The perspectives on membrane protein insertion and protein–bilayer interactions (J. Gen. Physiol. 129:351–377) made important points concerning amino acid residue hydrophobicity, interfacial propensity, and membrane partitioning (MacCallum et al., 2007; von Heijne, 2007; White, 2007; Wolfenden, 2007). It is particularly striking that the relative ordering of the tendencies for particular amino acids to be inserted into a transmembrane helix from a translocon (Hessa et al., 2005) is highly correlated with the relative ordering of both (a) the experimental free energies for bare side chain (RH) transfer from cyclohexane to water (Wolfenden, 2007) and (b) the calculated free energies for side chain transfer from water to the center of a DOPC membrane (MacCallum et al., 2007). The relative octanol/water partitioning provides a good indication of interfacial propensity. Because water-saturated 1-octanol contains 2.3 M water (Wolfenden, 2007), the issues of comparing dry versus wet octanol, and of partitioning into dry octanol, seem as yet unresolved.
The case of tryptophan indeed deserves “special scrutiny” (Wolfenden, 2007). Tryptophan is the largest amino acid, but not the most hydrophobic. To be sure, the “biological” (insertion from a translocon into a transmembrane helix), “experimental” (cyclohexane/water partition), and “computational” (computed water-to-membrane transfer energy) criteria all categorize Trp as less hydrophobic than Phe, Leu, Ile, Met, and Val. The same is true when considering partitioning of these respective side chains between water and either a vapor phase or chloroform (Wolfenden, 2007). Numerous other criteria also indicate that Trp is less hydrophobic than Phe or the aliphatic amino acids. For example, it is well known that substitution of only one of four gramicidin A tryptophans by Phe causes a much higher affinity (longer elution time) for reversed-phase chromatography stationary phases (Koeppe et al., 1985). Furthermore, the very short lifetime of [Trp1] gramicidin A channels suggests that it is unfavorable to place Trp near the center of a lipid bilayer (Mazet et al., 1984), whereas [Phe1], [Leu1], [Met1], [Ile1], and [Val1] gramicidin A channels have lifetimes within a “normal” range of 0.5–1.0 s (Russell et al., 1986). The short lifetime of [Trp1] gramicidin channels is additionally consistent with the positional dependence for Trp insertion from a translocon into a transmembrane helix (Hessa et al., 2005). Furthermore, if all tryptophans 9, 11, 13, and 15 of gramicidin are changed to Phe, allowing these sequence positions to become membrane buried, then the gramicidin folding preference changes from single stranded to double stranded (Durkin et al., 1992; Salom et al., 1998).
Recently, we have compared the properties of Trp indole rings near the N and C terminals of membrane-spanning peptides. α-Helices are asymmetric structures that possess a significant permanent dipole moment and exhibit important differences in the ways that side chains project from the N- and C-terminal regions of the backbone with respect to a reference surface such as a membrane/water interface. These differences become important when considering the role of the side chain position in the bilayer.
Based on solid-state 2H-NMR spectra of labeled Trp indole rings in acetyl-GWWLALALALALAWWA-ethanolamide (WALP16) and acetyl-GWWLALALALALALALWWA-ethanolamide (WALP19) incorporated into oriented hydrated lipid bilayers, we have deduced that Trp indole rings near the C terminus are significantly more mobile than rings near the N terminus (van der Wel et al., 2007). For a ring having a fixed orientation with respect to an external magnetic field, Ho, each ring deuteron yields a characteristic quadrupolar splitting that depends upon the angle between the C-2H bond and Ho (Hu et al., 1993). Ring motions will reduce the magnitudes of all of these quadrupolar splittings (Koeppe et al., 1994), eventually to zero in the case of isotropic motion. For a particular ring, therefore, the maximum observed quadrupolar splitting becomes a qualitative indicator of the extent of overall ring motion.
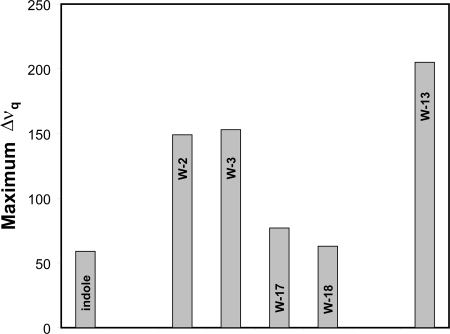
Fig. 1 illustrates the maximum observed quadrupolar splittings for several types of membrane-incorporated indole rings. A “free” indole ring, not linked to any peptide, is highly mobile yet still partially oriented within a membrane (Persson et al., 1998); the corresponding 2H-NMR spectrum shows a maximum quadrupolar splitting of ∼60 kHz (Fig. 1). At the other extreme of motional restriction, tryptophans 11, 13, and 15 of gramicidin A channels exhibit maximum 2H quadrupolar splittings >180 kHz. Within this framework, the Trps in WALP16 and WALP19 (van der Wel et al., 2007) experience intermediate motion between the extremes of unattached indole and gramicidin A. Surprisingly, nevertheless, the N-terminal and C-terminal Trps differ considerably in their dynamics; the N-terminal Trps are quite restricted in their motions, whereas the C-terminal Trps appear nearly as mobile as the “free” indole (Fig. 1). Indeed the N-terminal Trps appear more similar to the gramicidin Trps than to the C-terminal Trps on the same helix. Surveys and prediction of single-span membrane proteins indicate that Trp is favored at both the N- and C-interfacial boundaries of transmembrane segments (Landolt-Marticorena et al., 1993; Pilpel et al., 1999; Ulmschneider and Sansom, 2001). Nevertheless, transmembrane helices are asymmetric, and Trp indole rings have distinctly different properties at the two ends of such a helix, including different allowed regions for their side chain χ1 and χ2 angles (van der Wel et al., 2007). To be precise, the question of side chain localization should address not only the position relative to the bilayer midplane, but also whether a given side chain is near the N or C terminus of the α-helix. Although the difference in side chain dynamics at the two boundaries has to date been recognized only for Trp, examination of standard rotamer libraries (compare Dunbrack, 2002) suggests that the feature could be general in membrane proteins.
Figure 1.
Maximum 2H-NMR quadrupolar splittings observed for deuterated indole rings in bilayers of DMPC, either “free” to diffuse within the lipid or attached to a membrane-spanning peptide. From left to right: “free” indole (from Persson et al., 1998), four tryptophans in WALP19, and Trp13 of gramicidin A (from van der Wel et al., 2007).
Acknowledgments
Olaf S. Andersen served as editor.
References
- Dunbrack, R.L., Jr. 2002. Rotamer libraries in the 21st century. Curr. Opin. Struct. Biol. 12:431–440. [DOI] [PubMed] [Google Scholar]
- Durkin, J.T., L.L. Providence, R.E. Koeppe II, and O.S. Andersen. 1992. Formation of non-β6.3-helical gramicidin channels between sequence-substituted gramicidin analogues. Biophys. J. 62:145–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessa, T., H. Kim, K. Bihlmaier, C. Lundin, J. Boekel, H. Andersson, I. Nilsson, S.H. White, and G. von Heijne. 2005. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 433:377–381. [DOI] [PubMed] [Google Scholar]
- Hu, W., K.C. Lee, and T.A. Cross. 1993. Tryptophans in membrane proteins: indole ring orientations and functional implications in the gramicidin channel. Biochemistry. 32:7035–7047. [DOI] [PubMed] [Google Scholar]
- Koeppe, R.E., II, J.A. Killian, and D.V. Greathouse. 1994. Orientations of the tryptophan 9 and 11 side chains of the gramicidin channel based on deuterium NMR spectroscopy. Biophys. J. 66:14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeppe, R.E., II, J.A. Paczkowski, and W.L. Whaley. 1985. Gramicidin K, a new linear channel-forming gramicidin from Bacillus brevis. Biochemistry. 24:2822–2826. [DOI] [PubMed] [Google Scholar]
- Landolt-Marticorena, C., K.A. Williams, C.M. Deber, and R.A.F. Reithmeier. 1993. Non-random distribution of amino acids in the transmembrane segments of human type I single span membrane proteins. J. Mol. Biol. 229:602–608. [DOI] [PubMed] [Google Scholar]
- MacCallum, J.L., W.F.D. Bennett, and D.P. Tieleman. 2007. Partitioning of amino acid side chains into lipid bilayers: results from computer simulations and comparison to experiment. J. Gen. Physiol. 129:371–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazet, J.L., O.S. Andersen, and R.E. Koeppe II. 1984. Single- channel studies on linear gramicidins with altered amino acid sequences. A comparison of phenylalanine, tryptophan, and tyrosine substitutions at positions 1 and 11. Biophys. J. 45:263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson, S., J.A. Killian, and G. Lindblom. 1998. Molecular ordering of interfacially localized tryptophan analogs in ester- and ether-lipid bilayers studied by 2H-NMR. Biophys. J. 75:1365–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilpel, Y., N. Ben-Tal, and D. Lancet. 1999. KPROT: a knowledge-based scale for the propensity of residue orientation in transmembrane segments. Application to membrane protein structure prediction. J. Mol. Biol. 294:921–935. [DOI] [PubMed] [Google Scholar]
- Russell, E.W.B., L.B. Weiss, F.I. Navetta, R.E. Koeppe II, and O.S. Andersen. 1986. Single-channel studies on linear gramicidins with altered amino acid side chains. Effects of altering the polarity of the side chain at position 1 in gramicidin A. Biophys. J. 49:673–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salom, D., E. Pérez-Payá, J. Pascal, and C. Abad. 1998. Environment- and sequence-dependent modulation of the double-stranded to single-stranded conformational transition of gramicidin A in membranes. Biochemistry. 37:14279–14291. [DOI] [PubMed] [Google Scholar]
- Ulmschneider, M.B., and M.S.P. Sansom. 2001. Amino acid distributions in integral membrane protein structures. Biochim. Biophys. Acta. 1512:1–14. [DOI] [PubMed] [Google Scholar]
- van der Wel, P.C.A., N.D. Reed, D.V. Greathouse, and R.E. Koeppe II. 2007. Orientation and motion of tryptophan interfacial anchors in membrane-spanning peptides. Biochemistry. 46:7514–7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne, G. 2007. Formation of transmembrane helices in vivo—is hydrophobicity all that matters? J. Gen. Physiol. 129:353–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, S.H. 2007. Membrane protein insertion: the biology-physics nexus. J. Gen. Physiol. 129:363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfenden, R. 2007. Experimental measures of amino acid hydrophobicity and the topology of transmembrane and globular proteins. J. Gen. Physiol. 129:357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]