Abstract
Interactions between nontransmembrane domains and the lipid membrane are proposed to modulate activity of many ion channels. In Kir channels, the so-called “slide-helix” is proposed to interact with the lipid headgroups and control channel gating. We examined this possibility directly in a cell-free system consisting of KirBac1.1 reconstituted into pure lipid vesicles. Cysteine substitution of positively charged slide-helix residues (R49C and K57C) leads to loss of channel activity that is rescued by in situ restoration of charge following modification by MTSET+ or MTSEA+, but not MTSES− or neutral MMTS. Strikingly, activity is also rescued by modification with long-chain alkyl-MTS reagents. Such reagents are expected to partition into, and hence tether the side chain to, the membrane. Systematic scanning reveals additional slide-helix residues that are activated or inhibited following alkyl-MTS modification. A pattern emerges whereby lipid tethering of the N terminus, or C terminus, of the slide-helix, respectively inhibits, or activates, channel activity. This study establishes a critical role of the slide-helix in Kir channel gating, and directly demonstrates that physical interaction of soluble domains with the membrane can control ion channel activity.
INTRODUCTION
For many ion channels, lipid membrane composition is proposed to be an important regulator of channel gating (Hilgemann and Ball, 1996; Shyng and Nichols, 1998; Baukrowitz et al., 1998; Runnels et al., 2002; Rohacs et al., 2005; Suh and Hille, 2005) but physical evidence for direct interactions of channel domains with lipids is lacking. In the inwardly rectifying K (Kir) channels, a large and widespread channel family that modulates excitability throughout the organism, channel gating is proposed to be controlled by the “slide-helix,” a novel feature seen in the crystal structure of the bacterial Kir homologue KirBac 1.1 (Kuo et al., 2003). The slide-helix is an α-helical segment immediately preceding the first transmembrane segment, and is predicted to lie parallel to the membrane, in the vicinity of the phospholipid headgroups (Fig. 1 A). It has been proposed that the slide-helix interacts with the phospholipid headgroups and forms a link between the binding site for gating molecules (such as ATP in Kir6.2) and the ligand-dependent channel gate (Kuo et al., 2003), which is likely to be at the crossing point of the second transmembrane helix bundle. Mutations of Kir channels within the region of the slide-helix are known causes of inherited Kir channel disorders (Schulte et al., 1999; Plaster et al., 2001; Schulze et al., 2003; Gloyn et al., 2004).
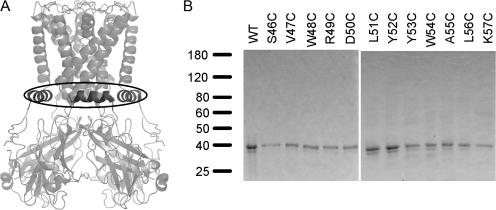
Figure 1.
(A) Ribbon model of KirBac1.1 crystal structure, demonstrating the location of the slide-helices, emphasized in dark gray. (B) SDS-PAGE of purified WT KirBac1.1 protein and cysteine-substituted mutants, stained with Coomassie blue.
In this study, we examine the role of interactions between the lipid membrane and the slide-helix in control of Kir channel gating in a pure channel–lipid system. The results provide direct demonstration of control of channel activity by physical interaction of a nontransmembrane domain of the channel with the lipid membrane.
MATERIALS AND METHODS
Methods are essentially as described previously (Enkvetchakul et al., 2004). KirBac1.1 was cloned from genomic DNA of Burkholderia pseudomallei by PCR and subcloned into the pQE60 vector (QIAGEN) as a C-terminal six histidine–tagged construct. Single cysteine mutations were made using the Quikchange Site-directed Mutagenesis Kit (Stratagene). All mutants were verified by DNA sequencing.
For protein purification, KirBac1.1 in pQE60 was expressed in BL21* (DE3) cells induced with isopropyl β-d-thiogalactopyranoside. Bacteria were lysed by a freeze–thaw cycle, incubated for 2–4 h in resuspension buffer (50 mM Tris-HCl, pH 8.0, 150 mM KCl, 250 mM sucrose, 10 mM MgSO4) with 30 mM decylmaltoside (Anatrace), and then centrifuged at 30,000 g for 30 min. The supernatant was mixed with ∼0.2–0.4 ml cobalt beads, washed with 40 bed volumes of wash buffer (50 mM Tris-HCl, pH 7.4, 150 mM KCl, 10 mM imidazole, and 5 mM decylmaltoside), and KirBac1.1 was eluted with 2 ml of wash buffer containing 500 mM imidazole. Proteins were concentrated using 30-kD centrifugal filters (Millipore) to 0.5–5 mg/ml.
For Rb+ flux assay, disposable polystyrene columns (Pierce Chemical Co.) were packed with Sephadex G-50 (fine) beads (1 ml), swollen overnight in buffer A or B (buffer A: 450 mM KCl, 10 mM HEPES, 4 mM NMG, pH 7; buffer B: 450 mM sorbitol, 10 mM HEPES, 4 mM NMG, 50 μM KCl, pH 7.0). 2–3 μg of purified protein per mg of total lipid was added to a CHAPS (37 mM) solubilized mixture of phosphatidylethanolamine:phosphatidylglycerol (3:1, Avanti Polar Lipids, Inc., 10 mg total lipid per ml) in buffer A and incubated 30 min. Column A (with Sephadex beads in buffer A) were partially dehydrated by spinning at 3,000 rpm in a Beckman TJ6 centrifuge. Liposomes were formed by spinning 100 μl of detergent-solubilized lipid/protein mixture through the partially dehydrated column A at 2,500 rpm. Extraliposomal solution was exchanged for buffer B by centrifugation through a partially dehydrated column B. The assay was initiated by adding 400 μl of buffer B with 1–5 μM 86Rb+. 50–60-μl aliquots of the radioactive mixture were taken at time points indicated, and extraliposomal 86Rb+ removed by passage over a 0.5-ml Dowex cation exchange column in the NMGH+ form. Samples were mixed with scintillation fluid and counted in a liquid scintillation counter. Valinomycin was used to assay maximal 86Rb+ uptake. MTS modification of KirBac1.1 cysteine mutants was prepared by incubating purified mutant protein in 100 μM MTS reagent for ∼30 min before reconstitution.
In all figures, values indicated are mean ± SEM, except Fig. 3, which shows values ± 95% confidence intervals.
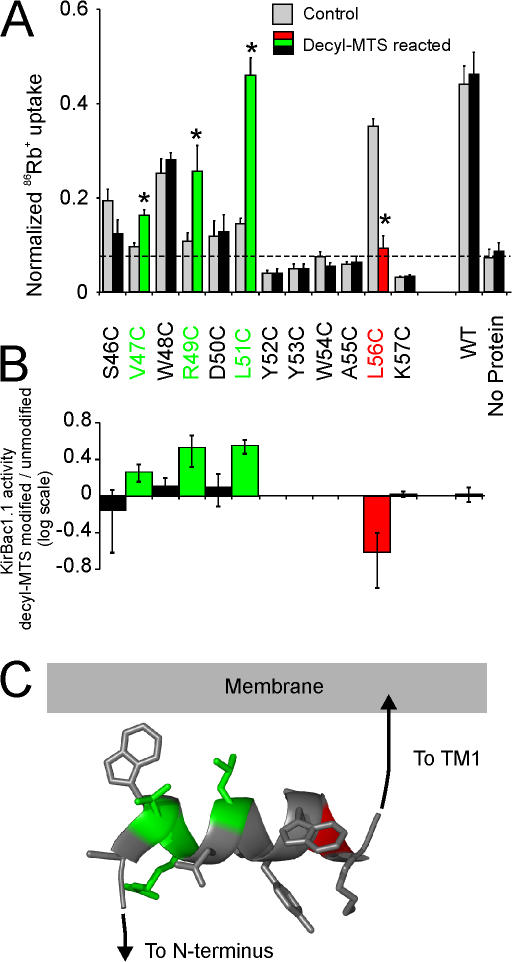
Figure 3.
KirBac1.1 channel activity modulated by covalent attachment of hydrophobic decyl anchors to the slide-helix. (A) Rubidium uptake of KirBac1.1 cysteine mutants (gray bars), or mutants reacted with decyl-MTS (red/green/black bars). KirBac1.1 protein was incubated with 100 μM decyl-MTS for 30 min before incorporation into liposomes. Significantly altered activity is indicated by asterisks (n = 3–6; *, P > 0.05, versus unmodified). (B) Log ratio of channel activity before and after modification with decyl-MTS. Mutants that have no channel activity before and after modification (as compared with liposomes) are excluded. Error bars represent 95% confidence interval. (C) Ribbon model of the KirBac1.1 slide-helix, with side chains shown in stick format. Residues are colored either green or red to represent increased or decreased channel activity after decyl-MTS modification, or as gray for mutants that had unchanged or no activity.
RESULTS AND DISCUSSION
Charged Residues in the Slide-Helix Control Channel Activity
To probe the role of the slide-helix in controlling channel activity, we systematically mutated each slide-helix residue in KirBac1.1 to cysteines, and examined the consequences of these mutations and subsequent in situ modification on channel activity. Of initial interest were the positively charged residues R49 and K57, since positive charges in this vicinity might regulate channel activity by interaction with negatively charged headgroups on the membrane lipids, or with negative charges in the cytoplasmic domain. Purified wild-type KirBac1.1 (which is cysteineless) (Fig. 1 B) shows robust channel activity when reconstituted into 3:1 POPE:POPG membranes (Enkvetchakul et al., 2004) (Fig. 2), but when R49 and K57 were mutated to cysteine, reconstituted KirBac1.1[R49C] and KirBac1.1[K57C] channel activity was negligible (Fig. 2). However, activity was restored following covalent modification by the positively charged MTSEA+, or MTSET+ (Fig. 2), indicating that a positive charged side chain is necessary and sufficient for channel activity. By contrast, channel activity was not restored by negatively charged MTSES or neutral methyl-MTS (Fig. 2). These results are entirely consistent with the side chains of R49 and K57 interacting electrostatically with negative charges located either within the channel or with negatively charged lipid headgroups to maintain channel activity.
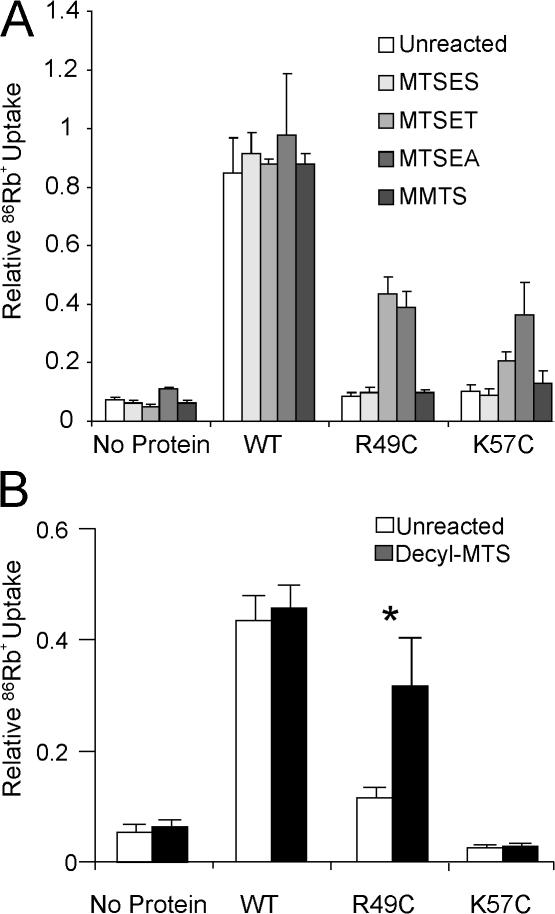
Figure 2.
(A) Rescue of mutant KirBac1.1 channels R49C or K57C, by restoration of positive charges after modification with MTSEA+ or MTSET+, but not with MTSES− or MMTS. (B) Rescue of R49C channel activity after modification with decyl-MTS. In both panels, 86Rb+ uptake was measured at 2 min in liposomes reconstituted with purified KirBac1.1 protein. Proteins were incubated with 100 μM MTS reagents for 30–60 min before incorporation into liposomes. Rubidium uptake was normalized to maximal uptake as measured by valinomycin (n = 3–6; *, P > 0.05, versus unmodified).
Lipid Tethering Can Rescue Charge Neutralization
Control of gating by channel–membrane interactions has been postulated for multiple eukaryotic channels (Enkvetchakul and Nichols, 2003; Suh and Hille, 2005). Implicit in this is the simple, but untested, concept that electrostatic “anchoring” of cytoplasmic, nontransmembrane domains to the membrane provides the energetic push or pull to stabilize open or closed states. We hypothesized that if a physical interaction of the slide-helix with the lipid membrane is needed for channel activity, then tethering of the slide-helix through hydrophobic anchors that favorably partition into the lipid membrane should have the same effect. The cysteine mutation allows us to take advantage of MTS reagents to attach just such a hydrophobic anchor. Modification by alkyl MTS reagents attaches a long hydrophobic side chain to the cysteine, which in turn will be expected to partition into, and hence “tether” the side chain to, the lipid membrane. KirBac[R49C] protein was modified by decyl-MTS before reconstitution into liposomes. A striking rescue of R49C activity was indeed achieved by modification with decyl-MTS (Fig. 2 B).
We then systematically mutated each residue of the slide-helix to cysteine to assess the effect on channel activity of attaching a lipid tether at different points along the length of the slide-helix. Cysteine substitution was tolerated at many residues, without loss of channel activity, but activity was abolished for nearly all residues in the N-terminal end of the helix (residues 52–55 and 57, Fig. 3 A). We then examined channel activity after modification of these residues with decyl-MTS before reconstitution into liposomes. Decyl-MTS modification caused significant channel activation at the predicted exposed residues V47C, R49C, and L51C in the N-terminal segment of the slide-helix (Fig. 3). Strong inhibition by decyl-MTS modification was seen for L56C, which, in contrast to the activated residues, is normally buried within the protein interior at the C-terminal end of the slide-helix (Fig. 3).
Channel Activity Is Proportional to Degree of Tethering
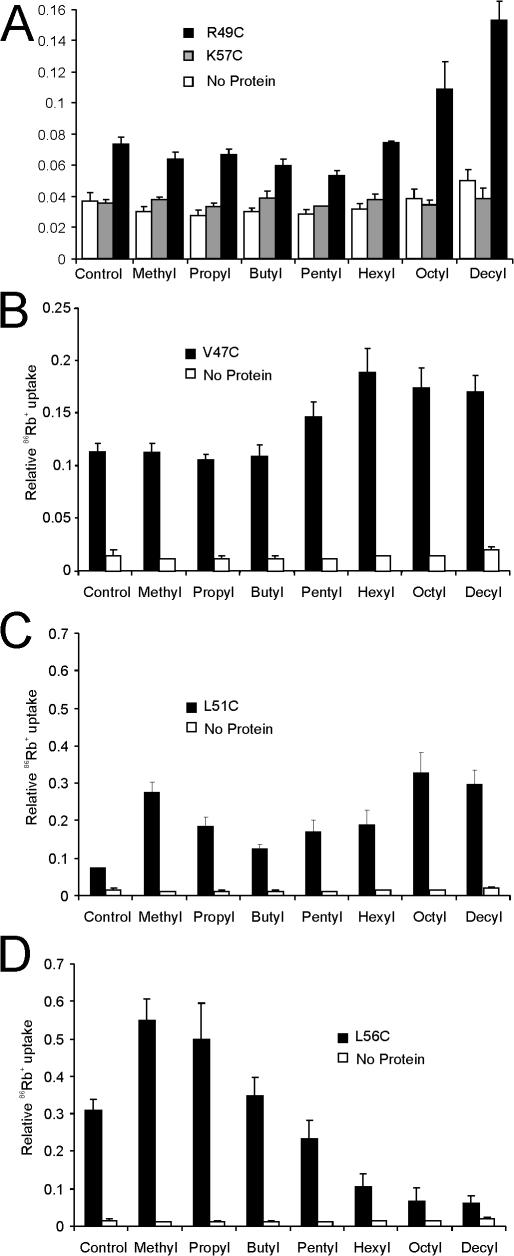
If tethering of residues on the slide-helix to the lipid membrane is indeed responsible for modulation of channel activity, this effect is expected to vary with the hydrophobicity of the attached tether. Tethers with increased hydrophobicity are expected to partition more strongly into the lipid membrane, in proportion to the length of the attached alkyl chain. In accordance with the prediction, alkyl-MTS modification of R49C (Fig. 4 A) or V47C (Fig. 4 B) demonstrated an essentially monotonic increase in channel activity with increasing chain length. In contrast, no effect was seen with alkyl-MTS modification of K57C (Fig. 4 A), consistent with the notion that lipid tethering of residue 57 does not promote channel opening. L51C (Fig. 4 C) and L56C (Fig. 4 D) demonstrated a more complex behavior. L51C exhibited progressively greater activation with alkyl chains longer than butyl, consistent with channel activity depending on the degree of lipid tethering of this residue to the lipid membrane (Fig. 4 C). However, weak activation of channel activity is also seen in both L51C and L56C with shorter (methyl and propyl) chains. This might be expected, however, considering that the native residue is leucine, and addition of a short alkyl chain will effectively restore the hydrophobic bulk normally provided by the leucine side chain. Longer chain alkyl-MTS modification resulted in monotonic inhibition of L56C, indicating that tethering of this residue at the C-terminal end of the slide-helix causes closure of the channel. In each case, for V47C, R49C, L51C, and L56C, progressive activation or inhibition was seen, dependent on the length of the attached alkyl chain and on the increased octanol-water partition coefficient of longer alkyl groups (Khadikar et al., 2003). This argues strongly that the effect on channel activity is directly proportional to the ability of the alkyl side chain to partition into the lipid, and thereby to tether the residue to the membrane.
Figure 4.
KirBac1.1 channel activity dependence of alkyl side chain length. Purified KirBac1.1 channel activity was measured in a rubidium uptake assay for (A) R49C, K57C, (B) V47C, (C) L51C, or (D) L56C. Proteins were incubated with 100 μM alkyl-MTS reagent for 30 min before incorporation into liposomes.
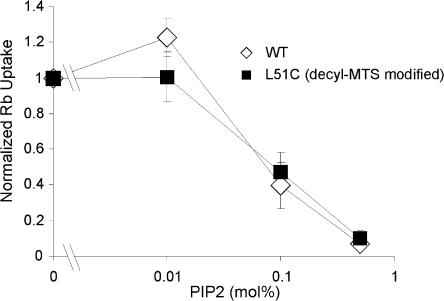
PIP2 is a potent inhibitor of KirBac1.1 channel activity (Enkvetchakul et al., 2005), and as hypothesized for eukaryotic Kir channels, PIP2 may interact through its negatively charged headgroup with residues in the N and C termini. We investigated the effect of lipid tethering of the slide-helix to the membrane on the potency of PIP2 inhibition of KirBac1.1 channel activity, and focused on decyl-MTS–modified L51C, which demonstrated the greatest degree of activation (Fig. 3 A). L51C was incubated with decyl-MTS before reconstitution, and the activity of L51C and WT KirBac1.1 channels were measured in liposomes formed with varying concentrations of PIP2. The effect of lipid tethering of L51C did not alter PIP2 sensitivity, and both WT and L51C were half-maximally inhibited by membrane PIP2 at ∼0.1 mol% (Fig. 5).
Figure 5.
PIP2 inhibition of KirBac1.1 WT or L51C channel activity. Purified KirBac protein was reconstituted into liposomes formed with varying percentage of PIP2. Uptake was measured at 30 s. L51C protein was incubated with 100 μM decyl-MTS for 30 min before incorporation into liposomes.
Role of the Slide-Helix in Kir Channel Gating
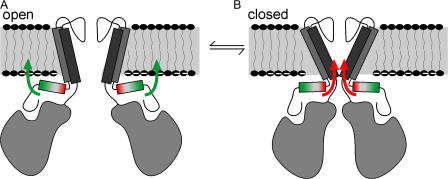
The KirBac1.1 crystal (Kuo et al., 2003) provides a structural model for understanding the molecular basis of gating in Kir channels as a whole. In this structure, KirBac1.1 channels are presumed to be in the closed state, with the central ion permeation pathway occluded by the side chain of the phenylalanine at position 146. Gating has been proposed to occur by coupling of the movement of the cytoplasmic domains to the transmembrane domains through the amphipathic slide- helices, which are predicted to lie along the plane of the lipid membrane. Consistent with this idea, mutation of residues in the slide-helix region of eukaryotic Kir channels can have a profound effect on channel gating, and can underlie human disease (Schulte et al., 1999; Plaster et al., 2001; Schulze et al., 2003; Gloyn et al., 2004). Potentially this results from altering slide-helix interactions with phospholipid headgroups of the lipid membrane (Plaster et al., 2001; Schulze et al., 2003). However, the complex system of the eukaryotic cell membrane precludes any rigorous assessment of channel– lipid interactions, and these ideas have remained essentially unaddressable. The above experiments show clearly that manipulations of the slide-helix can control KirBac1.1 channel activity, and that tethering of slide-helix residues to the membrane can control channel activity directly. Furthermore, the data suggest that specific movements of the slide-helix are involved. KirBac1.1 channels are activated by lipid tethering of residues in the N terminus of the slide-helix to the membrane by addition of alkyl side chains, and are inhibited by lipid tethering of a normally buried residue at the C terminus of the slide-helix. Based on this, we propose that slide-helix movements are involved in Kir channel gating (Fig. 6), whereby movement of the N- or C-terminal ends with respect to the lipid membrane results in increased stability of the open or closed state of the channel, respectively. The slide-helix is intimately linked with the cytoplasmic C-terminal domain through a β-sheet association of the N terminus (Kuo et al., 2003). This makes it intuitively clear how movement of the cytoplasmic domain, through binding of ligands, could result in movement of the slide-helix, and we propose that binding of ligands to Kir channel cytoplasmic domains will be coupled to gating movements in the transmembrane domains through similar movement of the slide-helix.
Figure 6.
Cartoon model of the role of the slide-helix in Kir channel gating. Movements of the slide-helix with respect to the membrane are associated with increased stability of the open or closed state of the channel. (A) Tethering of the C-terminal end to the membrane, which may involve rotation, displacement, or tilting (green arrows), facilitates opening of the channel. In contrast, (B) tethering of the C-terminal end (red arrows) favors the closed state of the channel.
We have shown that the positively charged R49 residue is necessary for channel activity, and that restoration of charge in the R49C mutant by MTS modification rescues activity. We suggest that R49 may interact with membrane phospholipids, consistent with the finding that lipid tethering of this residue and of nearby residues can activate the channel. Interestingly, primary sequence alignment of the KirBacs reveals that the position homologous to R49 in KirBac1.1 tends to be conserved as a basic residue. However, KirBac3.1 has a neutral leucine or positively charged aspartate (depending on the alignment) at the homologous position, and is an inactive/low activity channel (Sun et al., 2006), consistent with the findings that loss of a positive charge at residue 49 results in decreased KirBac1.1 channel activity.
The residue homologous to position K57 of KirBac1.1, however, is poorly conserved within the KirBac family, and a positively charged residue at this position is not necessary for activity for other members of the family, at least as assayed by growth rescue in K auxotrophic bacteria (Sun et al., 2006). We speculate that this positively charged residue may be important for structural integrity of the channel through its interaction with other charged residues within the KirBac1.1 protein in addition to the lipid membrane.
The findings of this paper are not inconsistent with our previously proposed mechanisms of PIP2 inhibition (Enkvetchakul et al., 2005). PIP2 likely binds to several residues in the cytoplasmic domain of eukaryotic Kir channels, including residues located outside of the slide-helix in the N and C termini. This binding has been hypothesized to cause a generalized movement of the cytoplasmic domain toward the membrane, but the net effect is likely more complex, yielding a yet unpredictable motion of the slide-helix. Lipid tethering of the slide-helix to the membrane at residue L51C activates the channel and would be expected to antagonize PIP2 inhibition if PIP2 also acted through a slide-helix displacement. The fact that lipid tethering does not alter PIP2 sensitivity suggests that PIP2 may have alternate transduction pathways outside of the slide-helix.
Covalent modification by hydrophobic moieties is a recognized mechanism for protein modulation in general, and palmitoylation may serve to tether various proteins, including ion channels (Hurley et al., 2000; Gubitosi-Klug et al., 2005; Resh, 2006), to lipid membranes, thereby influencing their location within the cell (Zhang and Casey, 1996; Boccuni et al., 2000; Peters et al., 2004; Smotrys and Linder, 2004). Direct changes of protein function as a result of such interactions have received less consideration. The present study demonstrates that for one particular class of ion channel, interaction of a specific domain, the slide- helix, with the membrane can directly control channel activity, and may illustrate a general principle by which cytoplasmic domain interactions with the membrane control gating of other channel types (Hilgemann and Ball, 1996; Baukrowitz et al., 1998; Shyng and Nichols, 1998; Runnels et al., 2002; Rohacs et al., 2005; Suh and Hille, 2005).
Acknowledgments
This work was supported by National Institutes of Health grant HL54171 (to C.G. Nichols) and DK69424 (to D. Enkvetchakul).
Lawrence G. Palmer served as editor.
References
- Baukrowitz, T., U. Schulte, D. Oliver, S. Herlitze, T. Krauter, S.J. Tucker, J.P. Ruppersberg, and B. Fakler. 1998. PIP2 and PIP as determinants for ATP inhibition of KATP channels. Science. 282:1141–1144. [DOI] [PubMed] [Google Scholar]
- Boccuni, P., L. Del Vecchio, R. Di Noto, and B. Rotoli. 2000. Glycosyl phosphatidylinositol (GPI)-anchored molecules and the pathogenesis of paroxysmal nocturnal hemoglobinuria. Crit. Rev. Oncol. Hematol. 33:25–43. [DOI] [PubMed] [Google Scholar]
- Enkvetchakul, D., and C.G. Nichols. 2003. Gating mechanism of KATP channels: function fits form. J. Gen. Physiol. 122:471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enkvetchakul, D., J. Bhattacharyya, I. Jeliazkova, D.K. Groesbeck, C.A. Cukras, and C.G. Nichols. 2004. Functional characterization of a prokaryotic Kir channel. J. Biol. Chem. 279:47076–47080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enkvetchakul, D., I. Jeliazkova, and C.G. Nichols. 2005. Direct modulation of Kir channel gating by membrane phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 280:35785–35788. [DOI] [PubMed] [Google Scholar]
- Gloyn, A.L., E.R. Pearson, J.F. Antcliff, P. Proks, G.J. Bruining, A.S. Slingerland, N. Howard, S. Srinivasan, J.M. Silva, J. Molnes, et al. 2004. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N. Engl. J. Med. 350:1838–1849. [DOI] [PubMed] [Google Scholar]
- Gubitosi-Klug, R.A., D.J. Mancuso, and R.W. Gross. 2005. The human Kv1.1 channel is palmitoylated, modulating voltage sensing: identification of a palmitoylation consensus sequence. Proc. Natl. Acad. Sci. USA. 102:5964–5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgemann, D.W., and R. Ball. 1996. Regulation of cardiac Na+, Ca2+ exchange and KATP potassium channels by PIP2. Science. 273:956–959. [DOI] [PubMed] [Google Scholar]
- Hurley, J.H., A.L. Cahill, K.P. Currie, and A.P. Fox. 2000. The role of dynamic palmitoylation in Ca2+ channel inactivation. Proc. Natl. Acad. Sci. USA. 97:9293–9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadikar, P.V., D. Mandloi, A.V. Bajaj, and S. Joshi. 2003. QSAR study on solubility of alkanes in water and their partition coefficients in different solvent systems using PI index. Bioorg. Med. Chem. Lett. 13:419–422. [DOI] [PubMed] [Google Scholar]
- Kuo, A., J.M. Gulbis, J.F. Antcliff, T. Rahman, E.D. Lowe, J. Zimmer, J. Cuthbertson, F.M. Ashcroft, T. Ezaki, and D.A. Doyle. 2003. Crystal structure of the potassium channel KirBac1.1 in the closed state. Science. 300:1922–1926. [DOI] [PubMed] [Google Scholar]
- Peters, C., A. Wolf, M. Wagner, J. Kuhlmann, and H. Waldmann. 2004. The cholesterol membrane anchor of the Hedgehog protein confers stable membrane association to lipid-modified proteins. Proc. Natl. Acad. Sci. USA. 101:8531–8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaster, N.M., R. Tawil, M. Tristani-Firouzi, S. Canun, S. Bendahhou, A. Tsunoda, M.R. Donaldson, S.T. Iannaccone, E. Brunt, R. Barohn, et al. 2001. Mutations in Kir2.1 cause the developmental and episodic electrical phenotypes of Andersen's syndrome. Cell. 105:511–519. [DOI] [PubMed] [Google Scholar]
- Resh, M.D. 2006. Palmitoylation of ligands, receptors, and intracellular signaling molecules. Sci. STKE. 2006:re14. [DOI] [PubMed]
- Rohacs, T., C.M. Lopes, I. Michailidis, and D.E. Logothetis. 2005. PI(4,5)P2 regulates the activation and desensitization of TRPM8 channels through the TRP domain. Nat. Neurosci. 8:626–634. [DOI] [PubMed] [Google Scholar]
- Runnels, L.W., L. Yue, and D.E. Clapham. 2002. The TRPM7 channel is inactivated by PIP(2) hydrolysis. Nat. Cell Biol. 4:329–336. [DOI] [PubMed] [Google Scholar]
- Schulte, U., H. Hahn, M. Konrad, N. Jeck, C. Derst, K. Wild, S. Weidemann, J.P. Ruppersberg, B. Fakler, and J. Ludwig. 1999. pH gating of ROMK (K(ir)1.1) channels: control by an Arg-Lys-Arg triad disrupted in antenatal Bartter syndrome. Proc. Natl. Acad. Sci. USA. 96:15298–15303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze, D., T. Krauter, H. Fritzenschaft, M. Soom, and T. Baukrowitz. 2003. Phosphatidylinositol 4,5-bisphosphate (PIP2) modulation of ATP and pH sensitivity in Kir channels. A tale of an active and a silent PIP2 site in the N terminus. J. Biol. Chem. 278:10500–10505. [DOI] [PubMed] [Google Scholar]
- Shyng, S.L., and C.G. Nichols. 1998. Membrane phospholipid control of nucleotide sensitivity of KATP channels. Science. 282:1138–1141. [DOI] [PubMed] [Google Scholar]
- Smotrys, J.E., and M.E. Linder. 2004. Palmitoylation of intracellular signaling proteins: regulation and function. Annu. Rev. Biochem. 73:559–587. [DOI] [PubMed] [Google Scholar]
- Suh, B.C., and B. Hille. 2005. Regulation of ion channels by phosphatidylinositol 4,5-bisphosphate. Curr. Opin. Neurobiol. 15:370–378. [DOI] [PubMed] [Google Scholar]
- Sun, S., J.H. Gan, J.J. Paynter, and S.J. Tucker. 2006. Cloning and functional characterization of a superfamily of microbial inwardly rectifying potassium channels. Physiol. Genomics. 26:1–7. [DOI] [PubMed] [Google Scholar]
- Zhang, F.L., and P.J. Casey. 1996. Protein prenylation: molecular mechanisms and functional consequences. Annu. Rev. Biochem. 65:241–269. [DOI] [PubMed] [Google Scholar]