELISA as described in Materials and Methods. Experiments were performed in triplicate. Error bars equal 1 standard error; asterisks indicate statistical significance as measured by a t test (P < 0.0001).
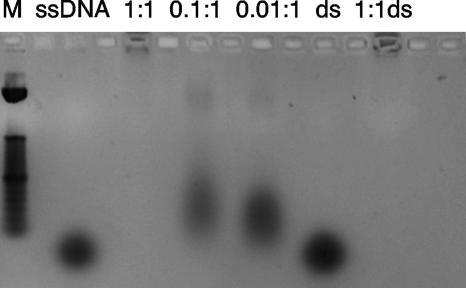
To further address the structure-function relationship mPE shares with other antimicrobial peptides, we investigated its ability to bind DNA both in silico and in vitro. A computational model was used first in an attempt to identify the most energetically feasible binding site. Using AutoDock, we observed a potential binding site in the major groove of the DNA (see Fig. S2 in the supplemental material). This was confirmed by observing the retardation of both single- and double-stranded DNA when coincubated with mPE in gel migration assays (Fig. 5). The results obtained using a GC-rich oligonucleotide demonstrated retardation of migration at a 1:1 molar ratio. Similar results were observed for AT-rich oligonucleotides. When a molar excess of single-stranded DNA (30 nM) was preincubated with mPE, it had a small inhibitory effect on the MIC (increasing from 1.0 to 2.5 μg/ml), suggesting that the affinity for bacterial membranes exceeds that for DNA.
FIG. 5.
DNA binding activity of mPE. A single-stranded GC-rich oligonucleotide (ssDNA) was incubated at 37°C in the absence or presence of mPE at a molar ratio (mPE to DNA) of 1:1, 0.1:1, or 0.01:1, followed by agarose gel electrophoresis. The gel was visualized after ethidium bromide staining and UV irradiation. Double-stranded DNA was created by annealing of the single-stranded oligonucleotide at 65°C in isotonic saline buffer. The double-stranded oligonucleotide was incubated with mPE at a molar ratio of 1:1 (1:1ds) or in buffer alone (ds). The results shown are representative of three experiments. M, molecular marker.
DISCUSSION
Previous studies have reported low MIC ranges for antimicrobial peptide mimetics against other pathogens (29). The findings in this study report similar concentration ranges against oral pathogens, both aerobic and anaerobic. Antimicrobial peptides have been reported to have their activity neutralized in environments containing serum or tryptone, the potential antagonist being free proteins (11). The MICs we observed have been verified in multiple types of media, and the stability and activity of mPE are maintainted in saliva, demonstrating its potential for use in vivo. Furthermore, as the MIC assays for S. mutans are performed static, in sealed, humidified chambers for up to 2 days, they mimic the microaerophilic environment in which these bacteria are found under cariogenic conditions (34). This suggests that molecules such as mPE have high potential as therapeutic antibiotics. The activity against S. mutans is significantly greater than that for magainin, which we have observed to be 1 μg/ml, which equals 41 nM. The rapidity of the activity we observe correlates with magainin's ability to rapidly aggregate and destabilize cellular membranes, forming toroidal pores within seconds of binding (16). A search of the PubChem database as of March 2007 resulted in few matches at the lowest homology setting (80%), showing the uniqueness of these types of compounds in relation to their structure-function activity. The few compounds that emerged matched only based on the backbone, but none were listed as possessing the same type of activity.
A potential oral use that has been evaluated is the application of mPE against biofilms. Biofilms are a diverse and complex aggregate of bacteria that exhibit over 100-fold resistance to conventional antibiotics (18). Once a biofilm is established, the live cells are typically buried beneath the surface or between layers of dead cells and encased by a glycocalyx, an extracellular matrix of carbohydrates, proteoglycans, DNA, and other cellular constituents. Not only does this complex of biological molecules inhibit diffusion due to steric hindrance, but its constituents are believed to carry charges that have been known to interfere with the diffusion of other antibiotics (17). In the oral environment, biofilms of S. mutans are most associated with dental caries (2). We have examined two main phases of biofilms in this work: preformed biofilms, where bacteria are preparing to adhere or have just adhered to a surface, and already established biofilms. Our results show that mPE has the same MIC for bacteria in the preformed biofilm stage as it does against planktonic cells, as they are both very similar in nature. It also shows activity against 1- and 2-day-old biofilms when they are treated continuously for identical 3-day periods. Treatment with a concentration of only 5× MIC of planktonic bacteria (2.5 μg/ml) was sufficient to mediate bacteriostatic activity against an established biofilm. In comparison, a concentration of over 100 μg/ml magainin is required to result in an inhibition of S. mutans biofilm growth (33). The significance of the 2-day biofilm is that it contains twice the cellular mass of the 1-day biofilm yet still succumbs to the same concentration of mPE. While factors such as biofilm structural heterogeneity and diffusion remain unknown, it is assumed that a combination of an advantageous molar ratio, limited passive diffusion, and continuous exposure favors the microbicidal activity of this compound.
Since mPE was designed to possess the physiochemical characteristics of magainin, the structure-function mechanisms of activity were also investigated. The main mechanism of antimicrobial peptides involves their binding to the bacterial membrane and then their disrupting the phospholipid bilayer, leading to subsequent cellular death (24). The positively charged proteins are strongly attracted to the negatively charged constituents of the bacterial membrane, specifically lipoteichoic acid and LPS (32). Magainin has also been extensively studied for its ability to bind both membrane-bound and free LPSs. Our results demonstrated that mPE inhibits the LPS-mediated activation of macrophages at concentrations as low as 1 nM. In comparison, incubation of the same concentration of LPS with 10 μM magainin results in a 30% reduction in TNF activation (26). This suggests that mPE's ability to bind LPS, both free and in bacterial membranes, is similar to that of magainin, probably due to its amphipathic nature and overall charge. It remains unknown whether mPE competes with LPS for binding of the CD14 receptor as with cathelicidin (22).
Antimicrobial peptides are known to bind to intracellular targets after having disrupted cellular osmolarity (1). For example, indolicidin, an antimicrobial peptide from bovine neutrophils, has been shown to bind and retard DNA movement on a gel based upon weight/weight ratio (12). Computational docking models have shown that mPE binds DNA, most likely in the GC-rich area of the major groove (see the supplemental material). Experimentally, our results demonstrate that mPE similarly binds and retards the electrophoretic mobilities of both single- and double-stranded DNA in a dose-dependent fashion with great affinity. This mobility retardation was observed only in the absence of ethidium bromide, suggesting that mPE competed for binding of the DNA. While mPE showed a greater affinity for GC-rich oligomers, it also inhibited AT-rich oligomers better than pentamidine (data not shown), a known DNA binding agent with a similar backbone (7). Interference with DNA replication could lead to a second bactericidal mechanism. This could support our observation of dramatic synergy with chlorhexidine. This common oral antiseptic agent permeates microbial membranes and disrupts membrane potential (15). Together with the potential intracellular activity of mPE, this supports the development of mPE as an antibiotic for oral applications that could be used in conjunction with lower doses of chlorhexidine, thus reducing the unwanted side effects of that agent. While mPE is not cytotoxic at the levels used here (29), a possible danger of the binding of mPE to DNA is the induction of carcinogenesis, which must be examined further.
In summary, our results support the development of inexpensive, broad-spectrum peptide mimetics such as mPE for use as therapeutic anti-infectives in the treatment of oral infectious diseases. mPE has shown rapid activity against several prolific oral pathogens and efficacy in treating oral biofilms. Further investigation of its activity against other biofilm sources is warranted, as its efficacy may extend to other commercial applications.
Supplementary Material
Acknowledgments
We thank John Kerrigan for assistance with computational studies, David Lagunoff and Peter Carroll for assistance with confocal microscopy, N. Ramasubbu for help with molecular modeling, and Daniel Kadouri for helpful advice.
This work was supported by grants from Polymedix, Inc. and from the U.S. Public Health Service (NIH 1R01DE14897) to G.D. G.N.T. thanks the NIH and the ONR for generous support (grants RO1-GM-65803 and N00140310503).
Footnotes
Published ahead of print on 4 September 2007.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Brogden, K. A. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3:238-250. [DOI] [PubMed] [Google Scholar]
- 2.Burne, R. A. 1998. Oral streptococci…products of their environment. J. Dent. Res. 77:445-452. [DOI] [PubMed] [Google Scholar]
- 3.Chen, X., H. Tang, M. A. Even, J. Wang, G. N. Tew, and Z. Chen. 2006. Observing a molecular knife at work. J. Am. Chem. Soc. 128:2711-2714. [DOI] [PubMed] [Google Scholar]
- 4.Cole, A. M., R. O. Darouiche, D. Legarda, N. Connell, and G. Diamond. 2000. Characterization of a fish antimicrobial peptide: gene expression, subcellular localization, and spectrum of activity. Antimicrob. Agents Chemother. 44:2039-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole, A. M., P. Weis, and G. Diamond. 1997. Isolation and characterization of pleurocidin, an antimicrobial peptide in the skin secretions of winter flounder. J. Biol. Chem. 272:12008-12013. [DOI] [PubMed] [Google Scholar]
- 6.Edgerton, M., and S. E. Koshlukova. 2000. Salivary histatin 5 and its similarities to the other antimicrobial proteins in human saliva. Adv. Dent. Res. 14:16-21. [DOI] [PubMed] [Google Scholar]
- 7.Edwards, K. J., T. C. Jenkins, and S. Neidle. 1992. Crystal structure of a pentamidine-oligonucleotide complex: implications for DNA-binding properties. Biochemistry 31:7104-7109. [DOI] [PubMed] [Google Scholar]
- 8.Ferguson, A. D., V. Braun, H. P. Fiedler, J. W. Coulton, K. Diederichs, and W. Welte. 2000. Crystal structure of the antibiotic albomycin in complex with the outer membrane transporter FhuA. Protein Sci. 9:956-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gasteiger, J., and J. Marsili. 1980. Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron 36:3219-3228. [Google Scholar]
- 10.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 11.Hancock, R. E., and G. Diamond. 2000. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 8:402-410. [DOI] [PubMed] [Google Scholar]
- 12.Hsu, C. H., C. Chen, M. L. Jou, A. Y. Lee, Y. C. Lin, Y. P. Yu, W. T. Huang, and S. H. Wu. 2005. Structural and DNA-binding studies on the bovine antimicrobial peptide, indolicidin: evidence for multiple conformations involved in binding to membranes and DNA. Nucleic Acids Res. 33:4053-4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kerrigan, J. E., D. S. Pilch, A. L. Ruchelman, N. Zhou, A. Liu, L. Liu, and E. J. LaVoie. 2003. 5H-8,9-Dimethoxy-5-(2-N,N-dimethylaminoethyl)dibenzo[c,h][1,6]naphthyridin-6-ones and related compounds as TOP1-targeting agents: influence of structure on the ternary cleavable complex formation. Bioorg. Med. Chem. Lett. 13:3395-3399. [DOI] [PubMed] [Google Scholar]
- 14.Kim, S. S., S. Kim, E. Kim, B. Hyun, K. K. Kim, and B. J. Lee. 2003. Synergistic inhibitory effect of cationic peptides and antimicrobial agents on the growth of oral streptococci. Caries Res. 37:425-430. [DOI] [PubMed] [Google Scholar]
- 15.Kuyyakanond, T., and L. B. Quesnel. 1992. The mechanism of action of chlorhexidine. FEMS Microbiol. Lett. 79:211-215. [DOI] [PubMed] [Google Scholar]
- 16.Leontiadou, H., A. E. Mark, and S. J. Marrink. 2006. Antimicrobial peptides in action. J. Am. Chem. Soc. 128:12156-12161. [DOI] [PubMed] [Google Scholar]
- 17.Lewis, K. 2001. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45:999-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mah, T. F., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 19.Merritt, J. H., D. Kadouri, and G. A. O'Toole. 2005. Growing and analyzing static biofilms, p. 1-17. In R. Coico, T. Kowalik, J. Quarles, B. Stevenson, and R. Taylor (ed.), Current protocols in microbiology, vol. 1. John Wiley & Sons, Hoboken, NJ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, M. A. 2002. Chemical database techniques in drug discovery. Nat. Rev. Drug Discov. 1:220-227. [DOI] [PubMed] [Google Scholar]
- 21.Morris, G. M., D. S. Goodsell, R. S. Halliday, R. Huey, W. E. Hart, R. K. Belew, and A. J. Olson. 1998. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comp. Chem. 19:1639-1662. [Google Scholar]
- 22.Nagaoka, I., S. Hirota, F. Niyonsaba, M. Hirata, Y. Adachi, H. Tamura, and D. Heumann. 2001. Cathelicidin family of antibacterial peptides CAP18 and CAP11 inhibit the expression of TNF-alpha by blocking the binding of LPS to CD14(+) cells. J. Immunol. 167:3329-3338. [DOI] [PubMed] [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. 2002. Reference method for broth dilution antifungal susceptibility testing of yeasts Approved standard M27-A2. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 24.Pazgier, M., D. M. Hoover, D. Yang, W. Lu, and J. Lubkowski. 2006. Human beta-defensins. Cell. Mol. Life Sci. 63:1294-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rennie, J., L. Arnt, H. Tang, K. Nusslein, and G. N. Tew. 2005. Simple oligomers as antimicrobial peptide mimics. J. Ind. Microbiol. Biotechnol. 32:296-300. [DOI] [PubMed] [Google Scholar]
- 26.Rosenfeld, Y., N. Papo, and Y. Shai. 2006. Endotoxin (lipopolysaccharide) neutralization by innate immunity host-defense peptides. Peptide properties and plausible modes of action. J. Biol. Chem. 281:1636-1643. [DOI] [PubMed] [Google Scholar]
- 27.Sheridan, R. P. 2002. The most common chemical replacements in drug-like compounds. J. Chem. Inf. Comput. Sci. 42:103-108. [DOI] [PubMed] [Google Scholar]
- 28.Steinberg, D. A., and R. I. Lehrer. 1997. Designer assays for antimicrobial peptides. Disputing the “one-size-fits-all” theory. Methods Mol. Biol. 78:169-186. [DOI] [PubMed] [Google Scholar]
- 29.Tew, G. N., D. Clements, H. Tang, L. Arnt, and R. W. Scott. 2006. Antimicrobial activity of an abiotic host defense peptide mimic. Biochim. Biophys. Acta 1758:1387-1392. [DOI] [PubMed] [Google Scholar]
- 30.Tew, G. N., D. Liu, B. Chen, R. J. Doerksen, J. Kaplan, P. J. Carroll, M. L. Klein, and W. F. DeGrado. 2002. De novo design of biomimetic antimicrobial polymers. Proc. Natl. Acad. Sci. USA 99:5110-5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Velliyagounder, K., J. B. Kaplan, D. Furgang, D. Legarda, G. Diamond, R. E. Parkin, and D. H. Fine. 2003. One of two human lactoferrin variants exhibits increased antibacterial and transcriptional activation activities and is associated with localized juvenile periodontitis. Infect. Immun. 71:6141-6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vorland, L. H., H. Ulvatne, O. Rekdal, and J. S. Svendsen. 1999. Initial binding sites of antimicrobial peptides in Staphylococcus aureus and Escherichia coli. Scand. J. Infect. Dis. 31:467-473. [DOI] [PubMed] [Google Scholar]
- 33.Wei, G. X., A. N. Campagna, and L. A. Bobek. 2006. Effect of MUC7 peptides on the growth of bacteria and on Streptococcus mutans biofilm. J. Antimicrob Chemother. 57:1100-1109. [DOI] [PubMed] [Google Scholar]
- 34.Yamada, T., S. Takahashi-Abbe, and K. Abbe. 1985. Effects of oxygen on pyruvate formate-lyase in situ and sugar metabolism of Streptococcus mutans and Streptococcus sanguis. Infect. Immun. 47:129-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389-395. [DOI] [PubMed] [Google Scholar]
- 36.Zasloff, M. 1987. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc. Natl. Acad. Sci. USA 84:5449-5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.