Abstract
Mupirocin resistance in Staphylococcus aureus is increasingly being reported in many parts of the world. This study describes the epidemiology and laboratory characterization of mupirocin-resistant methicillin-resistant S. aureus (MRSA) strains in Canadian hospitals. Broth microdilution susceptibility testing of 4,980 MRSA isolates obtained between 1995 and 2004 from 32 Canadian hospitals was done in accordance with CLSI guidelines. The clinical and epidemiologic characteristics of strains with high-level mupirocin resistance (HLMupr) were compared with those of mupirocin-susceptible (Mups) strains. MRSA strains were characterized by pulsed-field gel electrophoresis (PFGE) and typing of the staphylococcal chromosomal cassette mec. PCR was done to detect the presence of the mupA gene. For strains with mupA, plasmid DNA was extracted and subjected to Southern blot hybridization. A total of 198 (4.0%) HLMupr MRSA isolates were identified. The proportion of MRSA strains with HLMupr increased from 1.6% in the first 5 years of surveillance (1995 to 1999) to 7.0% from 2000 to 2004 (P < 0.001). Patients with HLMupr MRSA strains were more likely to have been aboriginal (odds ratio [OR], 3.7; 95% confidence interval [CI], 1.5 to 9.4; P = 0.006), to have had community-associated MRSA (OR, 2.2; 95% CI, 1.0 to 5.0; P = 0.05), and to have been colonized with MRSA (OR, 1.7; 95% CI, 1.0 to 3.0; P = 0.04). HLMupr MRSA strains were also more likely to be resistant to fusidic acid (21% versus 4% for mupirocin-susceptible strains; P < 0.001). All HLMupr MRSA strains had a plasmid-associated mupA gene, most often associated with a 9-kb HindIII fragment. PFGE typing and analysis of the plasmid profiles indicate that both plasmid transmission and the clonal spread of HLMupr MRSA have occurred in Canadian hospitals. These results indicate that the incidence of HLMupr is increasing among Canadian strains of MRSA and that HLMupr MRSA is recovered from patients with distinct clinical and epidemiologic characteristics compared to the characteristics of patents with Mups MRSA strains.
Mupirocin is a topical antimicrobial agent that interferes with protein synthesis by competitive inhibition of bacterial isoleucyl-tRNA synthetase (42). It has been used to treat skin and soft tissue infections and to eradicate staphylococcal carriage in health care workers and patients (7). Intranasal mupirocin has also been used preoperatively to prevent surgical site infections (17, 19, 28, 41) and to control the transmission of methicillin-resistant Staphylococcus aureus (MRSA) in health care facilities (2, 14, 18, 40). However, the prevalence of mupirocin resistance in MRSA has increased in settings with extensive use of this agent (8, 22, 38), and it has also been reported in community-associated MRSA strains (13). In Canada, high-level mupirocin resistance has recently been reported in more than 50% of community-associated strains identified in an outbreak in northern Saskatchewan (23).
Although no performance standards or interpretive criteria have been published for mupirocin susceptibility testing, mupirocin resistance in staphylococci is commonly defined as low-level resistance (MICs, 8 to 256 μg/ml) or high-level resistance (MICs, ≥512 μg/ml) (3, 16). Low-level resistance is usually associated with point mutations in the chromosomally encoded ileS gene (10, 36), whereas high-level resistance is generally due to a plasmid-mediated gene, mupA (also referred to as ileS2), which encodes an additional modified isoleucyl-tRNA synthetase (15, 36). Treatment with mupirocin is not likely to be effective in the presence of high-level mupirocin resistance (6, 34, 39), and there is some evidence to suggest that low-level resistance may also predict treatment failure (39). In one study involving patients undergoing long-term peritoneal dialysis, the development of mupirocin resistance was associated with an increased risk of staphylococcal infections (26).
In this report, we describe the epidemiology and clinical features of hospitalized patients with high-level mupirocin-resistant MRSA strains in a network of Canadian hospitals between 1995 and 2004. We also characterized these strains in order to determine the molecular epidemiology and mechanisms of mupirocin resistance.
MATERIALS AND METHODS
Surveillance for MRSA has been conducted by hospitals in Canada participating in the Canadian Nosocomial Infection Surveillance Program since January 1995. The surveillance methods used have been described previously (32, 33). When a new case of MRSA infection or colonization in an inpatient was identified, the infection control practitioner used a standardized data collection form to abstract demographic and clinical information from the medical records. The presence of infection caused by MRSA was determined by the infection control practitioner using standard definitions (11). The site of MRSA acquisition (health care facility or community) was determined by using previously published criteria (9). The designation of isolates as community acquired was based on epidemiologic data and the absence of established risk factors for health care-associated MRSA, prior to knowledge of the molecular strain typing results. Demographic, clinical, and epidemiologic data for patients with high-level mupirocin-resistant MRSA were compared with those for patients with mupirocin-susceptible MRSA (excluding those with low-level mupirocin resistance).
The first MRSA isolate from each patient was sent to a central laboratory for additional testing. The isolates were confirmed to be MRSA by detection of the mecA and nuc genes by a multiplex PCR assay (21). Antimicrobial susceptibility testing was done by broth microdilution methods in accordance with Clinical and Laboratory Standards Institute guidelines (5). Inducible resistance to clindamycin in macrolide-resistant strains of MRSA was detected by a standardized disk approximation test (5). MRSA strains were typed by pulsed-field gel electrophoresis (PFGE) with SmaI digests of genomic DNA; DNA profiles were digitized and analyzed with BioNumerics software, version 3.5 (Applied Maths, Austin, TX) (33). Typing of the staphylococcal chromosomal cassette mec (SCCmec) was done by PCR with primers and by the methods published previously (24).
The mupA gene was detected in DNA extracts by PCR with primers and by the methods described previously (1). Plasmid DNA was extracted by using a High Pure plasmid isolation kit (Roche Diagnostics, Laval, Quebec, Canada), but with a modification to the manufacturer's instructions, in which lysostaphin was added in the lysis step of the procedure. Purified plasmid DNA was eluted in 50 μl of TE (Tris-EDTA) buffer. The plasmids were restricted with HindIII for 1 h, separated on a 1% agarose gel in 0.5× TAE (Tris-acetate-EDTA) at 60 V for 3 h, and transferred onto a Hybond N+ membrane (GE Healthcare, Piscataway, NJ). The membrane was probed with a 458-bp PCR-amplified mupA gene probe by using an enhanced chemiluminescence direct nucleic acid labeling and detection system (GE Healthcare). HindIII restriction fragment length polymorphisms were examined and assigned profile descriptors.
Statistical analyses were done by Student's t test, the chi-square test, and Fisher's exact test, as appropriate. All statistical tests were two tailed, with a P value of ≤0.05 considered statistically significant. A multivariate logistic regression analysis was done and included variables with P values of <0.20 in the univariate analysis. All analyses were done with SPSS software, version 11.0.
RESULTS
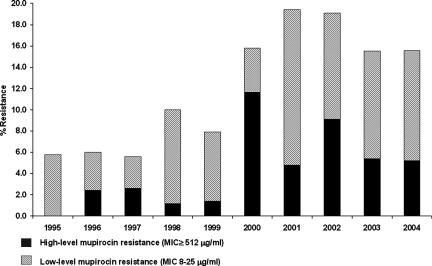
A total of 4,980 unique patient MRSA isolates recovered from 32 Canadian Nosocomial Infection Surveillance Program hospitals between 1995 and 2004 were available for antimicrobial susceptibility testing. Of these, 198 (4.0%) were found to have high-level resistance to mupirocin, 396 (8.0%) had low-level mupirocin resistance, and 4,386 were susceptible to mupirocin. The proportion of isolates that were resistant to mupirocin increased over time (Fig. 1). In the first 5 years of surveillance (1995 to 1999), 46 (1.6%) MRSA strains had high-level resistance, whereas the rates increased nearly fivefold to 7.0% among isolates recovered from 2000 to 2004 (P < 0.001). The rates of low-level mupirocin resistance also increased during this time, from 6.4% (1995 to 1999) to 10.0% (2000 to 2004) (P < 0.001). MRSA strains with high-level resistance were identified in 17 hospitals across the country (representing 53% of the hospitals with MRSA in the surveillance), with rates ranging from 0 to 26% (median, 3%) among the MRSA strains tested. Isolates from five hospitals from geographically diverse regions of the country accounted for 72% of all the MRSA isolates with high-level resistance to mupirocin; only 38% of all the MRSA isolates identified were reported from these five hospitals.
FIG. 1.
Annual rates of mupirocin resistance in MRSA strains recovered from Canadian hospitals, 1995 to 2004.
Complete clinical and epidemiologic data were available for 139 (70%) patients with high-level mupirocin resistant MRSA and for 3,187 (73%) patients with mupirocin-susceptible MRSA. The demographic and clinical characteristics of these patients are summarized in Table 1. In the multivariate analysis, the detection of high-level mupirocin-resistant MRSA strains was found to be associated with being a native aboriginal (odds ratio [OR], 3.71; 95% confidence interval [CI], 1.51 to 9.36; P = 0.006), with having a community-associated isolate (OR, 2.24; 95% CI, 1.02 to 4.96; P = 0.05), and with having been colonized rather than infected with MRSA (OR, 1.74; 95% CI, 1.02 to 2.99; P = 0.04) (Table 2).
TABLE 1.
Demographic and clinical characteristics of hospitalized patients with mupirocin-susceptible and mupirocin-resistant MRSA strains, 1995 to 2004a
| Characteristic | Patients with mupirocin-susceptible MRSA | Patients with mupirocin-resistant MRSA | OR (95% CI) | P value |
|---|---|---|---|---|
| No. of patients | 3,187 | 139 | ||
| Median age (yr) | 69.6 | 71.6 | 0.40 | |
| No. (%) males | 1,918 (60) | 91 (66) | 1.2 (0.9-1.8) | 0.26 |
| No. (%) of patients of aboriginal ethnicity | 92 (3) | 20 (18) | 6.3 (3.7-10.7) | <0.001 |
| No. (%) of patients with community-associated MRSA | 134 (6) | 13 (14) | 2.5 (1.4-4.6) | 0.003 |
| No. (%) of patients from the following region of country: | ||||
| Eastb | 162 (5) | 2 (1) | ||
| Centralc | 1,931 (61) | 94 (68) | ||
| Westd | 1,094 (34) | 43 (31) | 0.08 | |
| No. (%) of patients with MRSA infection | 1,064 (33) | 34 (24) | 0.7 (0.5-1.1) | 0.11 |
| No. (%) of patients from whom MRSA was recovered from the following anatomic site: | ||||
| Blood | 186 (6) | 2 (1) | 0.24 (0.06-0.96) | 0.03 |
| Sputum | 648 (20) | 18 (13) | 0.58 (0.35-0.96) | 0.03 |
| Urine | 281 (9) | 12 (9) | 0.98 (0.53-1.78) | 0.94 |
| Surgical site | 391 (12) | 14 (10) | 0.80 (0.46-1.40) | 0.44 |
| Skin or soft tissue | 921 (29) | 39 (28) | 0.96 (0.66-1.40) | 0.92 |
| Nose | 1,333 (42) | 60 (43) | 1.06 (0.75-1.49) | 0.75 |
| Perineum or groin | 433 (14) | 24 (17) | 1.33 (0.85-2.09) | 0.27 |
| Other site | 772 (24) | 25 (18) | 0.69 (0.44-1.07) | 0.09 |
Mupirocin susceptible, MIC ≤ 4 μg/ml; mupirocin resistant, MIC ≥ 512 μg/ml.
East, provinces of Nova Scotia, New Brunswick, and Newfoundland.
Central, provinces of Quebec and Ontario.
West, provinces of Manitoba, Saskatchewan, Alberta, and British Columbia.
TABLE 2.
Multivariate analysis of variables associated with high-level mupirocin resistance in MRSA strains
| Variable | OR (95% CI) | P value |
|---|---|---|
| Aboriginal ethnicity | 3.71 (1.51-9.36) | 0.006 |
| Community-associated MRSA | 2.24 (1.02-4.96) | 0.05 |
| MRSA colonization, without infection | 1.74 (1.02-2.99) | 0.04 |
The antimicrobial susceptibility test results for mupirocin-susceptible and mupirocin-resistant MRSA isolates are summarized in Table 3. Resistance to vancomycin or linezolid was not identified. Compared to mupirocin-susceptible strains of MRSA, strains with high-level mupirocin resistance were more likely to be susceptible to tetracycline (7% versus 23%; P < 0.001), trimethoprim-sulfamethoxazole (10% versus 40%; P < 0.001), and ciprofloxacin (75% versus 90%; P < 0.001). Mupirocin-resistant strains were more likely to be resistant to fusidic acid (21% versus 4%; P < 0.001).
TABLE 3.
Antimicrobial susceptibilities of mupirocin-susceptible and mupirocin-resistant MRSA strains recovered from hospitalized patients, 1995 to 2004
| Antimicrobial agent | Mupirocin-susceptible MRSA (n = 4,386)a
|
Mupirocin-resistant MRSA (n = 198)b
|
P value | |||
|---|---|---|---|---|---|---|
| MIC90 (μg/ml) | % Resistant | MIC90 (μg/ml) | % Resistant | |||
| Mupirocin | 0.5 | 0 | >512 | 100 | ||
| Erythromycin | >8.0 | 94 | >8.0 | 86 | ||
| Clindamycin | >8.0 | 86 | >8.0 | 85 | ||
| Linezolid | 1.0 | 0 | 2.0 | 0 | ||
| Tetracycline | >16.0 | 23 | <4.0 | 7 | <0.001 | |
| Trimethoprim-sulfamethoxazole | >8.0 | 40 | 2.0 | 10 | <0.001 | |
| Ciprofloxacin | >8.0 | 90 | >8.0 | 75 | <0.001 | |
| Rifampin | <0.5 | 2 | <0.5 | 4 | ||
| Fusidic acidc | 0.5 | 4 | >8.0 | 21 | <0.001 | |
| Vancomycin | 1.0 | 0 | 1.0 | 0 | ||
Mupirocin susceptible, MIC ≤ 4.0 μg/ml.
Mupirocin resistant, MIC ≥ 512 μg/ml.
Fusidic acid provisional susceptibility breakpoint, MIC ≤ 0.5 μg/ml.
Most (73%) strains with high-level resistance to mupirocin possessed SCCmec type II; and the predominant DNA profile, as determined by PFGE, was CMRSA-2, accounting for 30.3% of the isolates (Table 4). This PFGE profile is identical to or closely related to U.S. PFGE type USA100/800, sequence type 5 (ST5) (4, 33), and was also the most common among the mupirocin-susceptible strains of MRSA. A strain designated CMRSA-9 (SCCmec type II; ST8) was also relatively common, accounting for 20.7% of the strains with high-level resistance, although most of these were recovered from patients at two hospitals located in the same city. Compared to the mupirocin-susceptible strains, mupirocin-resistant MRSA strains were more likely to be CMRSA-9 (20.7% versus 0.5%; P < 0.001) and less likely to be CMRSA-1 (USA600; ST45) (10.1% versus 31.2%; P < 0.001). Clustering of strains, as determined by PFGE, occurred commonly within hospitals (data not shown).
TABLE 4.
Distribution of PFGE DNA profiles of MRSA isolates recovered in Canadian hospitals, 1995 to 2004
| PFGE profile (U.S. PFGE type; MLST)a | % of strains
|
|
|---|---|---|
| Mupirocin susceptible | High-level mupirocin resistant | |
| CMRSA-1 (USA600; ST45) | 31.2 | 10.1b |
| CMRSA-2 (USA100/800; ST5) | 38.4 | 30.3 |
| CMRSA-5 (USA500; ST8) | 5.4 | 7.4 |
| CMRSA-7 (USA400; ST1) | 0.9 | 7.6b |
| CMRSA-9 (ST8) | 0.5 | 20.7b |
| CMRSA-10 (USA300; ST8) | 1.5 | 0.5 |
| Others | 22.1 | 23.4 |
In total, only 7% of all isolates were thought to have been community acquired on the basis of epidemiologic criteria (9), and 14% of these isolates were found to have high-level mupirocin resistance. However, only 135 (2.7%) isolates had PFGE profiles of CMRSA-10 (USA300; ST8) or CMRSA-7 (USA400; ST1), the most commonly identified community-associated clones in North America. Only 1 of the 74 CMRSA-10 strains had high-level mupirocin resistance. However, 15 (25%) of the CMRSA-7 strains were mupirocin resistant, and mupirocin-resistant MRSA strains were more likely than mupirocin-susceptible strains to be CMRSA-7 (8% versus 1%; P < 0.001).
Of the 198 strains with high-level resistance to mupirocin, 144 (73%) were SCCmec type II, 44 (22%) were SCCmec type IV, six (3%) were SCCmec type III, and four were SCCmec type I. The predominant PFGE DNA profiles and corresponding SCCmec types are summarized in Table 5.
TABLE 5.
PFGE DNA patterns, SCCmec types, and mupA restriction fragment length polymorphism (HindIII restriction profiles) of predominant Canadian MRSA strains with high-level mupirocin resistance
| PFGE profile (U.S. PFGE type; MLST type; no. of isolates)a | SCCmec type (no. of isolates) | Plasmid profile (no. of isolates) |
|---|---|---|
| CMRSA-2 (USA100/800, ST5; 66) | II (53) | B (46) |
| IV (13) | H (2) | |
| Other (18) | ||
| CMRSA-9 (ST8; 54) | II (54) | H (44) |
| D (7) | ||
| Other (3) | ||
| CMRSA-1 (USA600, ST45; 20) | II (17) | A (17) |
| IV (3) | Other (3) | |
| CMRSA-7 (USA400, ST1; 15) | IV (15) | G (12) |
| Other (3) |
A total of 46 different plasmid profiles were identified in strains with high-level mupirocin resistance, as determined by HindIII restriction. Five plasmid types (designated plasmid profiles A, B, D, G, and H) accounted for 71% of all the isolates (Table 5; Fig. 2). These plasmid profiles had a wide distribution in hospitals across the country, although plasmid profile A was identified only in hospitals in Ontario and Quebec, whereas profile G, associated with CMRSA-7, was seen only in hospitals in western Canada.
FIG. 2.
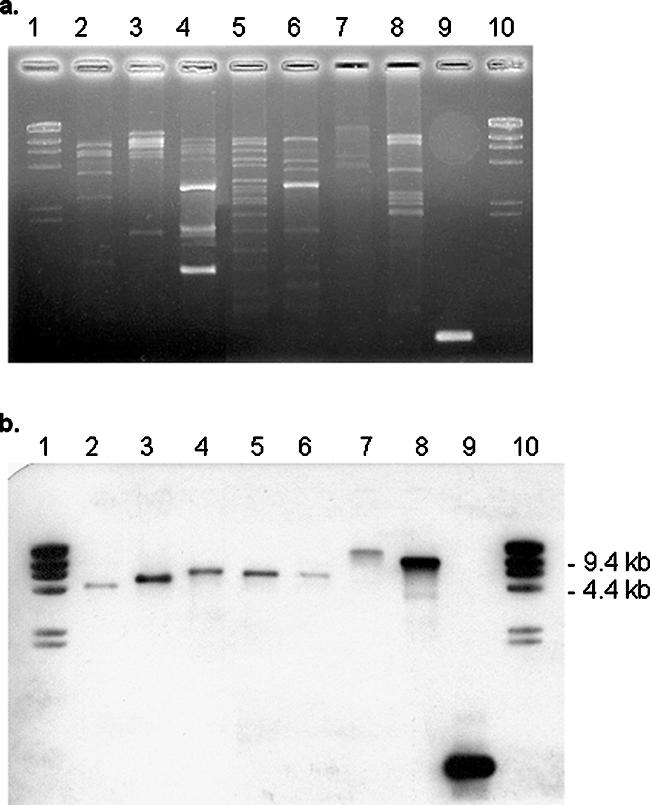
(a) HindIII-restricted plasmid profiles of representative MRSA isolates with (b) corresponding mupA hybridization.
The mupA gene was detected by PCR in total DNA extracted from the cells and from plasmid DNA in all of the 198 MRSA strains with high-level mupirocin resistance. The mupA gene probe most often hybridized with HindIII fragments of just under 9 kb in size (Fig. 2). However, all CMRSA-9 isolates had mupA HindIII-digested fragments approximately 12 kb in size, and most (11 of 15) CMRSA-7 (USA400) strains had fragments approximately 15 kb in size. The mupA gene was not detected in any of the 104 MRSA strains with low-level resistance that were assayed or in the 117 strains susceptible to mupirocin.
DISCUSSION
In the past few years, mupirocin resistance has been increasing among staphylococci in many parts of the world (6, 8, 27, 37, 43). The risk of the emergence of such resistance appears to be greater among methicillin-resistant strains of S. aureus than among methicillin-susceptible strains (3, 31) and is often associated with the widespread use of mupirocin (8, 22, 38). In this study, an increase in both high-level and low-level mupirocin resistance was identified over 10 years in MRSA isolates recovered from patients in Canadian hospitals. The isolates with high-level mupirocin resistance were characterized by PFGE and determination of mupA gene-associated plasmid profiles. We found that MRSA isolates with high-level mupirocin resistance were nearly four times more likely to be recovered from those with an aboriginal ethnicity and from patients who were colonized with MRSA without evidence of infection. Mupirocin resistance in MRSA was also more likely to be identified in strains thought to have been community acquired, based on epidemiologic criteria. It is important to note that this study included isolates obtained prior to 2005. The emergence and spread of community-associated clones (USA300 or USA400) in Canada has occurred only since 2004, and these strains are still not as prevalent in Canada as they are in many U.S. centers (4, 12, 23). Therefore, only a relatively small number of these community-associated strains were available for inclusion in this study. Although CMRSA-10 (USA300) strains were rarely mupirocin resistant, one-quarter of the CMRSA-7 (USA400) strains had high-level mupirocin resistance.
MRSA strains with mupirocin resistance were often found to be more susceptible to other antimicrobial agents, such as tetracycline and trimethoprim-sulfamethoxazole. This observation is also consistent with the association of mupirocin resistance in MRSA with community acquisition. In contrast, mupirocin-resistant isolates were more likely to be resistant to fusidic acid. It is tempting to speculate that the fusB determinant, which is responsible for fusidic acid resistance (25), is on the same plasmid as the mupA gene in isolates with high-level mupirocin resistance, but our study was not able to address this issue.
As in previous investigations done in the United States, we also found that high-level mupirocin resistance occurred in a variety of MRSA strains (as determined by PFGE) from different geographic regions of the country (30) and that transmission of the same strain was more likely to occur within a health care facility (3, 20, 29, 43). However, even within an institution, high-level mupirocin resistance often appeared to arise from multiple clones. We did not identify a chromosomal location of the mupA gene in any of our isolates, as has occasionally been described by Udo et al. (35). All Canadian strains of MRSA with high-level mupirocin resistance possessed a plasmid-associated mupA gene, but the plasmids were of various sizes and had various HindIII restriction digest profiles. Some plasmid profiles were associated with specific PFGE patterns and were more commonly found in certain geographic regions of the country. These findings suggest that both plasmid transmission and the clonal spread of mupirocin-resistant MRSA strains have occurred in Canadian hospitals, as has been reported previously in other countries (3, 20, 43). In this study, the mupA gene usually hybridized to a HindIII fragment of approximately 9 kb in size or less. In reports from Spain, Poland, South Korea, and the United States, the most common previously reported plasmid-derived HindIII fragments that contained mupA ranged in size from 4.5 kb to 10 kb (3, 20, 30, 43).
This surveillance for mupirocin resistance in MRSA included a large sample of both clinical and surveillance isolates recovered from patients in 32 hospitals across Canada over 10 years. However, the surveillance represented a convenience sample of hospital sites, and only the initial MRSA isolates recovered from hospitalized patients were included in the study. The results may not be representative of those for MRSA strains from outpatients or residents of long-term care facilities. A major limitation of the analysis of the variables associated with mupirocin resistance was the lack of information regarding the utilization of mupirocin or other antimicrobial agents. Complete clinical and epidemiologic data regarding the variables associated with mupirocin resistance were available for only 70% of the cases, although there is no reason to believe that the characteristics of patients with missing data were any different from those of the patients whose strains were included in the analysis. Although not all the MRSA isolates were available, a large number were characterized in this study and are likely to be representative of the MRSA strains from the participating hospitals in Canada.
In summary, the results of this study indicate that the rate of mupirocin resistance has been increasing among Canadian strains of MRSA. Continued surveillance for mupirocin resistance is important in order to retain the usefulness of this agent for the treatment and prevention of staphylococcal infections.
Acknowledgments
This study was funded, in part, by a CIHR New Emerging Team Grant.
We thank K. M. Chung and S. Boroumandi for technical assistance and the infection control practitioners and laboratory personnel of all the hospitals in the Canadian Nosocomial Infection Surveillance Program for their invaluable assistance with data collection.
The members of the Canadian Nosocomial Infection Surveillance Program are E. Bryce, Vancouver General Hospital, Vancouver, British Columbia; J. Conly, University of Calgary, Calgary, Alberta; J. Embil, Health Sciences Centre, Winnipeg, Manitoba; Joanne Embree, Health Sciences Centre, Winnipeg, Manitoba; S. Forgie, Stollery Children's Hospital, Edmonton, Alberta; C. Frenette, Hôpital Charles LeMoyne, Greenfield Park, Quebec; M. Gardam, University Health Network, Toronto, Ontario; D. Gravel, Centre for Infectious Disease Prevention and Control, Public Health Agency of Canada, Ottawa, Ontario; E. Henderson, Peter Lougheed Centre, Calgary, Alberta; J. Hutchinson, Health Sciences Centre, St. John's, Newfoundland; M. John, London Health Sciences Centre, London, Ontario; L. Johnston, Queen Elizabeth II Health Sciences Centre, Halifax, Nova Scotia; P. Kibsey, Victoria General Hospital, Victoria, British Columbia; M. Kuhn, The Moncton Hospital, Moncton, New Brunswick; J. Langley, I. W. K. Health Science Centre, Halifax, Nova Scotia; M. Loeb, Hamilton Health Sciences Corporation, Hamilton, Ontario; A. Matlow, Hospital for Sick Children, Toronto, Ontario; A. McGeer, Mount Sinai Hospital, Toronto, Ontario; S. Michaud, CHUS-Hôpital Fleurimont, Sherbrooke, Quebec; M. Miller, SMBD-Jewish General Hospital, Montreal, Quebec; D. Moore, Montreal Children's Hospital, Montreal, Quebec; M. Mulvey, Canadian Science Centre for Human and Animal Health, National Microbiology Laboratory, Winnipeg, Manitoba; M. Ofner-Agostini, Centre for Infectious Disease Prevention and Control, Public Health Agency of Canada, Ottawa, Ontario; S. Paton, Centre for Infectious Disease Prevention and Control, Public Health Agency of Canada, Ottawa, Ontario; V. Roth, The Ottawa Hospital, Ottawa, Ontario; A. Simor, Sunnybrook Health Sciences Centre, Toronto, Ontario; J. Stegenga, Centre for Infectious Disease Prevention and Control, Public Health Agency of Canada, Ottawa, Ontario; K. Suh, Children's Hospital of Eastern Ontario, Ottawa, Ontario; G. Taylor, University of Alberta Hospital, Edmonton, Alberta; E. Thomas, Children's and Women's Health Centre, Vancouver, British Columbia; C. Tremblay, Hôtel-Dieu de Quebec du CHUQ, Sherbrooke, Quebec; M. Vearncombe, Sunnybrook Health Sciences Centre, Toronto, Ontario; K. Weiss, Hôpital Maisonneuve-Rosemont, Montreal, Quebec; A. Wong, Royal University Hospital, Saskatoon, Saskatchewan; and D. Zoutman, Kingston General Hospital, Kingston, Ontario.
Footnotes
Published ahead of print on 27 August 2007.
REFERENCES
- 1.Anthony, R. M., A. M. Connor, E. G. M. Power, and G. L. French. 1999. Use of the polymerase chain reaction for rapid detection of high-level mupirocin resistance in staphylococci. Eur. J. Clin. Microbiol. Infect. Dis. 18:30-34. [DOI] [PubMed] [Google Scholar]
- 2.Cederna, J. E., M. S. Terpenning, M. Ensberg, S. F. Bradley, and C. A. Kauffman. 1990. Staphylococcus aureus nasal colonization in a nursing home: eradication with mupirocin. Infect. Control Hosp. Epidemiol. 11:13-16. [DOI] [PubMed] [Google Scholar]
- 3.Chaves, F., J. García-Martínez, S. de Miguel, and J. R. Otero. 2004. Molecular characterization of resistance to mupirocin in methicillin-susceptible and -resistant isolates of Staphylococcus aureus from nasal samples. J. Clin. Microbiol. 42:822-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christianson, S., G. R. Golding, J. Campbell, the Canadian Nosocomial Infection Surveillance Program, and M. R. Mulvey. 2007. Comparative genomics of Canadian epidemic lineages of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 45:1904-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2007. Performance standards for antimicrobial susceptibility testing; 17th informational supplement. CLSI document M100-S17. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.Cookson, B. D. 1998. The emergence of mupirocin resistance: a challenge to infection control and antibiotic prescribing practice. J. Antimicrob. Chemother. 41:11-18. [DOI] [PubMed] [Google Scholar]
- 7.Doebbeling, B. N., D. R. Reagan, M. A. Pfaller, A. K. Houston, R. J. Hollis, and R. P. Wenzel. 1994. Long-term efficacy of intranasal mupirocin ointment. A prospective cohort study of Staphylococcus aureus carriage. Arch. Intern. Med. 154:1505-1508. [PubMed] [Google Scholar]
- 8.dos Santos, K. R. N., L. de Souza Fonseca, and P. P. G. Filho. 1996. Emergence of high-level mupirocin resistance in methicillin-resistant Staphylococcus aureus isolated from Brazilian university hospitals. Infect. Control Hosp. Epidemiol. 17:813-816. [PubMed] [Google Scholar]
- 9.Fridkin, S. K., J. C. Hageman, M. Morrison, L. T. Sanza, K. Como-Sabetti, J. A. Jernigan, K. Harriman, L. H. Harrison, R. Lynfield, and M. M. Farley. 2005. Methicillin-resistant Staphylococcus aureus disease in three communities. N. Engl. J. Med. 352:1436-1444. [DOI] [PubMed] [Google Scholar]
- 10.Fujimura, S., Y. Tokae, and A. Watanabe. 2003. Isoleucyl-tRNA synthetase mutations in Staphylococcus aureus clinical isolates and in vitro selection of low-level mupirocin-resistant strains. Antimicrob. Agents Chemother. 47:3373-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garner, J. S., W. R. Jarvis, T. G. Emori, T. C. Horan, and J. M. Hughes. 1988. CDC definitions of nosocomial infections, 1988. Am. J. Infect. Control 16:128-140. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert, M., J. MacDonald, D. Gregson, J. Siushansian, K. Zhang, S. Elsayed, K. Laupland, T. Louie, K. Hope, M. Mulvey, J. Gillespie, D. Nielsen, V. Wheeler, M. Louie, A. Honish, G. Keays, and J. Conly. 2006. Outbreak in Alberta of community-acquired (USA300) methicillin-resistant Staphylococcus aureus in people with a history of drug use, homelessness, or incarceration. Can. Med. Assoc. J. 175:149-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han, L. L., L. K. McDougal, R. J. Gorwitz, K. H. Mayer, J. B. Patel, J. M. Sennott, and J. L. Fontana. 2007. High frequencies of clindamycin and tetracycline resistance in methicillin-resistant Staphylococcus aureus pulsed-field type USA300 isolates collected at a Boston ambulatory health center. J. Clin. Microbiol. 45:1350-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hill, R. L. R., G. J. Duckworth, and M. W. Casewell. 1988. Elimination of nasal carriage of methicillin-resistant Staphylococcus aureus with mupirocin during a hospital outbreak. J. Antimicrob. Chemother. 22:377-384. [DOI] [PubMed] [Google Scholar]
- 15.Hodgson, J. E., S. P. Curnock, K. G. H. Dyke, R. Morris, D. R. Sylvester, and M. S. Gross. 1994. Molecular characterization of the gene encoding high-level mupirocin resistance in Staphylococcus aureus J2870. Antimicrob. Agents Chemother. 38:1205-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janssen, D. A., L. T. Zarins, D. R. Schaberg, S. F. Bradley, M. S. Terpenning, and C. A. Kauffman. 1993. Detection and characterization of mupirocin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 37:2003-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalmeijer, M. D., H. Coertjens, P. M. van Nieuwland-Bollen, D. Bogaers-Hofman, G. A. J. de Baere, A. Stuurman, A. van Belkum, and J. A. J. W. Kluytmans. 2002. Surgical site infections in orthopedic surgery: the effect of mupirocin nasal ointment in a double-blind, randomized, placebo-controlled study. Clin. Infect. Dis. 35:353-358. [DOI] [PubMed] [Google Scholar]
- 18.Kauffman, C. A., M. S. Terpenning, X. He, L. T. Zarins, M. A. Ramsey, K. A. Jorgensen, W. S. Sottile, and S. F. Bradley. 1993. Attempts to eradicate methicillin-resistant Staphylococcus aureus from a long-term-care facility with the use of mupirocin ointment. Am. J. Med. 94:371-378. [DOI] [PubMed] [Google Scholar]
- 19.Kluytmans, J. A. J. W., J. W. Mouton, M. F. Q. VandenBergh, M. A. A. J. Manders, A. P. W. M. Maat, J. H. T. Wagenvoort, M. F. Michel, and H. A. Verbrugh. 1996. Reduction of surgical-site infections in cardiothoracic surgery by elimination of nasal carriage of Staphylococcus aureus. Infect. Control Hosp. Epidemiol. 17:780-785. [DOI] [PubMed] [Google Scholar]
- 20.Leski, T. A., M. Gniadkowski, A. Skoczyńska, E. Stefaniuk, K. Trzciński, and W. Hryniewicz. 1999. Outbreak of mupirocin-resistant staphylococci in a hospital in Warsaw, Poland, due to plasmid transmission and clonal spread of several strains. J. Clin. Microbiol. 37:2781-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Louie, L., J. Goodfellow, P. Mathieu, A. Glatt, M. Louie, and A. E. Simor. 2002. Rapid detection of methicillin-resistant staphylococci from blood culture bottles by using a multiplex PCR assay. J. Clin. Microbiol. 40:2786-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, M. A., A. Dascal, J. Portnoy, and J. Mendelson. 1996. Development of mupirocin resistance among methicillin-resistant Staphylococcus aureus after widespread use of nasal mupirocin ointment. Infect. Control Hosp. Epidemiol. 17:811-813. [DOI] [PubMed] [Google Scholar]
- 23.Mulvey, M. R., L. MacDougall, B. Cholin, G. Horsman, M. Fidyk, S. Woods, and the Saskatchewan CA-MRSA Study Group. 2005. Community-associated methicillin-resistant Staphylococcus aureus, Canada. Emerg. Infect. Dis. 11:844-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Neill, A. J., F. McLaws, G. Kahlmeter, A. S. Henricksen, and I. Chopra. 2007. Genetic basis of resistance to fusidic acid in staphylococci. Antimicrob. Agents Chemother. 51:1737-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pérez-Fontán, M., M. Rosales, A. Rodríguez-Carmona, T. G. Falcón, and F. Valdés. 2002. Mupirocin resistance after long-term use for Staphylococcus aureus colonization in patients undergoing chronic peritoneal dialysis. Am. J. Kidney Dis. 39:337-341. [DOI] [PubMed] [Google Scholar]
- 27.Pérez-Roth, E., F. Claverie-Martin, N. Batista, A. Moreno, and S. Méndez-Álvarez. 2002. Mupirocin resistance in methicillin-resistant Staphylococcus aureus clinical isolates in a Spanish hospital. Co-application of multiplex PCR assay and conventional microbiology methods. Diagn. Microbiol. Infect. Dis. 43:123-128. [DOI] [PubMed] [Google Scholar]
- 28.Perl, T. M., J. J. Cullen, R. P. Wenzel, M. B. Zimmerman, M. A. Pfaller, D. Sheppard, J. Twombley, P. P. French, and L. A. Herwaldt. 2002. Intranasal mupirocin to prevent postoperative Staphylococcus aureus infections. N. Engl. J. Med. 346:1871-1877. [DOI] [PubMed] [Google Scholar]
- 29.Rahman, M., S. Connolly, W. C. Noble, B. Cookson, and I. Phillips. 1990. Diversity of staphylococci exhibiting high-level resistance to mupirocin. J. Med. Microbiol. 33:97-100. [DOI] [PubMed] [Google Scholar]
- 30.Ramsey, M. A., S. F. Bradley, C. A. Kauffman, T. M. Morton, J. E. Patterson, and D. R. Reagan. 1998. Characterization of mupirocin-resistant Staphylococcus aureus from different geographic areas. Antimicrob. Agents Chemother. 42:1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rotger, M., A. Trampuz, K. E. Piper, J. M. Steckelberg, and R. Patel. 2005. Phenotypic and genotypic mupirocin resistance among staphylococci causing prosthetic joint infection. J. Clin. Microbiol. 43:4266-4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simor, A. E., M. Ofner-Agostini, E. Bryce, K. Green, A. McGeer, M. Mulvey, and S. Paton. 2001. The evolution of methicillin-resistant Staphylococcus aureus in Canadian hospitals: 5 years of national surveillance. Can. Med. Assoc. J. 165:21-26. [PMC free article] [PubMed] [Google Scholar]
- 33.Simor, A. E., M. Ofner-Agostini, E. Bryce, A. McGeer, S. Paton, and M. R. Mulvey. 2002. Laboratory characterization of methicillin-resistant Staphylococcus aureus in Canadian hospitals: results of 5 years of national surveillance, 1995-1999. J. Infect. Dis. 186:652-660. [DOI] [PubMed] [Google Scholar]
- 34.Simor, A. E., E. Phillips, A. McGeer, A. Konvalinka, M. Loeb, H. R. Devlin, and A. Kiss. 2007. Randomized controlled trial of chlorhexidine gluconate for washing, intranasal mupirocin, and rifampin and doxycycline versus no treatment for the eradication of methicillin-resistant Staphylococcus aureus colonization. Clin. Infect. Dis. 44:178-185. [DOI] [PubMed] [Google Scholar]
- 35.Udo, E. E., N. Al-Sweih, and B. C. Noronha. 2003. A chromosomal location of the mupA gene in Staphylococcus aureus expressing high-level mupirocin resistance. J. Antimicrob. Chemother. 51:1283-1286. [DOI] [PubMed] [Google Scholar]
- 36.Udo, E. E., L. E. Jacob, and B. Mathew. 2001. Genetic analysis of methicillin-resistant Staphylococcus aureus expressing high- and low-level mupirocin resistance. J. Med. Microbiol. 50:909-915. [DOI] [PubMed] [Google Scholar]
- 37.Upton, A., S. Lang, and H. Heffernan. 2003. Mupirocin and Staphylococcus aureus: a recent paradigm of emerging antibiotic resistance. J. Antimicrob. Chemother. 51:613-617. [DOI] [PubMed] [Google Scholar]
- 38.Vasquez, J. E., E. S. Walker, B. W. Franzus, B. K. Overbay, D. R. Reagan, and F. A. Sarubbi. 2000. The epidemiology of mupirocin resistance among methicillin-resistant Staphylococcus aureus at a Veterans' Affairs hospital. Infect. Control Hosp. Epidemiol. 21:459-464. [DOI] [PubMed] [Google Scholar]
- 39.Walker, E. S., J. E. Vasquez, R. Dula, H. Bullock, and F. A. Sarubbi. 2003. Mupirocin-resistant, methicillin-resistant Staphylococcus aureus: does mupirocin remain effective? Infect. Control Hosp. Epidemiol. 24:342-346. [DOI] [PubMed] [Google Scholar]
- 40.Wertheim, H. F. L., M. C. Vos, A. Ott, A. Voss, J. A. J. W. Kluytmans, C. M. J. E. Vandenbroucke-Grauls, M. H. M. Meester, P. H. J. van Keulen, and H. A. Verbrugh. 2004. Mupirocin prophylaxis against nosocomial Staphylococcus aureus infections in nonsurgical patients. A randomized study. Ann. Intern. Med. 40:419-425. [DOI] [PubMed] [Google Scholar]
- 41.Wilcox, M. H., J. Hall, H. Pike, P. A. Templeton, W. N. Fawley, P. Parnell, and P. Verity. 2003. Use of perioperative mupirocin to prevent methicillin-resistant Staphylococcus aureus (MRSA) orthopaedic surgical site infections. J. Hosp. Infect. 54:196-201. [DOI] [PubMed] [Google Scholar]
- 42.Yanagisawa, T., J. T. Lee, H. C. Wu, and M. Kawakami. 1994. Relationship of protein structure of isoleucyl-tRNA synthetase with pseudomonic acid resistance of Escherichia coli. A proposed mode of action of pseudomonic acid as an inhibitor of isoleucyl-tRNA synthetase. J. Biol. Chem. 269:24304-24309. [PubMed] [Google Scholar]
- 43.Yoo, J. I., E. S. Shin, J. O. Cha, J. K. Lee, Y. H. Jung, K. M. Lee, B. S. Kim, and Y. S. Lee. 2006. Clonal dissemination and mupA gene polymorphism of mupirocin-resistant Staphylococcus aureus isolates from long-term-care facilities in South Korea. Antimicrob. Agents Chemother. 50:365-367. [DOI] [PMC free article] [PubMed] [Google Scholar]