Abstract
The agouti signaling protein (ASIP) and its homolog, the agouti-related protein (AgRP), act as inverse agonists that control, respectively, pigmentation and metabolic function in mammals. NMR investigations find that the C-terminal domains of these proteins adopt a fold consistent with an Inhibitor Cystine Knot (ICK), previously identified in invertebrate toxins. Although these structural studies suggest that ASIP and AgRP define a new mammalian protein fold class, the results with ASIP are inconclusive. Here we apply direct chemical mapping to determine the complete set of disulfide linkages in ASIP. The results demonstrate unequivocally that ASIP adopts the ICK fold and thereby supports a recent evolution structure function analysis, which proposes that ASIP and AgRP arose from a common antagonist ligand.
Keywords: Agouti-Related Protein, Melanocortin Receptor, Pigmentation, tris(2-carboxyethyl)phosphine
1. Introduction
Binding of the agouti signaling protein (ASIP) to the melanocortin-1 receptor (MC1R) in the skin results in production of the yellow/red pigment pheomelanin [1,2]. Mice with the agouti lethal yellow mutation (Ay/a) express ASIP ubiquitously, and the interaction of ASIP with the MC4 receptor in the hypothalamus causes obesity and metabolic characteristics associated with type II diabetes, including insulin and leptin resistance [3].
There are several unique and important aspects of ASIP’s structure and biochemical behavior. First, melanocortin receptors are members of the G-protein coupled receptor (GPCR) family and ASIP, along with its homolog the agouti-related protein (AgRP), which is normally expressed in the brain, are the only known endogenous GPCR antagonists (or inverse agonists); both ASIP and AgRP act to block the action of the agonist α-MSH (reviewed in ref. [4]). Second, whereas many GPCR ligands are small, easily diffusible peptides or low molecular weight species, ASIP is a 110 residue (after removal of the signal peptide) glycoprotein with a folded, cysteine-rich C-terminal domain [5]. Third, because of the hyperphagia and metabolic characteristics associated with Ay/a mice, these animals, and those with similar mutations, are widely used as obesity models [1].
ASIP’s forty residue, Cys-rich, C-terminal domain alone contains the determinants for high affinity binding to MC1/4R [6,7]. Structural studies of this domain identify yet another potentially unique ASIP feature – the ten cysteines are thought to form a network of five disulfide bonds arranged in an inhibitor cystine knot (ICK), or knottin, fold motif previously found in invertebrate toxins [6,8,9]. Characterization of this fold in the homolog AgRP is now well established by both chemical mapping [10] and NMR structural studies [11].
The specific fold of ASIP, however, is less clear. NMR studies on ASIP(80-132) by our lab identified a low energy backbone conformation quite similar to that of AgRP and consistent with the ICK motif [6]. But studies on structurally related toxins demonstrate that backbone fold alone is insufficient to define ICK disulfide bond pairings (e.g., ref. [12]). NMR contacts in ASIP unambiguously identified disulfide bonds Cys93-Cys108, Cys116-Cys123 and Cys111-Cys132 by NOEs between specific side-chain β protons [6]. However, Cys side-chains 100, 107, 114 and 125 are all in near proximity thus confounding direct disulfide assignment by visual inspection or by side chain NOEs. The situation is further complicated by cis/trans isomerization of the Ala104-Pro105 imide backbone bond, which gives two distinct orientations of the ASIP(80-132) N-terminal loop, and structural heterogeneity for residues 100–106. Oftentimes energy calculations provide a reliable means for identifying the correct disulfide network. Specifically, DYANA calculations performed on all possible disulfide arrangements give the lowest target function only for the backbone conformation with the correct linkages [13]. This approach was attempted with ASIP and, unfortunately, failed to clearly identify a unique low energy fold, applicable to both cis and trans conformations [6].
A recent study using Evolution Structure Function analysis suggests that AgRP and ASIP arose from a common antagonist ligand that originally served to control both pigmentation and energy balance as a means for adapting to starvation [14]. In light of this proposal, and the fundamental structural issues discussed above, elucidating the exact ASIP fold is vital for establishing whether indeed AgRP and ASIP comprise a unique motif newly identified in mammals. To address these issues, we apply direct chemical mapping to determine the exact disulfide arrangement in ASIP. The method employs alkylation of nascent sulfhydryl groups in partially reduced forms of ASIP, followed by trypsin digestion and tandem MS sequencing. These data, in conjunction with the recently determined NMR structure, provide an unequivocal classification of the ASIP fold.
2. Materials and Methods
2.1 Solid phase protein synthesis
In previous work, C-terminal ASIP was prepared as two separate strands, which were then linked by native chemical ligation [6]. For these current studies, ASIP(80-132, Q115Y, S124Y), referred to as ASIP-YY, was prepared as a single strand using an Applied Biosystems 433A synthesizer and standard Fmoc chemistry. Fmoc-Cys(Trt)-OPfp was used to avoid enantiomerization of the α-carbon. Purification and oxidative folding followed the exact same procedures described previously [6].
2.2 Partial reduction and alkylation of nascent cysteine residues
Partial reduction was initiated by introducing ASIP-YY (1.0 mM) into a pH = 3 solution containing 40 mM tris(2-carboxyethyl)phosphine (TCEP) and 100 mM sodium citrate [15]. After 20 min the solution was diluted with distilled water and injected into an HPLC fitted with a C18 column (Alltech’s Alltima). Each partially reduced fraction was collected and lyophilized overnight for future usage. In partially reduced fractions, nascent cysteines were alkylated with N-ethylmaleimide (NEM) using a pH = 3 solution of 100 mM NEM and 100mM sodium citrate. The reaction time was typically 20 minutes. Products were repurified by HPLC.
2.3 Trypsin digestion and analysis by mass spectrometry
Samples were first treated dithiothreitol (DTT) to eliminate any remaining disulfide bonds. Specifically, peptides were treated for two hours with 5 mM DTT, 6.0 M guanidine hydrochloride (GuHCl) in 100 mM 2-amino-2-hydroxymethyl-1,3-propanediol (Tris) solution at pH = 8. Next, a 50 mM NH4HCO3 solution was added to dilute the GuHCl concentration to less than 1 M. Trypsin digestion used reagents and procedures from Promega. Trypsin was used in a protease to protein ratio of 1:20 and incubated at 37° C for 4 hours. The mixture was diluted with distilled water and purified by HPLC using a C18 column. Each fraction was collected for mass and sequence analysis. Mass spectrometry was performed using either a Micromass ZMD 4000 or a Thermo Finnigan LTQ. MSn experiments were performed with the LTQ.
3. Results and Discussion
3.1 Disulfide Mapping
Chemical disulfide mapping was performed on the stable double mutant ASIP(80-132, Q115Y, S124Y), referred to as ASIP-YY [6]. Under oxidizing conditions, ASIP-YY folds to a uniform product, as determined by both HPLC and MS, and is fully active at the MC1/3/4 receptors, with the same potency as the wild type protein [6]. TCEP was used to gently and selectively reduce disulfide bonds [10,15]. Reduction at acidic pH avoids disulfide rearrangement. Partially reduced species were then separated by HPLC. Next, treatment of each separated species with the alkylating agent NEM labeled the free sulfhydryl groups, and this was followed by complete reduction using DTT. The selectively labeled peptides were subjected to trypsin proteolysis, which cleaves after basic residues (primarily Arg in ASIP), and the resulting fragments were sequenced by tandem MS to locate each NEM labeled thiol group.
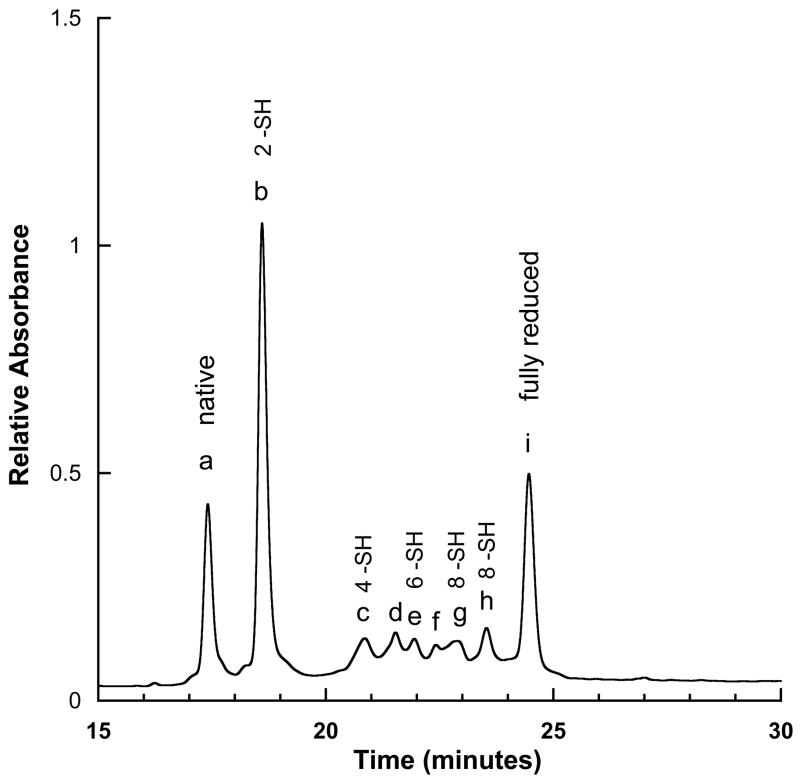
Figure 1 shows an HPLC trace following treatment with TCEP. Peaks b through h correspond to distinct partially reduced species. Starting with peak b, mass analysis of the NEM reacted product shows that this species contained two free sulfhydryl groups. Trypsin treatment, followed by MS analysis of the resulting fragments showed that the peptides ASIP-YY(98-117) and ASIP-YY(127-132) each contained a single NEM modification. The latter of these two peptides contains only a single Cys, thus locating one of the disulfide bond partners to Cys132. By contrast, ASIP-YY(98-117) contains six Cys, thus requiring direct sequencing to locate the modified residue.
Figure 1.
HPLC trace of partially reduced ASIP-YY. The number of free –SH groups in each fraction is labeled and was determined by reaction with NEM, followed by mass analysis.
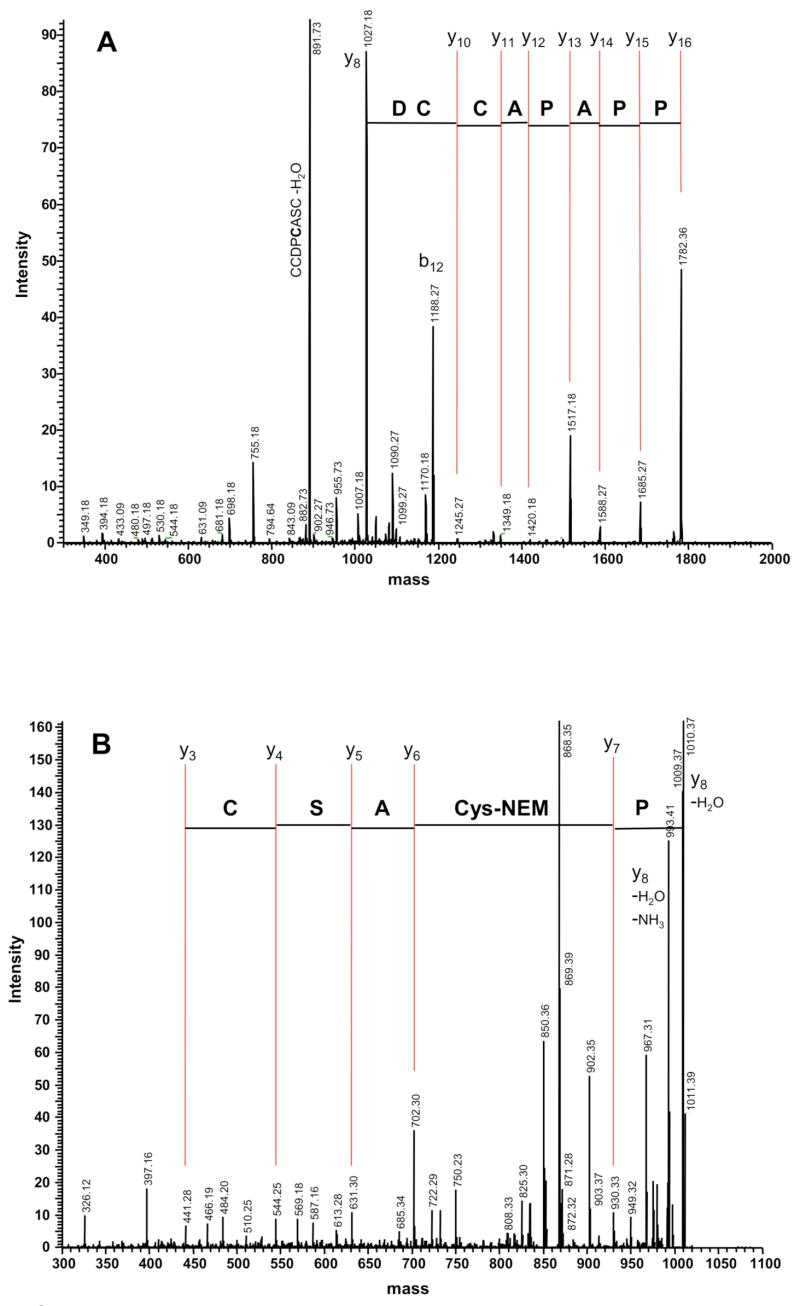
MS/MS of singly NEM modified ASIP-YY(98-117) from peak b, shown in Figure 2A, reveals a progression of signals consistent with a loss of single amino acids cleaved at the peptide bond. Typical of trypsin digests, y ions are enhanced due to the positively charged residue at their C-termini. Sequencing from the N-terminus through residue 109 showed that the modified Cys resided in the segment 110–117. Further fragmentation was not observed so the ASIP-YY(110-117) y8-ion (1027.18 amu) was maintained in the MS trap for further MS/MS/MS analysis, as shown in Figure 2B. Following loss of N-terminal Pro110, sequential fragmentation of the y7-ion gave a change in mass from 930.33 amu to 702.30 amu, corresponding to an NEM modified cysteine, thus locating the other disulfide bond partner to Cys111. Loss of additional amino acids was consistent with y-ion fragmentation of the unmodified peptide ASCYCR. Taken together, these data demonstrate that ASIP-YY contains a disulfide bond between Cys111-Cys132, as found in the NMR structure.
Figure 2.
Example MS sequencing: trypsin fragment ASIP-YY(98-117) from peak b. A) MS/MS gives the expected sequence from Pro102 and demonstrates that Cys107 and Cys108 are not modified by NEM. B) MS/MS/MS of the y8-ion shows that Cys111 is NEM modified and thus forms a disulfide bond with Cys132. Several b-ions and other MS peaks are identified.
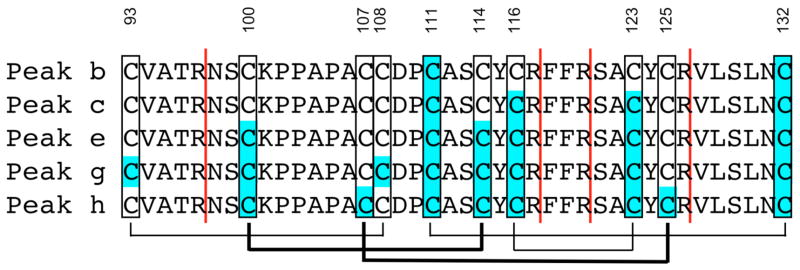
HPLC peaks c, e, g and h, with four, six, eight and eight NEM modifications, respectively, were subjected to the same digestion and MS analysis, thus locating all pairwise NEM labeled Cys residues. Peaks d and f were heterogeneous and therefore not used in our analysis. The results are summarized in Figure 3. Here, Cys residues colored with a blue background identify NEM labeling. The vertical red lines indicate observed trypsin cleavage sites. Peak c revealed four NEM modifications; two confirmed labeling at Cys111 and Cys132 and two additional labeling sites at Cys116 and Cys123. Thus, with comparison to peak b, a new disulfide bond is identified between Cys116-Cys123. Similarly, peak e builds on the previous results and finds a disulfide bond between Cys100-Cys114 and peak g identifies disulfide bond Cys93-Cys108. Peak h is complementary to g and reveals NEM modifications that locate the final disulfide bond between Cys107-Cys125, consistent with the unmodified cysteines in peak g, and vice versa. For all peaks, the results were unambiguous – there was no evidence of alternate NEM labeling or disulfide connectivities inconsistent with those reported in Figure 3.
Figure 3.
Summary of NEM modifications in each of the HPLC peaks. Vertical red lines indicate trypsin cleavage sites and Cys residues with a blue background identify NEM modifications. Peak b contains two modifications thus identifying the Cys111-Cys132 disulfide. Peak c contains two additional modifications demonstrating linkage between Cys116-Cys123, and so forth. The resulting disulfide linkage map is shown below the sequences. The thin lines are consistent with disulfides identified in the NMR structure; the thick lines are newly identified by chemical mapping here.
3.2 Disulfide Bonds and Characterization of the ICK Fold
The resulting network of disulfide bonds is diagrammed underneath the sequences in Figure 3. The connectivities are supported by a previously reported partial chemical analysis by in situ S-pyridylethylation followed by N-terminal sequencing that located N- vs C- disulfide bond partners [7]. The thin lines in Figure 3 represent those disulfide bonds that were unequivocally mapped by NMR. The thick lines are newly identified here and essential for determining whether ASIP belongs in the ICK fold class. Figure 4 shows the NMR structure of ASIP-YY [6]. Disulfide bonds found by NMR and confirmed here are yellow and the new disfulfide bonds found by chemical mapping are green. Examination of the ASIP-YY structure reveals a topological circle formed by peptide segments 93–100 and 108–114, connected by disulfide bonds Cys93-Cys108 and Cys100-Cys114. Disulfide bond Cys107-Cys125 threads from the front of the protein to the back through this topological circle, a defining feature of the ICK fold motif [8,9,16,17].
Figure 4.
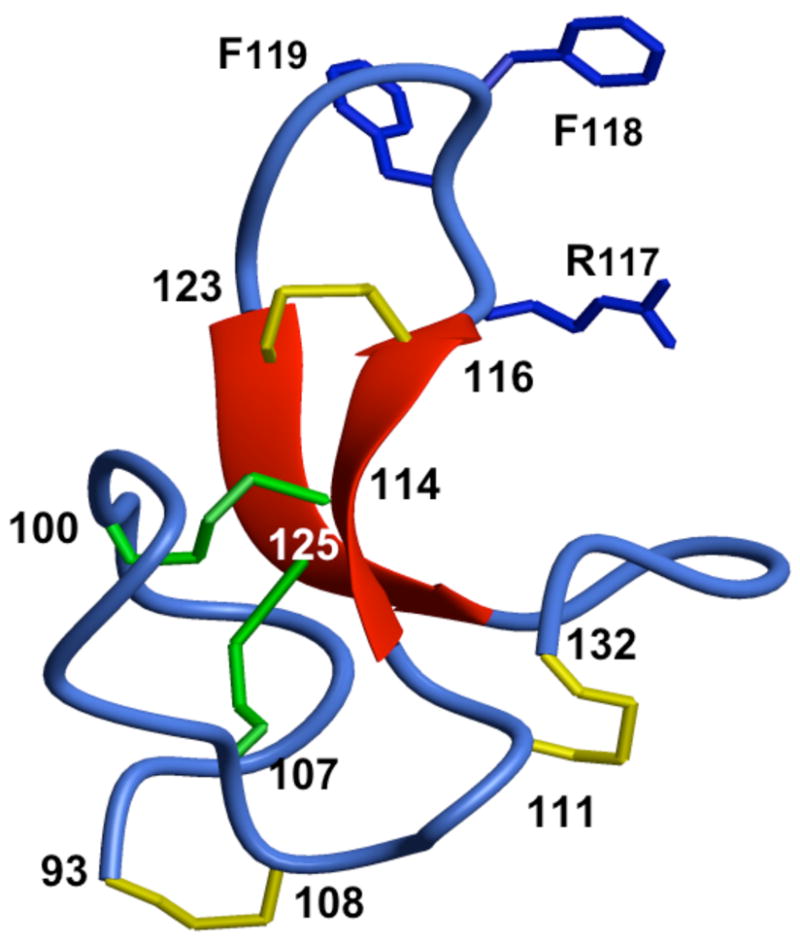
Structure of ASIP-YY showing the complete disulfide map. The yellow colored disulfide bonds are consistent with those identified in the NMR structure; the green bonds are identified here by chemical mapping here. The disulfide bond from Cys107 to Cys125 threads from the front to the back of the protein and demonstrates unequivocally that ASIP is a member of the inhibitor cystine knot family.
Homology models based on known toxin structures previously suggested that agouti and AGRP would adopt a toxin-like ICK fold [18]. Although such predictions seem reasonable by the criteria of homology models, the suggestion of a toxin fold for AGRP or ASIP was nevertheless remarkable since this fold type had never been observed in mammalian proteins. Over the last few years, the assignment of AgRP to the ICK fold has been conclusively demonstrated, both by chemical mapping and NMR. The ASIP sequence contains the appropriate cysteine spacing and the backbone geometry determined by NMR is consistent with an ICK fold. Our chemical mapping here demonstrates conclusively that ASIP indeed possesses the ICK disulfide links and, along with AgRP, defines a fundamentally new fold class for mammalian proteins.
The N-terminal domains of ASIP and AgRP exhibit little sequence homology. Moreover, recent studies clearly demonstrate they possess distinct physiological functions. The N-terminal domain of ASIP binds to the transmembrane protein Attractin, and this interaction is required for ASIP signaling in vivo [19]. In contrast, very recent work shows that the N-terminal domain of AgRP is actually a prodomain, the removal of which is required for high affinity MC4 receptor antagonism [14,20]. There are also fundamental sequence differences between the ASIP and AgRP Cys-rich C-terminal domains. The loops spanning the final two Cys residues (the so-called C-terminal loops) are different in length and only ASIP contains the distinct proline rich segment between the second and third Cys residues. Despite these differences, orthologs of both ASIP and AgRP, now found in a variety of lower vertebrates including fish, exhibit very similar intron-exon structures [6,21]. Phylogenetic analysis argues that ASIP and AgRP share a common ancestor that arose early in vertebrate evolution [14]. Although there are numerous sequence differences between ASIP and AgRP, as described above, our work shows that both proteins have identical C-terminal ICK disulfide crosslinks with superimposable backbone folds thus defining a new and unique mammalian fold motif.
Acknowledgments
This work was supported by grants from the National Institutes of Health (DK064265 to G.L.M and RR020939 in support of the LTQ mass spectrometer). The authors are grateful to Darren Thompson and Daniel Stevens for technical assistance and comments on the manuscript.
Abbreviations
- ASIP
agouti signaling protein
- MCR
melanocortin receptor
- TCEP
tris(2-carboxyethyl)phosphine
- NEM
N-ethylmaleimide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Wilson BD, Ollmann MM, Barsh GS. The role of agouti-related protein in regulating body weight. Molecular Medicine Today. 1999;5:250–256. doi: 10.1016/s1357-4310(99)01471-9. [DOI] [PubMed] [Google Scholar]
- 2.Bultman SJ, Michaud EJ, Woychik RP. Molecular characterization of the mouse agouti locus. Cell. 1992;71:1195–204. doi: 10.1016/s0092-8674(05)80067-4. [DOI] [PubMed] [Google Scholar]
- 3.Yen TT, Gill AM, Frigeri LG, Barsh GS, Wolff GL. Obesity, Diabetes, and Neoplasia in Yellow a(Vy)/- Mice - Ectopic Expression of the Agouti Gene. Faseb Journal. 1994;8:479–488. doi: 10.1096/fasebj.8.8.8181666. [DOI] [PubMed] [Google Scholar]
- 4.Adan RA, Tiesjema B, Hillebrand JJ, la Fleur SE, Kas MJ, de Krom M. The MC4 receptor and control of appetite. Br J Pharmacol. 2006;149:815–27. doi: 10.1038/sj.bjp.0706929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilczynski AM, Joseph CG, Haskell-Luevano C. Current trends in the structure-activity relationship studies of the endogenous agouti-related protein (AGRP) melanocortin receptor antagonist. Med Res Rev. 2005;25:545–56. doi: 10.1002/med.20037. [DOI] [PubMed] [Google Scholar]
- 6.McNulty JC, Jackson PJ, Thompson DA, Chai B, Gantz I, Barsh GS, Dawson PE, Millhauser GL. Structures of the agouti signaling protein. J Mol Biol. 2005;346:1059–70. doi: 10.1016/j.jmb.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 7.Willard DH, et al. Agouti structure and function: characterization of a potent alpha-melanocyte stimulating hormone receptor antagonist. Biochemistry. 1995;34:12341–6. doi: 10.1021/bi00038a030. [DOI] [PubMed] [Google Scholar]
- 8.Gelly JC, Gracy J, Kaas Q, Le-Nguyen D, Heitz A, Chiche L. The KNOTTIN website and database: a new information system dedicated to the knottin scaffold. Nucleic Acids Res. 2004;32(Database issue):D156–9. doi: 10.1093/nar/gkh015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Millhauser GL, McNulty JC, Jackson PJ, Thompson DA, Barsh GS, Gantz I. Loops and links: structural insights into the remarkable function of the agouti-related protein. Ann N Y Acad Sci. 2003;994:27–35. doi: 10.1111/j.1749-6632.2003.tb03159.x. [DOI] [PubMed] [Google Scholar]
- 10.Bures EJ, et al. Determination of disulfide structure in agouti-related protein (AGRP) by stepwise reduction and alkylation. Biochemistry. 1998;37:12172–7. doi: 10.1021/bi981082v. [DOI] [PubMed] [Google Scholar]
- 11.McNulty JC, Thompson DA, Bolin KA, Wilken J, Barsh GS, Millhauser GL. High Resolution NMR Structure of the Chemically-Synthesized Melanocortin Receptor Binding Domain AGRP(87 - 132) of the Agouti-Related Protein. Biochemistry. 2001;40:15520–15527. doi: 10.1021/bi0117192. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Connor M, Smith R, Maciejewski MW, Howden ME, Nicholson GM, Christie MJ, King GF. Discovery and characterization of a family of insecticidal neurotoxins with a rare vicinal disulfide bridge. Nat Struct Biol. 2000;7:505–13. doi: 10.1038/75921. [DOI] [PubMed] [Google Scholar]
- 13.Zahn R, Damberger F, Ortenzi C, Luporini P, Wuthrich K. NMR structure of the Euplotes raikovi pheromone Er-23 and identification of its five disulfide bonds. J Mol Biol. 2001;313:923–31. doi: 10.1006/jmbi.2001.5099. [DOI] [PubMed] [Google Scholar]
- 14.Jackson PJ, Douglas NR, Chai B, Binkley J, Sidow A, Barsh GS, Millhauser GL. Structural and molecular evolutionary analysis of Agouti and Agouti-related proteins. Chem Biol. 2006;13:1297–305. doi: 10.1016/j.chembiol.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray WR. Disulfide structures of highly bridged peptides: a new strategy for analysis. Protein Sci. 1993;2:1732–48. doi: 10.1002/pro.5560021017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craik DJ, Daly NL, Waine C. The cystine knot motif in toxins and implications for drug design. Toxicon. 2001;39:43–60. doi: 10.1016/s0041-0101(00)00160-4. [DOI] [PubMed] [Google Scholar]
- 17.Isaacs NW. Cystine knots. Current Opinion in Structural Biology. 1995;5:391–5. doi: 10.1016/0959-440x(95)80102-2. [DOI] [PubMed] [Google Scholar]
- 18.Tota MR, Smith TS, Mao C, MacNeil T, Mosley RT, Van der Ploeg LH, Fong TM. Molecular interaction of Agouti protein and Agouti-related protein with human melanocortin receptors. Biochemistry. 1999;38:897–904. doi: 10.1021/bi9815602. [DOI] [PubMed] [Google Scholar]
- 19.He L, Gunn TM, Bouley DM, Lu XY, Watson SJ, Schlossman SF, Duke-Cohan JS, Barsh GS. A biochemical function for attractin in agouti-induced pigmentation and obesity. Nat Genet. 2001;27:40–7. doi: 10.1038/83741. [DOI] [PubMed] [Google Scholar]
- 20.Creemers JW, et al. Agouti-related protein is post-translationally cleaved by proprotein convertase 1 to generate AGRP83-132: Interaction between AGRP83-132 and melanocortin receptors cannot be influenced by syndecan 3. Endocrinology. 2006;147:1621–1631. doi: 10.1210/en.2005-1373. [DOI] [PubMed] [Google Scholar]
- 21.Klovins J, Haitina T, Fridmanis D, Kilianova Z, Kapa I, Fredriksson R, Gallo-Payet N, Schioth HB. The melanocortin system in Fugu: determination of POMC/AGRP/MCR gene repertoire and synteny, as well as pharmacology and anatomical distribution of the MCRs. Mol Biol Evol. 2004;21:563–79. doi: 10.1093/molbev/msh050. [DOI] [PubMed] [Google Scholar]