Abstract
The process of DNA strand exchange during general genetic recombination is initiated within protein-stabilized synaptic filaments containing homologous regions of interacting DNA molecules. The RecA protein in bacteria and its analogs in eukaryotic organisms start this process by forming helical filamentous complexes on single-stranded or partially single-stranded DNA molecules. These complexes then progressively bind homologous double-stranded DNA molecules so that homologous regions of single- and double-stranded DNA molecules become aligned in register while presumably winding around common axis. The topological assay presented herein allows us to conclude that in synaptic complexes containing homologous single- and double-stranded DNA molecules, all three DNA strands have a helicity of approximately 19 nt per turn.
By characterizing the DNA structure adopted during the process of homologous recognition, it should be possible to decipher the actual molecular mechanism of DNA recombination. When double-stranded DNA (dsDNA) is covered with RecA protein, the former gets stretched by 50% and partially unwound to form a right-handed helix with 18.5 bp per right-handed turn (1–4). It has been proposed that this change of DNA structure is instrumental in the process of homologous recognition and strand exchange (5–8). However, to date there have been no measurements of DNA helicity during pairing, thus it was possible that DNA unwinding during the pairing stage is different than in the RecA–dsDNA complexes characterized earlier. For example, RecA could locally induce complete DNA unwinding during the stage of homologous pairing. Several earlier studies showed that there is extensive unwinding of duplex DNA during pairing with homologous single-stranded DNA (ssDNA) (9–11); however, these studies did not determine DNA helicity in the region of pairing. We demonstrate herein that during pairing between ssDNA and dsDNA, the duplex DNA conforms to the helical parameters of the RecA filament by adopting a structure in which both strands wind coaxially in right-handed manner and that it takes ≈19 nt of each strand to complete a turn.
MATERIALS AND METHODS
Unwinding Assay.
Single-stranded oligonucleotides, 72-mers, 94-mers, and 116-mers (MedProbe, Oslo) at a concentration of 50 μM nucleotides were incubated with 30 μM RecA protein (purified according to ref. 12) for 5 min at 37°C in 20 mM triethanolamine acetate, pH 7.4/1 mM magnesium acetate/10% glycerol/1 mM dithiothreitol/1 mM ATP. The complexes were then stabilized by the addition of adenosine 5′-[γ-thio]triphosphate (ATP[γS]) to a concentration of 2 mM. After 30 min of further incubation, 100-μl portions of the reaction mixtures were gel-filtered by using 1-ml spun columns containing Sepharose CL-4B preequilibrated with 20 mM triethanolamine acetate, pH 7.4/1 mM magnesium acetate/1 mM dithiothreitol/1 mM ATP[γS]. The flow-through fractions contained RecA-covered oligonucleotides but were free from unpolymerized RecA monomers. These fractions were then incubated with either singly nicked circular dsDNA molecules (by DNase I in the presence of ethidium bromide) or covalently closed dsDNA molecules relaxed by topoisomerase I (wheat germ; Promega). Typically, 11.2 nM pUC18 molecules (form II or IV) were mixed with a 5- to 10-fold molar excess of complexes (56 nM–112 nM ssDNA molecules) in 100 μl of column buffer, and then the magnesium acetate concentration was increased to 6 mM to stimulate the pairing reaction. The pairing reaction continued for 1 h at 37°C, and then either T4 DNA ligase (500 units/ml; Promega) supplemented with 1 mM ATP or wheat germ topoisomerase I (100 units/ml) was added and the reaction proceeded for 30 min at 37°C. Subsequently, the samples were deproteinized and analyzed on agarose gels (see Figs. 2 and 3).
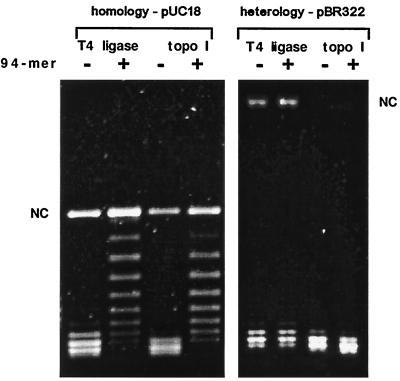
Figure 2.
Pairing induces a limited DNA unwinding. A 1% agarose gel was electrophoresed for 20 h at 2 V/cm in 0.5 × TBE buffer containing chloroquine at 0.4 μg/ml. At this concentration of chloroquine, there is a significant induction of positive supercoiling in the DNA molecules being analyzed so that incoming ssDNA is prevented from being retained by the covalently closed duplex DNA molecules (14, 15). Because dissociation of formed D-loops is strictly required for the correct estimation of the induced unwinding in the target duplex DNA, in separate experiments, we radiolabeled oligonucleotides to verify that under our gel conditions there was no label at the position of plasmid DNA. In the shown gel the DNA molecules that are unwound by the pairing are less positively supercoiled than the relaxed topoisomers from control reactions. Unwinding due to the pairing can be measured by counting bands separating the most abundant band in the samples containing control families of topoisomers and the most abundant band of the topoisomers that had their linking number locked in the stage of pairing. (Left) Incubation of RecA-covered 94-mers (complete homology to pUC18) induces unwinding by approximately 6 turns in pUC18. (Right) pBR322 is not unwound by the RecA-covered 94-mers (having no homology to pBR322). NC, position of nicked circular form of pUC18 and pBR322.
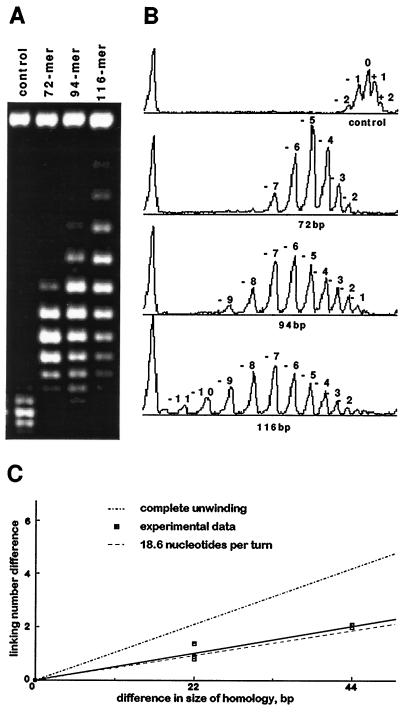
Figure 3.
Comparison of unwinding induced by different lengths of pairing regions. (A) Gel analysis of unwinding induced by pairing of the 72-mers, 94-mers, and 116-mers. (B) Corresponding densitometric scans of control reaction products without RecA-covered oligonucleotides and unwinding reaction products induced by pairing with respective RecA-covered oligonucleotides. Scans were produced and quantified with the National Institutes of Health image program. The 1.7% agarose gel was electrophoresed for 40 h at 2 V/cm in 0.5× TBE buffer containing chloroquine at 1 μg/ml. Unwinding assays were performed with only topoisomerase I (Fig. 1A). (C) Unwinding data obtained from three independent experiments are compared with the expected extent of unwinding for the model postulating a complete despiralization of the paired region (upper line) and for the model assuming that in the paired region each DNA strand participates in coaxial helical arrangements with 18.6 nt per turn (lower line). To find mean unwinding values for different experiments, Gaussian fits of the densitometric scans were calculated with the igor 1.24 program by WaveMetrics (Lake Oswego, OR).
Electron Microscopy.
The negatively stained specimens were prepared as described in ref. 13.
RESULTS
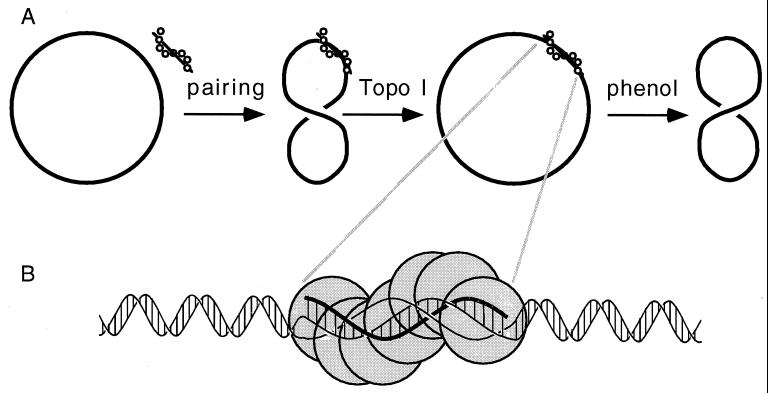
To measure the helical repeat of DNA in the region of pairing, we devised the following topological assay (Fig. 1). Short single-stranded oligomers (72-mers, 94-mers, or 116-mers) were complexed with RecA protein to form presynaptic filaments. These filaments upon separation from unbound RecA were then incubated with circular dsDNA containing sequences homologous to the ssDNA present in the presynaptic complexes. The circular dsDNA molecules were either in a covalently closed relaxed form produced by topoisomerase I (Fig. 1) or in a singly nicked form produced by DNase I. Upon pairing between RecA-covered oligonucleotides and homologous sequences in the circular DNA, either topoisomerase I was added to allow a change of the linking number of covalently closed DNA involved in pairing (Fig. 1) or T4 DNA ligase was added to close the nicked DNA. After deproteinization, DNA samples were analyzed by gel electrophoresis to measure the extent of unwinding.
Figure 1.
Schematic presentation of the unwinding assay where topoisomerase I changes the linking number of covalently closed dsDNA molecules (drawn with a single thick line) that are involved in the pairing reaction with RecA-covered oligonucleotides (A) and the proposed DNA structure within RecA-mediated synaptic filaments (B). In B, notice that all three strands in the pairing region are shown in a coaxial right-handed arrangement with approximately 18.6 bases per turn and that the incoming single strand (thick line) is base-paired with its complement in duplex DNA but the strand to be displaced in the reaction is not base-paired but still well structured within the helical RecA–DNA filament.
Fig. 2 Left shows that incubation of RecA-covered 94-mers with dsDNA containing one copy of the same sequence resulted in limited unwinding of the duplex DNA. The most abundant topoisomers produced upon pairing had their linking number decreased by 6 compared with the most abundant topoisomers produced in corresponding control reactions without RecA-covered 94-mers. This result was independent of the type of enzyme (topoisomerase or ligase) used in the assay whereby the 5:1 or 10:1 molar ratios of oligonucleotides to target plasmids produced the same unwinding (data not shown). Control reactions containing plasmids with no homology to the RecA-covered 94-mers showed that there was no appreciable unwinding (Fig. 2 Right).
When we started this project, we considered two possible outcomes of the unwinding assay. If the incoming single-strand pairs with its complement while the displaced strand is extrahelical (D-loop formation), one should observe a complete unwinding of dsDNA in the region of pairing. This should then lead to unwinding by one turn per each 10.4–10.5 bp of the pairing region (15) and thus to the expected decrease of the linking number by 9 for plasmids with a 94-bp homology to the ssDNA in the presynaptic filaments. The second possibility we considered was that in the region of pairing, all three participating strands are brought into a coaxial arrangement, by RecA, with 18.6 bases per turn (5). In such a case, the 94 bp of duplex DNA in the region of pairing would decrease its twist from almost 9 to almost 5, resulting in the reduction of the linking number by 4.
The observed reduction of the linking number by 6 was apparently too low for the classical D-loop formation and rather too high for the pairing within a helical structure with 18.6 bases per turn. However, it was observed earlier that when RecA-covered oligonucleotides interact with their target sequences on dsDNA, the regions extending 11–14 bp beyond the 5′ and 3′ ends of the paired oligonucleotide are protected from restriction enzymes (16). This suggests that RecA-covered oligonucleotides may affect the DNA structure over short stretches flanking the region of homologous pairing. It is possible that in these flanking regions, the DNA structure remains partially unwound resulting in a significant additional unwinding of plasmids undergoing pairing over a short length.
To compensate for these end effects and to verify that the observed unwinding does not depend on a particular sequence in the region of pairing, we took two additional oligonucleotides with different sizes (72-mer and 116-mer) that were homologous to other regions of the same plasmid. If instead of examining the absolute unwinding induced by the pairing with 72-mer, 94-mer, and 116-mer, one examines the differences between them, the “end effects” are compensated for (in each case one should expect a similar end effect). The observed difference of unwinding can then be attributed directly to the change of DNA helicity in the pairing region of 22 bp or 44 bp, respectively.
Fig. 3 A and B shows the difference of unwinding induced by the pairing with 94-mers and 72-mers or by pairing with 116-mers and 94-mers is almost exactly 1, which is the expected value of unwinding over 22 bp when DNA helicity changes from 10.4 bp per turn to 18.6 bp per turn (22/10.4 − 22/18.6 = 2.12 − 1.18 = 0.94). It follows that the difference of unwinding induced by pairing with 116-mer and 72-mer approaches a value of 2. In Fig. 3C, the unwinding results of three independent experiments are plotted and compared with the expected unwinding by assuming complete despiralization of the duplex DNA in the region of pairing (upper line) or assuming that each strand of the duplex winds around a common axis as it takes 18.6 bases per turn (lower line). It is visible that obtained experimental results are very close to the prediction of the second model.
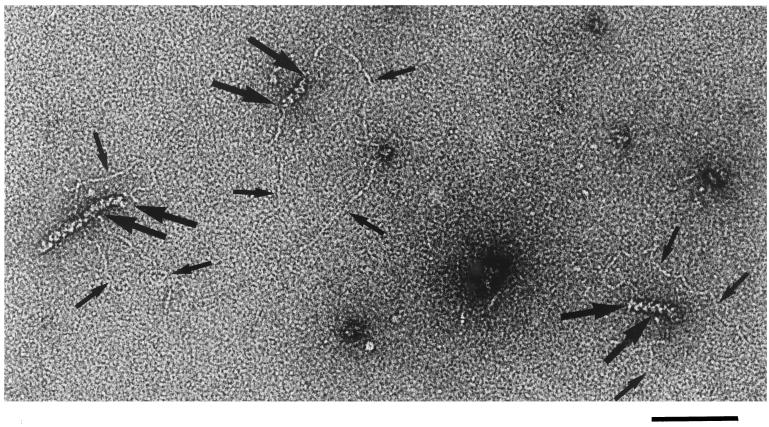
To obtain additional insight into the mechanism of the pairing reaction, we prepared electron microscopy specimens from ongoing pairing reactions. Fig. 4 shows interactions between RecA-covered 94-mers and relaxed plasmid molecules with a homologous target sequence. Interestingly, the observed RecA filaments are usually severalfold longer than the expected length of RecA-covered 94-mers. Because RecA–DNA complexes have a tendency for end-to-end stacking to form trains (17), the observed filaments are probably composed of several RecA-covered 94-mers. Only one 94-mer in the train can pair homologously with a given plasmid; however, 94-mers flanking the paired one may interact nonspecifically with the short regions of the duplex DNA flanking the target sequence. This may produce the additional unwinding observed herein and cause protection from restriction enzymes (16). However, as shown in Fig. 4, the unspecific interaction is rather limited because it appears that each dsDNA molecule enters the filament over a region corresponding to ≈100 bp, as if uptake of duplex DNA into presynaptic RecA–ssDNA complex would be attenuated beyond the region of homology.
Figure 4.
Electron microscopy visualization of the pairing reactions between nicked pUC18 and RecA-covered 94-mers. Notice three circular dsDNA molecules interacting with filaments of RecA. The filaments are frequently composed of several RecA-covered 94-mers (17). The molecule in the center apparently interacts with individual RecA-covered 94-mer. The molecule on the right side probably interacts with a train of two RecA-covered 94-mers, whereas the molecule on the left side interacts with a train of three or four individual filaments. However, the DNA enters into longer filaments only over a length that roughly corresponds to one RecA-covered 94-mer. Thin arrows are an aid to localize DNA contours, and thicker arrows indicate entry and exit points of DNA into RecA filaments. (Bar = 100 nm.)
DISCUSSION
We worked herein with RecA–ssDNA complexes stabilized by ATP[γS], a very slowly hydrolyzable analog of ATP (18). This allowed us to column-purify the presynaptic complexes from free RecA monomers that could otherwise unwind dsDNA by directly binding to duplex DNA (19). The use of ATP[γS] instead of ATP raises the question of the biological relevance of our findings. However, many RecA studies have demonstrated that RecA–DNA complexes formed in the presence of ATP[γS] or ATP are very similar in their activity and structure. For example, efficient homologous recognition occurs in the presence of ATP and ATP[γS] (20, 21). The process of strand exchange also proceeds efficiently in the presence of ATP[γS] (21, 22). The helicity of dsDNA upon covering by RecA is practically the same in complexes formed in the presence of ATP and ATP[γS] (4). At present it seems that ATP hydrolysis is only needed for the strand exchange between partially heterologous sequences and for releasing the RecA from DNA upon termination of strand exchange (23, 24). Thus the synaptic complexes formed in the presence of ATP[γS] are well suited for the investigation of pairing and strand exchange between fully homologous sequences.
Several reports have addressed the question of strand arrangement within RecA filaments containing homologous ssDNA and dsDNA molecules. There are strong indications that in such filaments formed in the presence of ATP[γS], the DNA strands are already switched; i.e., against chemical probes, the original single strand behaves like a strand of duplex DNA, whereas the strand to be displaced in the reaction behaves like a single strand (25). Other studies (21, 22) lead to the conclusion that in these complexes DNA strands are either already switched or at least strongly poised to be switched. On the basis of the unwinding data presented herein and the above-mentioned structural information, we propose that within synaptic complexes all three participating DNA strands form a coaxial helical arrangement with approximately 19 nt of each strand per helical turn (see Fig. 1B). Most likely the strands are already in the post-exchange arrangement; i.e., the incoming single strand is base-paired with its complement from the original duplex DNA, and the strand to be displaced in the reaction has a certain single-stranded character but is still wound around the newly formed duplex region.
Acknowledgments
We thank Prof. Jacques Dubochet for friendly support. This work was supported by Swiss National Science Foundation Grant 31-42158.94 to J. Dubochet and A.S.
ABBREVIATIONS
- ATP[γS]
adenosine [γ-thio]triphosphate
- dsDNA
double-stranded DNA
- ssDNA
single-stranded DNA
References
- 1.Stasiak A, Di Capua E, Koller T. J Mol Biol. 1981;151:557–564. doi: 10.1016/0022-2836(81)90010-3. [DOI] [PubMed] [Google Scholar]
- 2.Dunn K, Chrysogelos S, Griffith J. Cell. 1982;28:757–765. doi: 10.1016/0092-8674(82)90055-1. [DOI] [PubMed] [Google Scholar]
- 3.Stasiak A, Di Capua E. Nature (London) 1982;299:185–186. doi: 10.1038/299185a0. [DOI] [PubMed] [Google Scholar]
- 4.Pugh B F, Schutte B C, Cox M M. J Mol Biol. 1989;205:487–492. doi: 10.1016/0022-2836(89)90219-2. [DOI] [PubMed] [Google Scholar]
- 5.Howard-Flanders P, West S C, Stasiak A J. Nature (London) 1984;309:215–220. doi: 10.1038/309215a0. [DOI] [PubMed] [Google Scholar]
- 6.Zhurkin V B, Raghunathan G, Ulyanov N B, Camerini-Otero R D, Jernigan R L. J Mol Biol. 1994;239:181. doi: 10.1006/jmbi.1994.1362. [DOI] [PubMed] [Google Scholar]
- 7.Cluzel P, Lebrun A, Heller C, Lavery R, Viovy J-L, Chatenay D, Caron F. Science. 1996;271:792. doi: 10.1126/science.271.5250.792. [DOI] [PubMed] [Google Scholar]
- 8.Camerini-Otero R D, Hsieh P. Annu Rev Genet. 1995;29:509–552. doi: 10.1146/annurev.ge.29.120195.002453. [DOI] [PubMed] [Google Scholar]
- 9.Schutte B C, Cox M M. Biochemistry. 1988;27:7886–7894. doi: 10.1021/bi00420a046. [DOI] [PubMed] [Google Scholar]
- 10.Conley E C, West S C. J Biol Chem. 1990;265:10156–10163. [PubMed] [Google Scholar]
- 11.Rould E, Muniyappa K, Radding C M. J Mol Biol. 1992;226:127–139. doi: 10.1016/0022-2836(92)90129-8. [DOI] [PubMed] [Google Scholar]
- 12.Shibata T, Osber L, Radding C M. Methods Enzymol. 1983;100:197–209. doi: 10.1016/0076-6879(83)00056-7. [DOI] [PubMed] [Google Scholar]
- 13.Di Capua E, Engel A, Stasiak A, Koller T. J Mol Biol. 1982;157:87–103. doi: 10.1016/0022-2836(82)90514-9. [DOI] [PubMed] [Google Scholar]
- 14.Voloshin O N, Wang L, Camerini-Otero R D. Science. 1996;272:868–872. doi: 10.1126/science.272.5263.868. [DOI] [PubMed] [Google Scholar]
- 15.Beattie K L, Wiegand R C, Radding C M. J Mol Biol. 1977;116:783–803. doi: 10.1016/0022-2836(77)90271-6. [DOI] [PubMed] [Google Scholar]
- 16.Hsieh P, Camerini-Otero C S, Camerini-Otero R D. Proc Natl Acad Sci USA. 1992;89:6492–6496. doi: 10.1073/pnas.89.14.6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Register J C, Griffith J. Proc Natl Acad Sci USA. 1986;83:624–628. doi: 10.1073/pnas.83.3.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu X, Egelman E H. J Mol Biol. 1992;225:193–216. doi: 10.1016/0022-2836(92)91036-o. [DOI] [PubMed] [Google Scholar]
- 19.Pugh B F, Cox M M. J Mol Biol. 1988;203:479–493. doi: 10.1016/0022-2836(88)90014-9. [DOI] [PubMed] [Google Scholar]
- 20.Honigberg S M, Gonda D K, Flory J, Radding C M. J Biol Chem. 1985;260:11845–11851. [PubMed] [Google Scholar]
- 21.Rosselli W, Stasiak A. J Mol Biol. 1990;216:335–352. doi: 10.1016/S0022-2836(05)80325-0. [DOI] [PubMed] [Google Scholar]
- 22.Menetski J P, Bear D G, Kowalczykowski S C. Proc Natl Acad Sci USA. 1990;87:21–25. doi: 10.1073/pnas.87.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosselli W, Stasiak A. EMBO J. 1991;10:4391–4396. doi: 10.1002/j.1460-2075.1991.tb05017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J-I, Cox M M, Inman R B. J Biol Chem. 1992;267:16438–16443. [PubMed] [Google Scholar]
- 25.Adzuma K. Genes Dev. 1992;6:1679–1694. doi: 10.1101/gad.6.9.1679. [DOI] [PubMed] [Google Scholar]