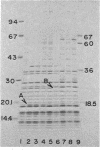
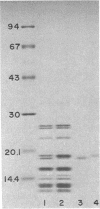
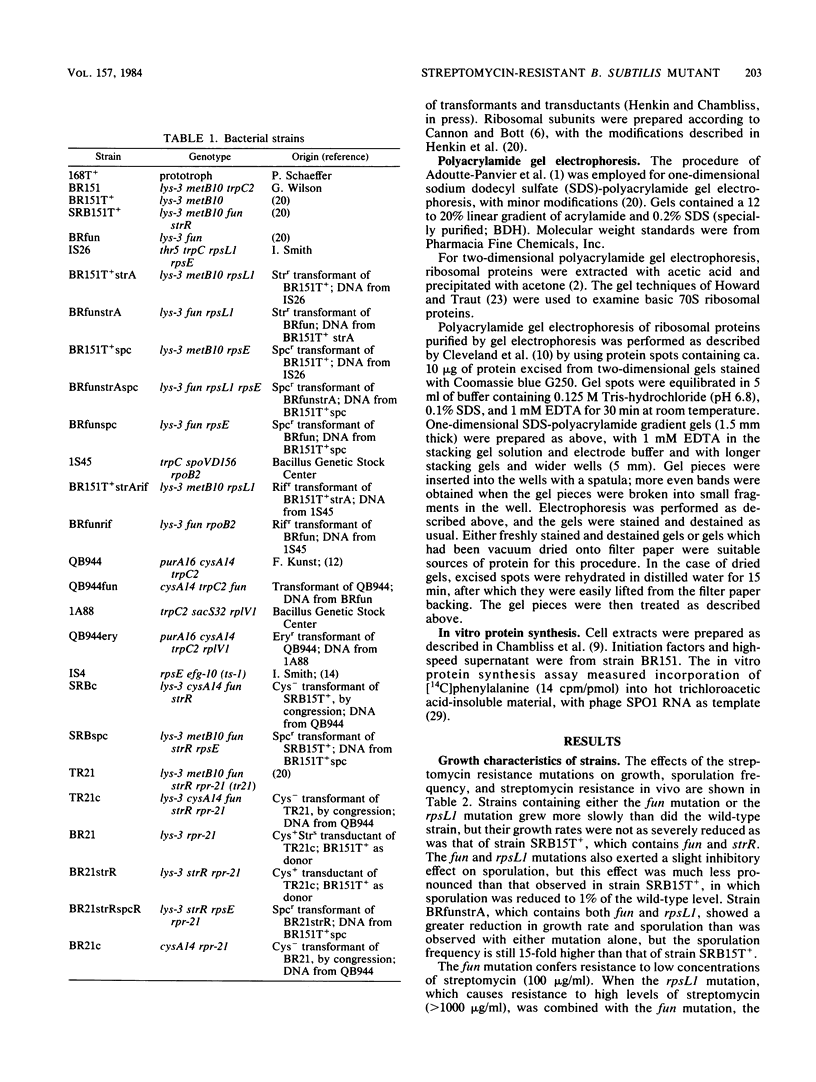
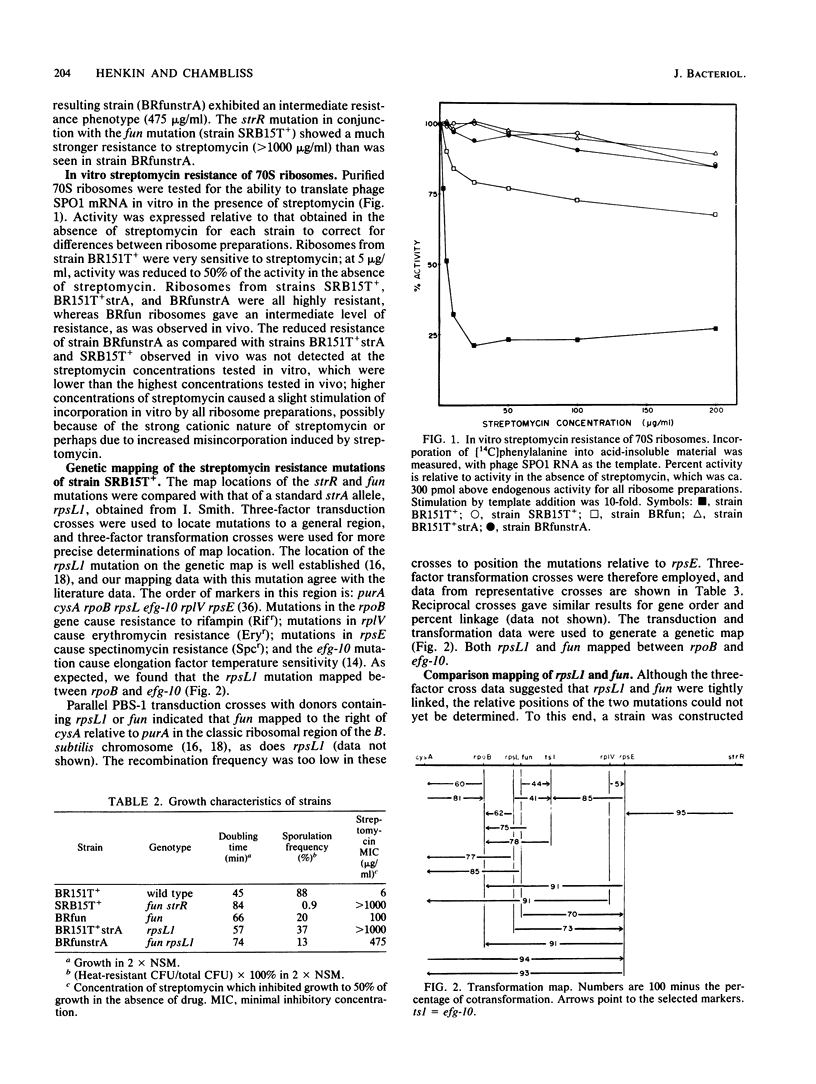
Abstract
Strain SRB15T+, a streptomycin-resistant, oligosporogenous mutant of Bacillus subtilis, contains two mutations, fun and strR. These mutations were mapped by PBS-1 mediated transduction and by transformation to two different sites in the cysA-linked region of the B. subtilis chromosome. The fun mutation mapped very close to rpsLl, a classic strA mutation, whereas strR mapped to a site distal to rpsE. The effects of these mutations on growth, sporulation, and streptomycin resistance in vivo and in vitro were determined. The fun mutation gave a different phenotype than did the rpsLl mutation and caused altered migration of a ribosomal protein which was identified as S12, the protein encoded by rpsL. It therefore appears that fun is an allele of the rpsL gene.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adoutte-Panvier A., Davies J. E., Gritz L. R., Littlewood B. S. Studies of ribosomal proteins of yeast species and their hybrids: gel electrophoresis and immunochemical cross-reactions. Mol Gen Genet. 1980;179(2):273–282. doi: 10.1007/BF00425454. [DOI] [PubMed] [Google Scholar]
- Barritault D., Expert-Bezancon A., Guérin M. F., Hayes D. The use of acetone precipitation in the isolation of ribosomal proteins. Eur J Biochem. 1976 Mar 16;63(1):131–135. doi: 10.1111/j.1432-1033.1976.tb10215.x. [DOI] [PubMed] [Google Scholar]
- Bjare U., Gorini L. Drug dependence reversed by a ribosomal ambiguity mutation, ram, in Escherichia coli. J Mol Biol. 1971 May 14;57(3):423–435. doi: 10.1016/0022-2836(71)90101-x. [DOI] [PubMed] [Google Scholar]
- Cabezón T., Herzog A., De Wilde M., Villarroel R., Bollen A. Cooperative control of translational fidelity by ribosomal proteins in Escherichia coli. III. A ram mutation in the structural gene for protein S5 (rpx E). Mol Gen Genet. 1976 Feb 27;144(1):59–62. doi: 10.1007/BF00277305. [DOI] [PubMed] [Google Scholar]
- Campbell K. M., Chambliss G. H. Streptomycin-resistant, asporogenous mutant of Bacillus subtilis. Mol Gen Genet. 1977 Dec 30;158(2):193–200. doi: 10.1007/BF00268313. [DOI] [PubMed] [Google Scholar]
- Cannon J. G., Bott K. F. Mutation affecting expression of spectinomycin resistance in Bacillus subtilis. J Bacteriol. 1980 Jan;141(1):409–412. doi: 10.1128/jb.141.1.409-412.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon J. G., Bott K. F. Spectinomycin-resistant mutants of Bacillus subtilis with altered sporulation properties. Mol Gen Genet. 1979 Jul 13;174(2):149–162. doi: 10.1007/BF00268352. [DOI] [PubMed] [Google Scholar]
- Carlton B. C. Fine-structure mapping by transformation in the tryptophan region of Bacillus subtilis. J Bacteriol. 1966 May;91(5):1795–1803. doi: 10.1128/jb.91.5.1795-1803.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambliss G. H., Henkin T. M., Leventhal J. M. Bacterial in vitro protein-synthesizing systems. Methods Enzymol. 1983;101:598–605. doi: 10.1016/0076-6879(83)01040-x. [DOI] [PubMed] [Google Scholar]
- Cleveland D. W., Fischer S. G., Kirschner M. W., Laemmli U. K. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem. 1977 Feb 10;252(3):1102–1106. [PubMed] [Google Scholar]
- DAVIES J., GILBERT W., GORINI L. STREPTOMYCIN, SUPPRESSION, AND THE CODE. Proc Natl Acad Sci U S A. 1964 May;51:883–890. doi: 10.1073/pnas.51.5.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedonder R. A., Lepesant J. A., Lepesant-Kejzlarová J., Billault A., Steinmetz M., Kunst F. Construction of a kit of reference strains for rapid genetic mapping in Bacillus subtilis 168. Appl Environ Microbiol. 1977 Apr;33(4):989–993. doi: 10.1128/aem.33.4.989-993.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis P. P. Transcription and translation in a pleiotropic streptomycin-resistant mutant of Escherichia coli. J Bacteriol. 1979 Jan;137(1):197–203. doi: 10.1128/jb.137.1.197-203.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau E., Pifko S., Sloma A., Cabane K., Smith I. Conditional mutations in the translational apparatus of Bacillus subtils. Mol Gen Genet. 1976 Aug 10;147(1):1–12. doi: 10.1007/BF00337929. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Branscomb E. W. Ribosome slowed by mutation to streptomycin resistance. Nature. 1976 Aug 12;262(5569):617–619. doi: 10.1038/262617b0. [DOI] [PubMed] [Google Scholar]
- Goldthwaite C., Dubnau D., Smith I. Genetic mapping of antibiotic resistance in markers Bacillus subtilis. Proc Natl Acad Sci U S A. 1970 Jan;65(1):96–103. doi: 10.1073/pnas.65.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorini L. Ribosomal discrimination of tRNAs. Nat New Biol. 1971 Dec 29;234(52):261–264. doi: 10.1038/newbio234261a0. [DOI] [PubMed] [Google Scholar]
- Harford N., Sueoka N. Chromosomal location of antibiotic resistance markers in Bacillus subtilis. J Mol Biol. 1970 Jul 28;51(2):267–286. doi: 10.1016/0022-2836(70)90142-7. [DOI] [PubMed] [Google Scholar]
- Hasenbank R., Guthrie C., Stöffler G., Wittmann H. G., Rosen L., Apirion D. Electrophoretic and immunological studies on ribosomal proteins of 100 Escherichia coli revertants from streptomycin dependence. Mol Gen Genet. 1973 Dec 14;127(1):1–18. doi: 10.1007/BF00267778. [DOI] [PubMed] [Google Scholar]
- Henkin T. M., Campbell K. M., Chambliss G. H. Revertants of a streptomycin-resistant, oligosporogenous mutant of Bacillus subtilis. Mol Gen Genet. 1982;186(3):347–354. doi: 10.1007/BF00729453. [DOI] [PubMed] [Google Scholar]
- Henner D. J., Hoch J. A. The Bacillus subtilis chromosome. Microbiol Rev. 1980 Mar;44(1):57–82. doi: 10.1128/mr.44.1.57-82.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K., Otaka E., Osawa S. Purification and characterization of 30S ribosomal proteins from Bacillus subtilis: correlation to Escherichia coli 30S proteins. Mol Gen Genet. 1982;185(2):239–244. doi: 10.1007/BF00330792. [DOI] [PubMed] [Google Scholar]
- Howard G. A., Traut R. R. Separation and radioautography of microgram quantities of ribosomal proteins by two-dimensional polyacrylamide gel electrophoresis. FEBS Lett. 1973 Jan 15;29(2):177–180. doi: 10.1016/0014-5793(73)80555-1. [DOI] [PubMed] [Google Scholar]
- Ito T., Kosugi H., Higo K., Osawa S. Ribosomal proteins from streptomycin-resistant and dependent mutants, and revertants from streptomycin-dependence to independence in Bacillus subtilis. Mol Gen Genet. 1975 Sep 8;139(4):293–301. doi: 10.1007/BF00267969. [DOI] [PubMed] [Google Scholar]
- Kreider G., Brownstein B. L. Ribosomal proteins involved in the suppression of streptomycin dependence in Escherichia coli. J Bacteriol. 1972 Feb;109(2):780–785. doi: 10.1128/jb.109.2.780-783.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwano M., Endo H., Ohnishi Y. Mutations to spectinomycin resistance which alleviate the restriction of an amber suppressor by streptomycin resistance. J Bacteriol. 1969 Feb;97(2):940–943. doi: 10.1128/jb.97.2.940-943.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LACKS S., HOTCHKISS R. D. A study of the genetic material determining an enzyme in Pneumococcus. Biochim Biophys Acta. 1960 Apr 22;39:508–518. doi: 10.1016/0006-3002(60)90205-5. [DOI] [PubMed] [Google Scholar]
- Legault-Demare L., Chambliss G. H. Natural messenger ribonucleic acid-directed cell-free protein-synthesizing system of Bacillus subtilis. J Bacteriol. 1974 Dec;120(3):1300–1307. doi: 10.1128/jb.120.3.1300-1307.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighton T. Sporulation-specific translational discrimination in Bacillus subtilis. J Mol Biol. 1974 Jul 15;86(4):855–863. doi: 10.1016/0022-2836(74)90358-1. [DOI] [PubMed] [Google Scholar]
- Matković B., Herzog A., Bollen A., Topisirović L. Translational fidelity in Escherichia coli: antagonistic effects of neaA and ramC gene products on the ribosome function. Mol Gen Genet. 1980;179(1):135–139. doi: 10.1007/BF00268455. [DOI] [PubMed] [Google Scholar]
- Osawa S., Tokui A., Saito H. Mapping by interspecies transformation experiments of several ribosomal protein genes near the replication origin of Bacillus subtilis chromosome. Mol Gen Genet. 1978 Aug 17;164(2):113–129. doi: 10.1007/BF00267376. [DOI] [PubMed] [Google Scholar]
- Ozaki M., Mizushima S., Nomura M. Identification and functional characterization of the protein controlled by the streptomycin-resistant locus in E. coli. Nature. 1969 Apr 26;222(5191):333–339. doi: 10.1038/222333a0. [DOI] [PubMed] [Google Scholar]
- Piepersberg W., Noseda V., Böck A. Bacterial ribosomes with two ambiguity mutations: effects of translational fidelity, on the response to aminoglycosides and on the rate of protein synthesis. Mol Gen Genet. 1979 Mar 9;171(1):23–34. doi: 10.1007/BF00274011. [DOI] [PubMed] [Google Scholar]
- Schaeffer P., Millet J., Aubert J. P. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci U S A. 1965 Sep;54(3):704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith I., Goldthwaite C., Dubnau D. The genetics of ribosomes in Bacillus subtilis. Cold Spring Harb Symp Quant Biol. 1969;34:85–89. doi: 10.1101/sqb.1969.034.01.013. [DOI] [PubMed] [Google Scholar]
- Spizizen J. TRANSFORMATION OF BIOCHEMICALLY DEFICIENT STRAINS OF BACILLUS SUBTILIS BY DEOXYRIBONUCLEATE. Proc Natl Acad Sci U S A. 1958 Oct 15;44(10):1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staal S. P., Hoch J. A. Conditional dihydrostreptomycin resistance in Bacillus subtilis. J Bacteriol. 1972 Apr;110(1):202–207. doi: 10.1128/jb.110.1.202-207.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strigini P., Gorini L. Ribosomal mutations affecting efficiency of amber suppression. J Mol Biol. 1970 Feb 14;47(3):517–530. doi: 10.1016/0022-2836(70)90319-0. [DOI] [PubMed] [Google Scholar]
- Topisirovic L., Villarroel R., De Wilde M., Herzog A., Cabezón T., Bollen A. Translational fidelity in Escherichia coli: contrasting role of neaA and ramA gene products in the ribosome functioning. Mol Gen Genet. 1977 Feb 28;151(1):89–94. doi: 10.1007/BF00446917. [DOI] [PubMed] [Google Scholar]
- Yates J. L., Gette W. R., Furth M. E., Nomura M. Effects of ribosomal mutations on the read-through of a chain termination signal: studies on the synthesis of bacteriophage lambda O gene protein in vitro. Proc Natl Acad Sci U S A. 1977 Feb;74(2):689–693. doi: 10.1073/pnas.74.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengel J. M., Young R., Dennis P. P., Nomura M. Role of ribosomal protein S12 in peptide chain elongation: analysis of pleiotropic, streptomycin-resistant mutants of Escherichia coli. J Bacteriol. 1977 Mar;129(3):1320–1329. doi: 10.1128/jb.129.3.1320-1329.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]