Abstract
ClC-0 is a chloride channel whose gating is sensitive to voltage, chloride, and pH. In a previous publication, we showed that the K149C mutation causes a +70-mV shift in the voltage dependence of ClC-0 fast gating. In this paper we analyze the effects of a series of mutations at K149 on the voltage and chloride dependence of gating. By fitting our data to the previously proposed four-state model for ClC-0 fast gating, we show which steps in fast-gate opening are likely to be affected by these mutations. Computational analysis of mutant ClC-0 homology models show electrostatic contributions to chloride binding that may partially account for the effects of K149 on gating. The analysis of gating kinetics in combination with the available structural information suggests some of the structural changes likely to underpin fast-gate opening.
INTRODUCTION
The structural changes involved in opening the ClC-0 fast gate are only partially understood. Several studies have attempted to identify residues or secondary structures that move during ClC-0 fast gating. Based on the prokaryotic CLC structures and mutations that greatly increase the open probability of ClC-0, Dutzler et al. (2003) suggested that the outward movement of E166, a residue on the extracellular side of the pore, is required to liberate a chloride-binding site in the pore (S ext) and to open the channel. By showing that inhibitors that bind from the cytoplasmic side and at sites distant from E166 can restore voltage-dependent gating to E166 mutants, Traverso et al. (2006) illustrated that other residues in the channel are likely to move during fast gating. Thus it is possible that E166 is not the sole element of the ClC-0 fast gate, and other residues are likely to play important roles in gating.
On the cytoplasmic side of the pore, several residues have been shown to affect fast-gate kinetics (Ludewig et al., 1997), the most well described being S123, K519, E127, and Y512 (Pusch et al., 1995; Ludewig et al., 1996; Middleton et al., 1996; Chen and Chen, 2003; Chen et al., 2003; Lin and Chen, 2003). Lin and Chen (2003) attempted to determine whether these residues move during fast gating by examining their state-dependent accessibility. They mutated these residues to cysteine and measured reaction rates of MTS reagents, using voltage or chloride to change the open probability. These data are difficult to interpret because accessibility to these residues can be affected by both the conformation of the protein and the chloride in the pore that could compete with the charged MTS reagents for access to these sites. Nonetheless the data support the hypothesis that there may be changes in the intracellular vestibule during fast gating, although there may not be a physical “gate” there.
Computational studies based on prokaryotic CLC X-ray crystal structures and ClC-0 homology models based on these crystal structures suggest additional residues that may have important roles in gating and permeation (Bostick and Berkowitz, 2004; Cohen and Schulten, 2004; Corry et al., 2004; Faraldo-Gómez and Roux, 2004; Miloshevsky and Jordan, 2004; Yin et al., 2004; Gervasio et al., 2006). K149 is one such residue, which is both strictly conserved (Fig. 1 A) and has been predicted to have a significant effect on permeation and gating (Corry et al., 2004; Faraldo-Gómez and Roux, 2004). In the prokaryotic structures, K131 (equivalent to K149 in ClC-0) does not directly coordinate bound chloride, but is part of a hydrogen bond network with G106 and S107, two residues that directly interact with chloride in the pore and lie in between two chloride-binding sites (S cen and S int) (Fig. 1, B–D). If the prokaryotic structures are similar to a closed state of ClC-0 (Estevez et al., 2003; Lin and Chen, 2003; Engh and Maduke, 2005), then these residues (G122 and S123 in ClC-0) must move in order for the channel to open so that chloride can pass between S cen and S ext. Thus, K149 is likely to play a key role in structural rearrangements required for ClC-0 to open. Moreover, electrostatic calculations indicate that the charge on K149 has a significant influence on the electrostatic profile of the pore (Faraldo-Gómez and Roux, 2004), suggesting that it is likely to play a key role in gating and permeation.
Figure 1.
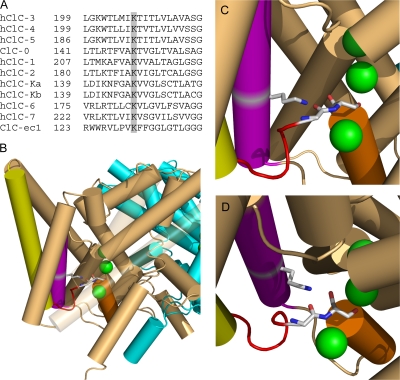
K149 conservation and structure. (A) K149 is highly conserved. ClC-0 (Torpedo), ClC-ec1 (E. coli), and the nine human CLCs were aligned using Clustal-W. The alignment of helix E is shown with K149 and equivalent positions shaded. (B–D) K131 interacts with chloride-coordinating residues on loop CD. (B) Structure of ClC-ec1 (1OTT) as viewed from within the membrane with the extracellular space at the top. Three bound chloride ions (from top to bottom, bound to sites S ext, S cen, S int) are shown in green. Subunits A and B are shown in light brown and cyan, respectively. Selected secondary structures on subunit A are colored as follows: helix C, yellow; loop CD, red; helix D, orange; helix E, purple. Helixes J and M and loop LM are transparent. To illustrate the interaction between K149 (on helix E) and residues on loop CD, residues corresponding to K149 (K131), G122 (G106), and S123 (S107) (ClC-0 sequence numbers, with ClC-ec1 numbers in parentheses) are shown as sticks and in CPK coloring. (C) Close up of image in (B). (D) Image in C rotated to better show G106 and S107. In both C and D helixes J and M and loop LM are removed. Structural images were created using Pymol (DeLano, 2002).
Previous work has shown that mutation at K149 causes significant effects on gating and permeation in ClC-0. Using cysteine scanning and mutagenesis, we screened over 50 sites predicted to be in or near the intracellular vestibule of ClC-0 (Engh and Maduke, 2005). Except for S123C, which severely disrupts voltage-dependent gating and kinetics, K149C caused the largest shift in voltage-dependent gating out of all the mutants tested. Gating kinetics and the shape of the open probability versus voltage curve for K149C look similar to wild type. The +70-mV shift in fast gating is the largest measurable mutation-induced shift yet recorded for ClC-0. This shift indicates that it is harder to open this mutant using voltage. Reaction of K149C with MTS reagents was slow (0.003 s−1) and caused almost complete inhibition of current. Although we were unable to determine whether this inhibition was caused by additional effects on gating or by the MTS reagents directly blocking the open pore, the slow rate of reaction suggests that K149 is likely to be buried (and not pore lining). Zhang et al. (2006) also assessed effects of mutation at K149. They showed that mutation to A, R, or L resulted in large shifts (+37 to +70 mV) in voltage-dependent gating as well as decreased selectivity for chloride relative to other anions. The magnitude of the effect of selectivity paralleled the magnitude of the effect on gating, consistent with the tight coupling between permeation and gating in ClC-0. Additionally, they showed that mutation at K149 affects the rate at which the pore-lining residue Y512C reacts with a negatively charged but not with a neutral MTS reagent. This result is consistent with the idea that the electrostatic field originating from the charge on K149 is significant at the position of Y512, a residue whose equivalent in the ClC-ec1 (Y445) directly coordinates a bound chloride ion (S cen) (Dutzler et. al., 2003). This suggests that K149 could affect permeation and gating via electrostatic interactions either with Y512 or with the permeating ions. It also confirms that K149 is protonated (i.e., positively charged), in spite of its location embedded within the transmembrane region of the channel.
In this paper we further describe the effects of K149 on the voltage and chloride dependence of fast gating in ClC-0. We show the range of effects caused by mutating K149 to 15 different amino acids. Of these, we characterized four mutants (R, M, S, L) more extensively by measuring the opening rate constant as a function of voltage and external chloride, and fitting this data to the four-state gating model described by Chen and Miller (1996). This allowed us to determine which steps in opening are most likely affected by the side chain at position 149. To understand these results in light of the available structural information, we created and analyzed ClC-0 homology models for both wild-type and K149L. Computational analyses of these models suggest that K149 affects chloride binding to all three chloride-binding sites in the pore, having the greatest effects on binding to S cen and S int. These analyses help us to begin to unravel the structural changes that underlie ClC-0 gating.
MATERIALS AND METHODS
Mutagenesis and Channel Expression
ClC-0 mutant constructs were made in a plasmid derived from the pBluescript vector (Stratagene) (Maduke et al., 1998). All constructs contained the point mutation C212S, which removes voltage-dependent slow-gate inactivation and has no other measurable effect on ClC-0 function (Lin et al., 1999). Mutations were introduced using PCR methods, and mutations were confirmed by sequencing through the cloning cassette. Channel expression and inside-out excised patch recording were performed as described in Engh et al. (see p. 335 of this issue).
Determining Open Probability and Gating Kinetics
The voltage protocols, data collection, correction, and analysis were performed as described in the companion paper Engh et al. (2007) to obtain open probability (P o), opening rate constant (α), and closing rate constant (β) as a function of external chloride and voltage (V).
The use of macroscopic recordings to derive opening rate constants (α values) is based on the assumption that the maximum P o is equal to one. Although single-channel recordings have shown this to be true for wild-type ClC-0 over a large range of external chloride concentrations (4–600 mM) (Chen and Miller, 1996), it is not certain if this is the case for all the mutants tested. However, this only poses a serious problem if the external chloride concentration affects P o, max; if P o, max is less than one but is unaffected by external chloride, the analysis will simply underestimate the effects of mutations on the opening rate constant.
Fitting α(V,[Cl]ext) to the Gating Models
The opening rate constant as a function of voltage and external chloride activity (α(V,[Cl]ext) was globally fit to the four-state model (Scheme 1) or the five-state model (Scheme 2) using Method 2 described in Engh et al. (2007). The errors on the gating parameters determined from these fits were also derived as previously described (Engh et al., 2007).
Online Supplemental Material
The supplemental material is available at http://www.jgp.org/cgi/content/full/jgp.200709760/DC1. Figs. S1–S3 depict the analysis of K149 mutations according to the five-state kinetic model. Figs. S4–S6 elaborate on the homology model analysis. Fig. S4 illustrates the effect of the dielectric constant on the calculated electrostatic contribution to chloride binding. Fig. S5 shows the electrostatic interaction energies between the bound chloride ions and side chains. Fig. S6 depicts the results of the electrostatic calculations made on an ensemble of WT and K149 conformations derived by allowing the residues in the vicinity of the chloride binding sites to fluctuate in the 0.3–0.5-Å range.
RESULTS
Side-Chain Identity at K149 Affects the Voltage Sensitivity of the ClC-0 Fast Gate
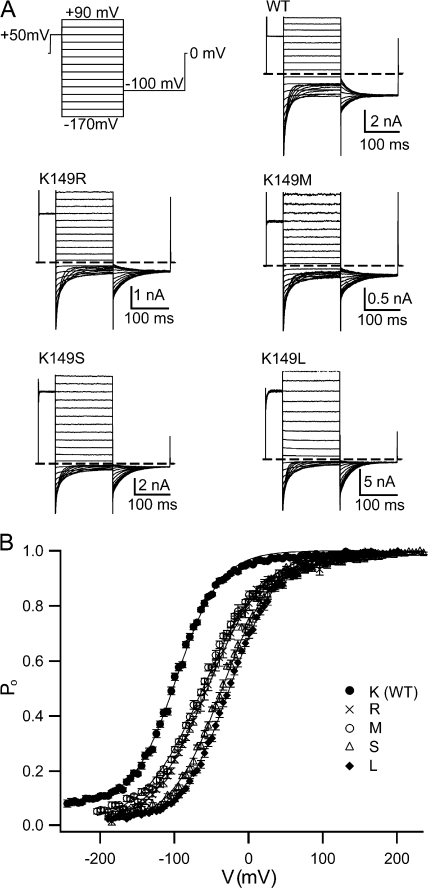
To better understand our previous observation that the K149C mutation causes a large right shift in the voltage dependence of ClC-0 fast gating, we examined (a) whether the effect is unique to cysteine substitution, and (b) what features of the side chain at K149 caused the observed effect. To address these questions, we made a series of mutations at K149 and measured their effects on ClC-0 fast gating. The effects of these mutations on voltage-dependent gating are shown in Fig. 2 and Table I. Large aromatic (Y, W) or negatively charged (D, E) side chains yielded no current, either because the channels were closed, had very small conductance, or were not expressed properly or transported to the plasma membrane. Amide-containing side chains (N, Q) yielded low current, which was detectable but insufficient for accurate characterization of fast gating. Of the 15 mutants tested, 9 gave enough current to allow for characterization of the voltage-dependent fast gating. All 9 mutations caused a right shift in the plot of open probability (P o) versus voltage (Fig. 2 B; Table I). The extent of this effect was side-chain dependent. It does not correlate with side-chain volume, pKa, or the ability to form hydrogen bonds (Table I). Consistent with the results presented in Zhang et al. (2006), the charge on K149 appears to be of some functional importance. The role of charge at K149 is not clear-cut, however; although negatively charged side chains are not tolerated at this position (K149D and E yield no current), many neutral side chains produce functional channels, and one neutral side chain causes an effect similar to that observed with a positively charged side chain of comparable length (compare K149R to K149M). When the results for whole series of mutations are considered, it is clear that the particular shape of the side chain at K149, and not the charge alone, is important for determining the voltage dependence of the ClC-0 fast gate. The extent of the effect corresponds to the side chain shape as follows: long linear (R, M) < short linear (A, G, S) < branched (T, L, N, Q) or bulky (W, Y). The one exception to this rule is K149C, since the shape of the cysteine side chain is similar to that of serine, and yet the V o lies in between that of the two branched side chains (T, L). Despite this one exception, it is reasonable to conclude that the specific shape of the side chain at position 149 plays an important role in determining the effect on fast gating.
Figure 2.
Effects of K149 side chain identity on voltage-dependent gating. (A) Example of a voltage pulse protocol (top left) and of current responses from a selection of K149 mutants. Such current responses were used to derive open probability (P o) as a function of voltage (V). (B). Plots of open probability were fit as previously described (Engh et al., 2007) (solid lines), and the resulting gating parameters are shown in Table I.
TABLE I.
Voltage-dependent Gating of K149 Mutants
| Side chain at 149 | V o | z | P min | Volume | pKa | H-bond donors | n |
|---|---|---|---|---|---|---|---|
| mV | Å3 | ||||||
| WT(K) | −95 ± 1.1 | 0.90 ± 0.01 | 0.070 ± 0.008 | 167 | 10.4 | 3 | 25 |
| M | −56 ± 1.7 | 0.66 ± 0.02 | 0.017 ± 0.005 | 167 | 18 | ||
| R | −55 ± 2.1 | 0.71 ± 0.02 | 0.015 ± 0.005 | 194 | ∼12 | 5 | 20 |
| A | −44 ± 0.7 | 0.84 ± 0.01 | 0.014 ± 0.004 | 90 | 13 | ||
| G | −43 ± 1.2 | 0.71 ± 0.02 | 0.009 ± 0.005 | 65 | 20 | ||
| S | −37 ± 1.0 | 0.82 ± 0.01 | 0.031 ± 0.007 | 95 | 1 | 45 | |
| T | −33 ± 1.0 | 0.86 ± 0.01 | 0.014 ± 0.003 | 122 | 1 | 16 | |
| C | −30 ± 1.1 | 0.84 ± 0.02 | 0.017 ± 0.006 | 103 | 9.1–9.5 | 13 | |
| L | −24 ± 1.3 | 0.85 ± 0.02 | 0.028 ± 0.007 | 164 | 13 | ||
| N, Qa | 125, 149 | 2 | |||||
| D, Eb | 117, 142 | ∼4.5 | 0 | ||||
| Wb | 228 | 1 | |||||
| Yb | 197 | 9.6 | 1 |
Plots of apparent open probability (P o) versus voltage were fit as described in Materials and methods to derive the voltage at the midpoint in the voltage-activation curve (V o), the apparent gating charge (z), and the minimum open probability (P min). The mean ± SEM is given for n patches. K149 mutants are listed in order of the severity of their effect along with the corresponding amino acid side-chain volume (V) (from Table 7 of Tsai et al, 1999), side-chain pKa, and number of side-chain hydrogen bond donors.
Mutants that expressed but had low current.
Mutants that did not give measurable current.
Despite the significant shifts in voltage-dependent gating caused by the K149 mutations, the other features of fast gating are preserved. For those mutants whose gating can be characterized, the shape of open probability (P o) versus voltage (V) plots is similar to wild type (Fig. 2 B and some mutants not depicted). Furthermore, these mutants are all activated by external chloride ([Cl]ext) in a fashion similar to wild type (compare Fig. 3 to Fig. 1 in Engh et al., 2007). These similarities indicate that in the mutant channels the fast gate is operating like wild type, but with altered energetics for one or more of the steps in the gating mechanism.
Figure 3.
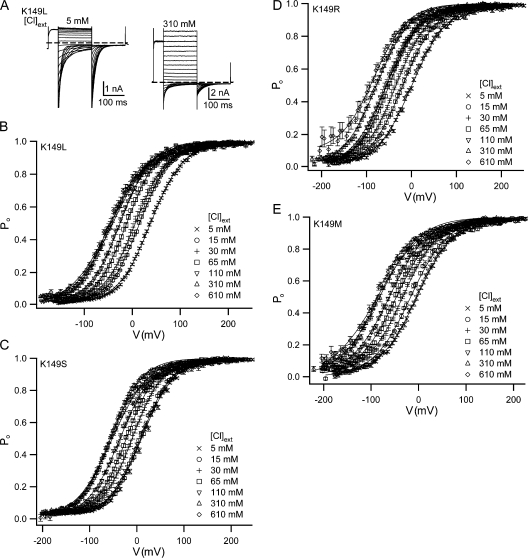
K149 mutants have chloride dependences similar to wild type. (A) Examples of K149L currents in 5 and 310 mM external chloride. Voltage protocols used were similar to the example shown in Fig. 2 A. Such current responses were used to derive open probability (P o) versus voltage (V) (B–E). P o(V) plots were fit as previously described (Engh et al., 2007) (solid lines), and gating parameters are shown in Table S1. Points show mean values ± SEM for 5–21 patches for all external chloride concentrations except 610 mM, for which 3–9 patches were used.
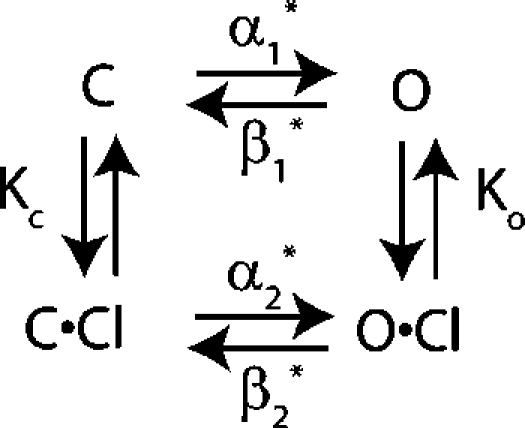
The Four- and Five-State Models for Fast Gating
What steps are involved in fast gating? Chen and Miller (1996) proposed a five-state model for fast-gate opening in ClC-0 (Scheme 2). This model is an extension of a simpler four-state model (Scheme 1).
 |
(SCHEME 1) |
 |
(SCHEME 2) |
Both these models explain the effect of voltage and external chloride on the kinetics of fast-gate opening. In the four-state model, the channel can open either before or after extracellular chloride has bound. Opening before chloride is bound is hyperpolarization activated; opening after chloride has bound is depolarization activated. In the five-state model, the channel can open via the hyperpolarization-activated pathway even after chloride has bound. In the companion paper (Engh et al., submitted), we showed that the wild-type data fit well to either the four- or five-state models. In this paper, we aim to understand how K149 affects gating by determining what step(s) in the channel opening process are affected by mutations at K149. To do this we measured opening rate constants for a series of K149 mutants and analyzed them using the conceptually simpler four-state model. Because the models are similar to one another (both have a chloride-binding step, hyperpolarization- activated opening, and depolarization-activated opening), the results are qualitatively similar. Results for the five-state model are in the online supplemental material (Figs. S1–S3, available at http://www.jgp.org/cgi/content/full/jgp.200709760/DC1).
As described in the companion paper for wild-type ClC-0 (Engh et al., 2007), plotting the opening rate constant (α) as a function of voltage (V) gives a nearly V-shaped curve (Fig. 4), like a chevron plot (Chan and Dill, 1998). The lefthand arm of the V is governed by the hyperpolarization-activated step (the α1 * step in the four-state model). The steepness of this arm is described by the z-value (z 1 *), and the position described by α1 *(0), the α1 * value at 0 mV. The righthand arm of the V is dominated by both the chloride-binding step (the K c step) and the depolarization-activated step (the α2 * step). The steepness of this righthand arm depends on both z 2 * and z c, and the position on α2 *(0), the α2 * value at 0 mV. How much this arm shifts in response to chloride is determined by K c, the chloride dissociation constant.
Figure 4.
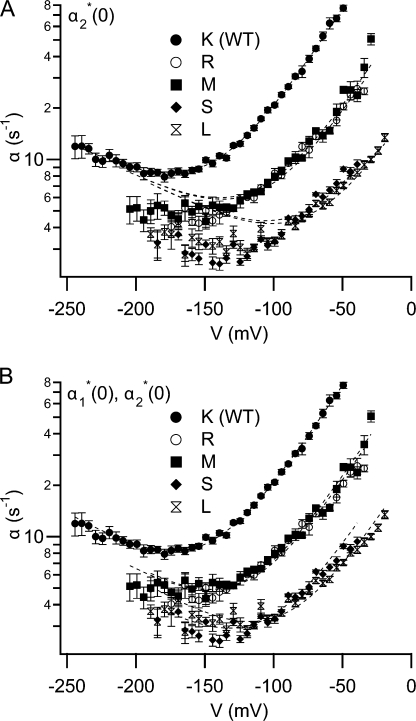
Effects of K149 mutations on the opening rate constant: initial fits to the four-state model. The voltage dependence of the opening rate constant (α) for wild type and selected K149 mutants at 110 mM external chloride were fit to the four-state model shown in Scheme 1. Fits are shown as dashed lines. Wild-type data were fit by letting all six parameters in the model vary. For mutant data, fits were derived by letting vary only α2 *(0) (A), or only α1 *(0) and α2 *(0) (B), while holding all other parameters at their wild-type best-fit values. The fits in (B) yielded α2 *(0) values for wild type, K149R, M, S, L, in s−1: 501, 135, 143, 55, 39; and corresponding α1 *(0) values: 0.96, 0.77, 0.74, 0.52, 0.67. For each voltage, the opening rate constant (α) was derived using data from at least five patches. These opening rate constants were averaged and the error bars show the SEM.
Predicting which Step(s) Are Affected by Mutations at 149
By inspection of the shifts in the α(V) plots caused by mutations at K149, it can be seen which gating parameters might be affected. For all the mutants, although the slope of the righthand arm of the α(V) curve is similar to wild type (Fig. 4), the positions of both arms are shifted relative to wild type, and the slope of the lefthand arm may also be affected. In the four-state model, the slope and position of the lefthand arm are determined largely by z 1 * and α1 *(0), respectively; the slope of the righthand arm by both z 2 * and z c, and the position by both α2 *(0) and K c(0). Therefore, changes in the slope of the lefthand arm suggest that z 1 * has changed; changes in the position of the lefthand arm suggest that α1 *(0) has changed; and a shift in the position of the righthand arm may either be caused by changes in α2 *(0) or K c(0) or both. Fig. 5 illustrates that increasing the external chloride concentration can compensate for the effects of these mutations on the position of the righthand arm of the curve. This shows that the mutations may affect chloride affinity, or if they affect α2 *(0), that these effects can be compensated for by adding chloride, since in the four-state model chloride binding precedes the α2 * step.
Figure 5.
Increasing the external chloride concentration can compensate for the effects of K149 mutations. For mutants K149M, R, S, and L, seven external chloride concentrations were tested. For each mutant the chloride concentration that yielded the α(V) curve most similar to wild type at 5 mM external chloride is shown.
To further examine the effect of the K149 mutations, we fit the α(V) plots to see if the mutation-induced shifts could be accounted for by changing the value of only one parameter in the four-state model (and holding the other five parameters to their wild-type values). For all parameters tested, the fits do not match the mutant data (see example where α2 *(0) was allowed to vary, Fig. 4 A). Letting two parameters vary yielded better fits, with the best fits resulting when the two parameters were from the following combinations of steps (in the four-state model): (a) α1 * and α2 *, or (b) α1 * and K c. Fig. 4 B shows one example, where α2 *(0) and α1 *(0) were allowed to vary. In every case, good fits to the mutant data required an ∼10-fold change in the wild-type value of the α2 *-step or K c-step parameter and a less than twofold change in the α1 * step parameter. This suggests that mutations at K149 affect at least two of the steps in the four-state model, and have a greater effect on the depolarization-activated opening (the α2 * and K c steps) than on the hyperpolarization-activated opening (the α1 * step).
Depolarization-activated Opening Is Affected by K149 Mutations
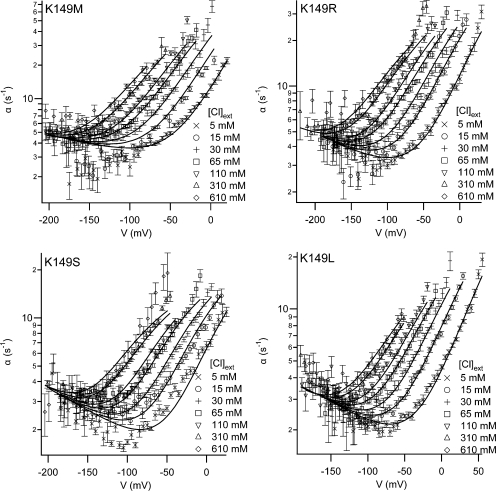
To more accurately determine which parameters in the gating models are affected by mutations at K149, we selected four mutants (M, R, S, L), whose effects on voltage-dependent gating span the range of severity (for those mutants where gating is measurable), and we measured their dependences on external chloride concentration ([Cl]ext). For each mutant, we determined the opening rate constant at seven external chloride concentrations (in mM: 5, 15, 30, 65, 110, 310, 610) over a range of voltages, and then globally fit the data to the four-state model (Fig. 6). The best-fit values for the parameters are shown in Fig. 7, along with errors determined as previously described (Engh et al., 2007). These errors are not simply the errors calculated by the fitting algorithm but are 95% confidence limits determined independently.
Figure 6.
Effects of K149 mutations on the opening rate constant: external chloride dependence and fits to the gating model. The opening rate constant (α) as a function of voltage (V) and external chloride ([Cl]ext) is shown for mutants K149 M, R, S, and L. Each dataset was globally fit to the four-state (Scheme 1) model proposed by Chen and Miller (1996) (solid lines). The resulting parameter values are shown in Fig. 7. For each condition (V, [Cl]ext), the opening rate constant (α) was derived using data from at least five patches. These opening rate constants were averaged and the error bars show the SEM.
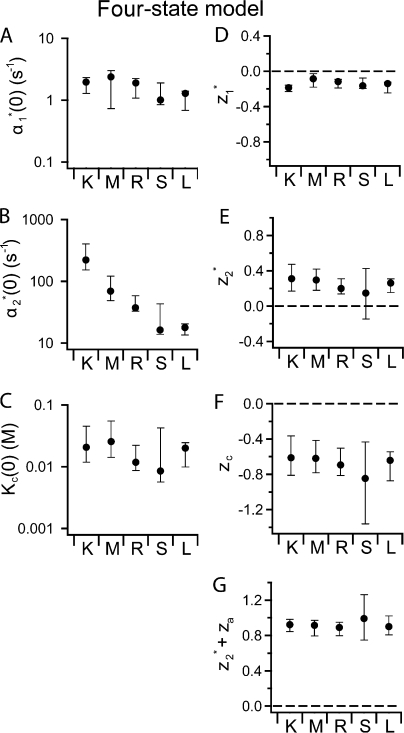
Figure 7.
Best-fit values for the four-state model gating parameters. (A–F) The opening rate constant as a function of voltage and chloride (α(V, [Cl]ext)) was globally fit to the four-state model (see Fig. 6), and the best fit values for the six parameters in the model are plotted for wild type (K) and the four K149 mutants (M, R, S, L). K c(0) is given in terms of chloride activity. (G) The total charge that moves during depolarization-activated gating (the K c step followed by the α2 * step) was calculated by adding z 2 * (the charge that moves during the α2 * step) to z a (the charge that moves during the K c step, which is equal to −z c). Error bars show the 95% confidence limits determined as described in Engh et al. (2007).
Although our preliminary analysis shown in Fig. 4 indicated that mutations at 149 likely affect two parameters in the gating model, our error analysis shows that for our datasets only one parameter is significantly affected, namely α2 *(0) (Fig. 7). Other parameters may also be affected (as suggested by our preliminary analysis in Figs. 4 and 5); however, if such effects exist they must be smaller than the error in the fits. This means that the K149 mutants mainly affect gating by affecting the barrier to depolarization-activated opening.
Interdependencies between Parameters Contribute to Error in their Determination
As discussed in the companion paper (Engh et al., 2007), fits to either gating model yield some unexpected interdependencies between some of the parameters. Specifically, all the parameters associated with the K c step and depolarization-activated opening step (α2 *) are interdependent with one another (α2 *(0)-K c(0), z 2 *-K c(0), α2 *(0)-z c, and z 2 *- z c). This partially accounts for the large error bars on these parameters (Fig. 7). The interdependencies seen with the K149 mutant data (unpublished data) are similar to those observed with the wild-type data (Engh et al., 2007). This means that parameters that covary with α2 *(0) may also be affected by mutations at 149. Such effects could be hidden within the large error bars on these covarying parameters (K c(0) and z c). Therefore, although the major effect of mutations at K149 on fast gating is on α2 *(0), which decreases by ∼10– 40-fold, K c(0) may also be affected up to ∼10-fold in either direction.
Effect of K149 on the Gating Charge
Since z 2 * and z c are also interdependent, changes in these parameters may also be hidden within the large error bars. However, a closer look at the data indicates that K149 mutations are not affecting voltage-dependent gating by affecting the gating charge. By considering together the z 2 * and z c values (derived from globally fitting the opening rate constant as a function of voltage and chloride), it can be seen that K149 mutations do not affect the total amount of charge that moves during depolarization-activated gating. As mentioned above, z 2 * and z c are interdependent in fits to wild type and all the mutant datasets. In accord with this interdependence, we see that upon mutation of K149 these two parameters covary; when moving from left to right across the series of mutants shown in Fig. 7 (E and F), z 2 * and z c go up and down together. This covariance means that the total charge that moves in the K c plus α2 * steps remains unchanged. To illustrate this, we need to point out that K c is the equilibrium constant for chloride dissociation, and z c is therefore the voltage dependence of the chloride dissociation equilibrium. To determine the charge that moves during depolarization-activated gating (the K c step followed by the α2 * step), we changed the sign on z c to convert it to z a, the charge that moves during chloride association before channel opening. We then added z 2 * and z a and found that this sum was always very near to one (Fig. 7 G), and that the uncertainty of this sum is much less than the uncertainty of the individual parameters. Therefore, for wild type and the four K149 mutants, the total charge moved during depolarization-activated fast gating, z 2 * + z a, is always ∼1. The value of ∼1 is expected based on the analysis of the P o(V) curves, which indicates that the z value for gating is ∼1 for wild type and all the mutants (Table I). Therefore, although α2 *(0) is interdependent with z 2 * and z c, we conclude that K149 mutations do not affect the gating charge of depolarization-activated fast gating.
Predicted Effects of K149 Mutations on Pore Electrostatics
Since many of the mutations tested are charge neutralizing, we wondered if the effects on gating were partially due to changes in electrostatics in the pore. In the companion paper (Engh et al., 2007) we described how we used the ClC-ec1 X-ray crystal structures to build homology models of ClC-0 and analyze the electrostatics of chloride binding and movement in the pore. We used a similar procedure to create a K149L ClC-0 homology model inspired by the E148Q mutant ClC-ec1 structure (Dutzler et al., 2003), a structure that has been proposed to resemble the conducting state of ClC-0. In this ClC-0 homology model, E166 (the E148 equivalent) is protonated and rotated out of the S ext chloride-binding site. The effects of this manipulation on our electrostatic calculations are discussed in the companion paper (Engh et al., 2007), and are the same for both the wild-type and K149L backgrounds. Using the E148Q ClC-ec1 structure as a basis for our K149L homology model has the benefit of allowing us to make calculations for the S ext site.
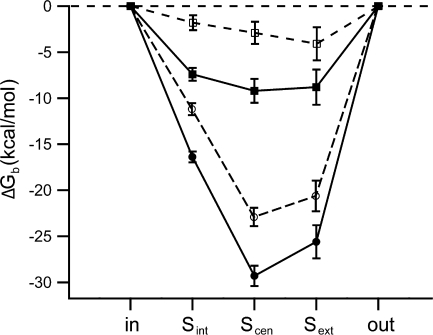
Our analysis of the K149L homology models suggests that this mutation affects the electrostatic component of chloride binding (Fig. 8, compare ΔG b for wild type and K149L). The K149L mutation causes a significant decrease in the electrostatic component of chloride affinity for all three binding sites, S int, S cen, and S ext, and has the greatest effect on the affinity for Si nt and S cen. K149L has lower affinity for chloride than wild type regardless of the number of chlorides in the pore (Fig. 8, compare the difference between the open and closed circles and the difference between the open and closed squares) and regardless of the dielectric constant of the protein used in the calculations (see Fig. S4). Although the K149L mutation affects the calculated chloride binding affinity for all three binding sites, this mutation does not affect the electrostatic interactions between chloride and other residues in the channel (see Fig. S5).
Figure 8.
Predicted effects of K149 mutants on chloride binding to ClC-0: homology model analysis. (A) Mutations at K149 decrease the affinity of chloride. Poisson-Boltzmann theory was used to calculate the electrostatic contribution to the free energy of chloride binding (ΔG b) to each site in the pore (starting from the cytoplasmic side: S int, S cen, S ext), for wild type (filled symbols, solid lines) and K149L (open symbols, dashed lines) ClC-0 homology models. The binding of the first chloride to the pore is shown in circles and the binding of the third chloride (two chlorides are already bound) is shown in squares. The methodology for structural modeling and electrostatics analysis is analogous to that previously reported (Engh et. al., 2007), except that here the side chain of E166 was neutralized and rotated away of the pore, so as to accommodate a chloride ion in S ext. A value of 4 was used as the dielectric constant of the protein (see supplemental material for further analysis). Lines connect data points for ease in viewing, and error bars represent an estimate of the discretization error arising from the grid-based Poisson-Boltzmann calculations.
Our homology model analysis also indicates that K149L does not affect how the transmembrane field falls across the pore. We calculated the effect of K149L on the fraction of transmembrane potential a chloride ion traverses as it moves through the pore (V/V tm). This measurement is mostly sensitive to the shape of the pore and insensitive to the charges lining the pore. Since minimal adjustments were made to create these two homology models, and thus the shapes of their pores are similar, the V/V tm curve looks the same for K149L (unpublished data) and wild type (Fig. 6 A in Engh et al., 2007). This suggests that the calculated changes in the chloride binding affinities (Fig. 8) are not a result of changes in position of the chloride-binding sites with respect to the transmembrane field.
DISCUSSION
K149 Mutants Affect Gating by Affecting α2 *(0) and Possibly Kc(0)
We found that all the mutations at K149 we tested caused a rightward shift in the P o(V) curve for fast gating. Since the effects of K149 mutations on V o (Fig. 2 B; Table I) are largely due to changes in the opening rate α rather than the closing rate β (unpublished data), dissection of fast-gate opening provides insight into how the mutations affect gating. Our results suggest that mutations at K149 alter α by affecting the depolarization-activated arm of the gating models (C→C·Cl→O). In the four-state model, this corresponds to the K c and α2 * steps. Error analysis on the fits to the four-state model indicates that α2 *(0) is the only parameter significantly affected by mutations at K149, with changes in the range of up to ∼30-fold for the most severe mutations (Fig. 7). The large size of the error bars, which denote 95% confidence limits, indicates that mutations at K149 could also have smaller effects on chloride binding affinity, since K c(0) could vary up to ∼8-fold between mutants and wild type and still fall within the calculated error bars (Fig. 7 C). Likewise, mutations at K149 could have even smaller effects on hyperpolarization-activated fast-gate opening (the α1 * step), since α1 *(0) could vary up to two to threefold and still fall within the calculated error bars (Fig. 7 A). Whereas our data clearly show that mutations at K149 affect fast gating by slowing the rate constant of depolarization-activated opening, they do not show whether mutations at K149 affect chloride binding affinity or hyperpolarization-activated opening. Further work is needed to examine possible subtle effects of K149 mutations on these other two processes.
In the companion paper (Engh et. al., 2007), analysis of the interdependence of gating parameters in the four-state model suggests that the large size of the error bars on K c(0) is in part due to the interdependence of K c(0) and α1 *(0). The interdependence between the α2 * step and the K c step has two sources: (1) the lack of features in the α(V[Cl]ext) data needed to determine all the parameters uniquely and (2) the possibility that the chloride-binding step is not in rapid equilibrium, such that K c is not the microscopic chloride binding equilibrium, but rather an apparent chloride affinity, defined as K c = (α2 * + k −c)/k c, where k c and k −c are the microscopic chloride association and dissociation rate constants, respectively. K c corresponds to the microscopic chloride affinity (k −c/k c) only if k −c ≫ α2 *. Although ion-binding steps are often assumed to be in rapid equilibrium, no independent measurement of chloride binding kinetics in ClC-0 has been made. If the kinetics of chloride binding and of the α2 * step are similar, K c will necessarily be affected by changes in α2 *, even though the actual chloride-binding step may not be affected.
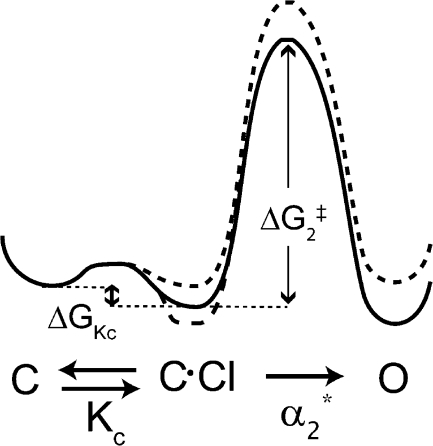
The effects of K149 mutants on α2 *(0) and possible effects on K c(0) are illustrated in the free-energy diagram shown in Fig. 9. This diagram only shows depolarization-activated fast gating. Since K149 mutants decrease the rate constant α2 *(0), the height of the α2 * step barrier (ΔG 2 ‡) is greater for the mutants (dashed line) than for wild type (solid line). Since K149 mutants may also increase or decrease the equilibrium constant K c(0), the difference between the C and the C·Cl states (ΔG Kc) could be either smaller or bigger for the mutants than for wild type.
Figure 9.
Hypothetical free energy diagram for depolarization-activated fast gating in wild-type (thick solid line) and K149 mutants (thick dashed lines). Since K149 mutants decrease α2 *(0), they increase the height of the C·Cl→O transition, ΔG 2 ‡. Since K149 mutants may also affect K c(0) (either lowering or raising the affinity for chloride, but most likely lowering), the change in free energy for the C to C·Cl transition, ΔG Kc, is different for the mutants than for wild type. This energy diagram is for a voltage that favors opening, so the open state (O) has lower free energy than the closed state (C). For the K149 mutants, this difference is smaller, since opening is less favorable.
Predicted Effects of Charge-neutralizing Mutations on Electrostatics of Chloride Binding
Our homology model analysis predicts that charge- neutralizing mutations at K149 (such as K149L) decrease the electrostatic contribution to chloride binding affinity at all three sites in the pore. If chloride binding is part of the mechanism by which mutations at K149 affect fast gating, then the effects of mutations at K149 on the electrostatics of chloride binding are likely to be relevant. As explained in the previous sections, chloride binding could be part of the mechanism of fast-gate opening if either (a) K c(0) is affected by mutations at K149 but this effect is hidden within the large error bars or (b) the kinetics of the various steps in the gating mechanism are such that one of the microscopic chloride-binding steps contributes to α2 *(0). If either of these is true, then electrostatic contributions to chloride binding could partially account for the effects of K149 on gating. This helps us in thinking about the results from the kinetic analysis, which suggest that mutations at K149 might either increase or decrease K c(0). Since our homology model analysis indicates a decrease in the electrostatic contribution to chloride binding, this suggests that an increase in K c(0) (corresponding to a decrease in the chloride binding affinity) is more likely.
The relevance of our electrostatic calculations also relies on the assumption that other contributions to the mutation-induced change in the chloride binding affinity do not counteract the electrostatic contribution. These additional contributions to the free energy of chloride binding include the nonpolar interactions between chloride and protein as well as the binding entropy. Assuming no significant changes in the structure and dynamics of the chloride binding sites, these nonelectrostatic contributions are likely to be comparable for the wild type and K149 mutants. As described in the supplemental material, we examined the effect of such small fluctuations in the structure on the calculated electrostatic contribution to chloride binding affinities. We found that the effect of the K149L mutation on the electrostatic contribution to chloride binding affinity is similar whether the single model or an ensemble of conformations (within 0.5 Å) is used in the calculations. However, further work is required to explore whether larger structural or dynamic changes contribute to the effect of K149 mutations on other contributions to chloride binding affinity.
Two Models for Depolarization-activated Fast-Gate Opening
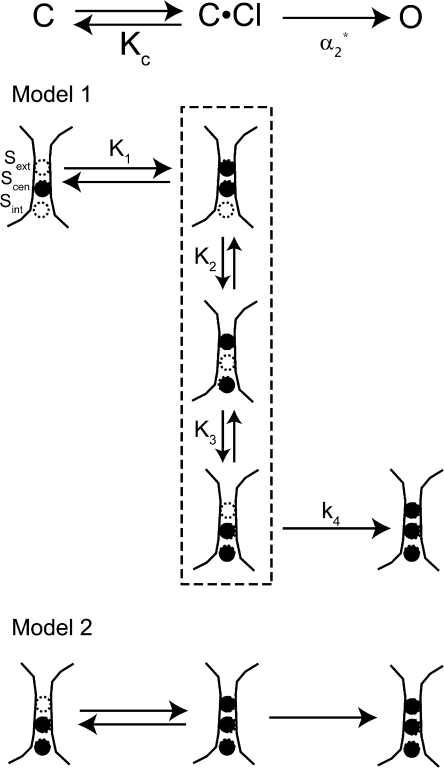
In the companion paper (Engh et al., 2007) we proposed two models for how chloride binds to and moves in the pore during depolarization-activated fast gating (Models 1 and 2, Fig. 10). In Model 1, the closed state before chloride binding (C) has only one chloride bound, shown in S cen because this site has the highest chloride binding affinity based on our electrostatic calculations. The microscopic chloride-binding step, with equilibrium constant K 1, involves chloride binding to S ext, and therefore must involve the rotation of the E166 side chain out of this site. The closed state after chloride binding consists of three microstates in equilibrium, each with two chlorides bound. The equilibrium constants K 2 and K 3 describe the movement of chloride between sites in the pore, and the K 3 step could also include the movement of the E166 side chain back into S ext. The second chloride-binding step, with rate constant k 4, could also include the rotation of the E166 side chain out of the pore. All of the steps in the model could involve other changes, including the binding, unbinding, or movement of protons, and/or protein structural rearrangements.
Figure 10.
Two models for chloride movement during depolarization-activated fast gating. As described in the companion paper (Engh et. al., 2007), kinetic and structural analyses were used to develop two possible models that describe chloride binding and movement in the pore during fast-gate opening. In Model 1, the dashed line outlines the microstates that are all part of C·Cl. K 1, K 2, and K 3 are microscopic equilibrium constants, and k 4 is a microscopic rate constant. In Model 2, the second step has to involve some conformational change that opens the channel but does not change chloride occupancy.
Model 2 is different than Model 1 in that the starting state (C) has two chlorides bound instead of one. The two bound chlorides are shown in S cen and S int, as seen in the structures of wild-type ClC-ec1. The chloride binding step (C→C·Cl) involves the movement of the E166 side chain out of the pore, and the binding of chloride to S ext. It could also involve other structural rearrangements in the protein. In Model 2 the second step (C·Cl→O) must involve protein structural rearrangements, since there is no change in chloride occupancy. Therefore, Model 1 and Model 2 differ not only in which chloride occupancy states correspond to the kinetically observed states (C, C·Cl, O), but also in that Model 2 requires additional structural rearrangements (in addition to chloride movement and E166 side-chain rotation) whereas Model 1 does not.
Hypotheses for How Mutations at K149 Affect Gating
These models provide two different frameworks for explaining how mutations at 149 can change α2 *(0) and indicate whether changes in K c(0) would also be expected. In Model 1, effects on K 1 would manifest themselves as effects on the calculated K c, but not as effects on α2 *. Since all the steps are coupled, depending on their relative rates, effects on K 2, K 3, or k 4 could manifest themselves as effects on K c or α2 * or both. In Model 2, effects on chloride binding affinity would only affect K c, but effects on the protein conformational change that occurs during the second step could affect α2 * alone or both α2 * and K c. Although without more information we cannot determine whether Model 1 or Model 2 is more likely, we can evaluate three simple hypotheses that explain how mutations at K149 affect α2 *(0) and possibly K c(0). In order for a hypothesis to be deemed satisfactory it must explain the effect of K149 mutations on α2 *(0). Since at this point no effects of K149 mutations on K c(0) have been shown definitively, a hypothesis does not have to explain possible effects of K149 on chloride binding affinity to be considered valid.
One hypothesis (Hypothesis 1) is that mutations at K149 affect gating by weakening chloride binding to S ext. Our homology model analysis supports the idea that K149L weakens chloride binding to S ext. This effect on chloride binding affinity is observed both when there is one and when there are multiple chloride ions in the pore (Fig. 8). Could a decrease in chloride affinity to S ext decrease α2 *(0)? For Model 1 (Fig. 10, top) the answer is yes because a decrease in chloride binding affinity to S ext would affect α2 * by affecting k 4. It would also affect K c by affecting K 1 and k 4. For Model 2 (Fig. 10, bottom) the answer is no because an effect on binding to S ext would only be expected to affect K c, and not α2 *. Therefore the hypothesis that mutations at K149 affect gating by affecting chloride affinity to S ext is reasonable for Model 1 but not Model 2.
A second hypothesis (Hypothesis 2) is that mutations at K149 affect gating by weakening chloride binding to S int and/or S cen. Our homology model analysis suggests that K149L weakens chloride binding to S cen and S int, both when there is one and when there are multiple chloride ions in the pore. Could a decrease in chloride affinity to S cen and/or S int decrease α2 *(0)? For Model 1 the answer is yes because a decrease in chloride binding affinity to S cen and S int would affect K 3 and K 2, respectively. Either of these effects would change α2 * and K c. For Model 2 the answer is no because the α2 * step does not involve binding to S cen or S int. K c(0) is also not expected to be affected because it also does not involve chloride binding to either of these sites. The hypothesis that mutations at K149 affect gating by affecting chloride affinity to S int and/or S cen is therefore reasonable for Model 1 but not Model 2.
A third hypothesis (Hypothesis 3) is that mutations at K149 affect gating by affecting some other protein conformational change that is required for opening. For example, mutations at K149 may affect the barriers to chloride movement between the binding sites. Could a change in the barriers to chloride movement between binding sites decrease α2 *(0)? For Model 1 the answer is yes because an effect on the energetic barriers to chloride movement in the pore would affect K 2 and/or K 3, and therefore would change α2 * and K c. For Model 2 the answer is yes because the second step represents some conformational change required for opening that does not change chloride occupancy. If this conformational change involves lowering the barriers to chloride movement between sites, and if K149 affects these barriers, then K149 mutations would affect α2 *. Since the calculated K c and α2 * are interdependent (see discussion above), K c could also be affected. Therefore the hypothesis that K149 affects gating by affecting a protein conformational change (as opposed to a chloride occupancy change) is reasonable for both Model 1 and Model 2.
Finally, there is the possibility that wild type behaves according to Model 2, but that mutations at 149, by reducing chloride affinity, make the channel behave according to Model 1. Both α2 * and K c would then be affected since the underlying chloride movements for each of these steps would be different for wild type versus the mutant.
How the K149 Side-Chain Features Affect Fast Gating
To assess these above three hypotheses, we considered the chemical and physical properties of the side-chain substitutions at position 149, since these may give clues to the mechanism by which K149 mutations affect gating. Although negatively charged side chains are not tolerated, an array of neutral side chains give shifted but otherwise normal fast gating (Table I). This adds to a body of evidence that in ClC-0, sequence conservation does not correlate with the degree of the effect on gating; K149 falls into the same category as other highly conserved residues (e.g., Y512 and K165), the mutation of which causes mild effects on fast gating (i.e., only cause small changes in V o) (Lin and Chen, 2000, 2003; Accardi and Pusch, 2003; Engh and Maduke, 2005), in contrast to the severe effects on gating observed upon mutation of the highly conserved residues E166 and S123 (i.e., in addition to effects on V o also alter kinetics and P min, or abolish gating) (Ludewig et al., 1996; Accardi and Pusch, 2003; Chen and Chen, 2003; Dutzler et al., 2003; Lin and Chen, 2003; Traverso et al., 2003; Engh and Maduke, 2005). Most revealing is the finding that for a variety of neutralizing mutations the gating charge for depolarization-activated opening is similar to that of wild type (Fig. 7 G). Thus, the charge on K149 cannot be part of the gating charge. Assuming that the binding and/or movement of chloride in the pore contribute to the voltage dependence of depolarization-activated opening (Pusch et. al., 1995), this finding is consistent with our electrostatic calculations that suggest that the mutations at K149 do not change the electrostatic field in the pore and do not change the positions of the chloride binding sites in the electrostatic field.
It was previously proposed that the positive charge and/or the hydrogen bond donors on K149 help facilitate the movement of chloride between S cen and S int, the two chloride-binding sites closest to this residue in the ClC-ec1 structures. Our previously published results on K149C hinted that the pKa of this residue may play a role in fast gating. Since we have now shown that the degree of the effect on fast gating does not directly correlate to any of these three side-chain properties (charge, hydrogen-bond donors, pKa), then if these features are important then their effects must be short ranged, requiring the precise placement of the side chain. Although the current mutational analysis cannot fully separate the influence of each of these properties, the similarity in shape to lysine seems to be the most critical determinant of the extent of the effect of K149 mutations. Specifically, extended linear side chains display the smallest effect, followed by short linear side chains, short branched side chains, and lastly bulky side chains. The notion that the precise placement of the K149 charge is crucial is supported by the observation that the K149R mutant displays altered fast-gating properties, comparable to K149M. This suggests that the longer and bulkier arginine side chain cannot be accommodated within the protein environment in the same manner as the lysine side chain. Without additional experiments, such as introducing an unnatural amino acid to make a neutralizing mutation that preserves the shape of K149, it is not possible to determine the influence of the positive charge alone. Nonetheless, the finding that negative substitutions have severe effects on channel function and/or folding suggests that the charge on K149 is important.
Altogether, our analysis suggests that the packing of K149 with its neighbors is an important factor contributing to the barrier height of the α2 * step. The packing could be important because it determines how well chloride is coordinated when it is bound to S cen and S int, and how well it is coordinated as it moves between these two sites. Our data therefore indicate that mutations of K149 influence gating either by weakening chloride binding to S cen or S int (Hypothesis 2) and/or by affecting the movement of chloride between these two sites (Hypothesis 3).
Conclusion
In summary, mutations at K149 affect voltage-dependent fast gating in ClC-0. Side-chain shape seems to be a critical determinant of the severity of the effect. This argues that short-range, specific interactions between K149 and its neighbors are important for fast gating. Detailed analysis of fast-gate opening pinpoints the major effect of K149 mutations as being on the depolarization-activated gating step that follows binding of extracellular chloride. These results, together with previously published results and our analysis of ClC-0 homology models, suggest that K149 mutations are likely to affect gating by lowering chloride binding affinity to one or more sites in the pore or by affecting the barriers to chloride movement between sites. We suggest two models for chloride movement during depolarization-activated fast-gate opening, one which requires some conformational change other than chloride binding or movement, and one which does not. Further exploration is needed to distinguish these models.
Supplemental Material
Acknowledgments
We thank Gilbert Martinez and Drs. Richard Aldrich, Kimberly Matulef, and Joseph Mindell for comments on the manuscript. We also thank Dr. Benoît Roux for helpful discussions and Dr. Dan Cox for advice on data analysis.
This work has been supported by National Institutes of Health grant R01 GM070773 and by the Mathers Foundation. A. Engh was supported in part by a Predoctoral Fellowship from the American Heart Association.
Olaf S. Andersen served as editor.
References
- Accardi, A., and M. Pusch. 2003. Conformational changes in the pore of ClC-0. J. Gen. Physiol. 122:277–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick, D.L., and M.L. Berkowitz. 2004. Exterior site occupancy infers chloride-induced proton gating in a prokaryotic homolog of the ClC chloride channel. Biophys. J. 87:1686–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, H.S., and K. Dill. 1998. Protein folding in the landscape perspective: chevron plots and non-arrhenius kinetics. Proteins. 30:2–33. [DOI] [PubMed] [Google Scholar]
- Chen, M.-F., and T.Y. Chen. 2003. Side-chain charge effects and conductance determinants in the pore of ClC-0 chloride channels. J. Gen. Physiol. 122:133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T.-Y., M.-F. Chen, and C.-W. Lin. 2003. Electrostatic control and chloride regulation of the fast gating of ClC-0 chloride channels. J. Gen. Physiol. 122:641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T.Y., and C. Miller. 1996. Nonequilibrium gating and voltage dependence of the ClC-0 Cl- channel. J. Gen. Physiol. 108:237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, J., and K. Schulten. 2004. Mechanism of anionic conduction across ClC. Biophys. J. 86:836–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corry, B., M. O'Mara, and S.-H. Chung. 2004. Conduction mechanisms of chloride ions in ClC-type channels. Biophys. J. 86:846–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano, W.L. 2002. The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA. http://www.pymol.org
- Dutzler, R., E.B. Campbell, and R. Mackinnon. 2003. Gating the selectivity filter in ClC chloride channels. Science. 300:108–112. [DOI] [PubMed] [Google Scholar]
- Engh, A.M., and M. Maduke. 2005. Cysteine accessibility in ClC-0 supports conservation of the ClC intracellular vestibule. J. Gen. Physiol. 125:601–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engh, A.M., J. Faraldo-Gómez, and M. Maduke. 2007. The mechanism of fast-gate opening in ClC-0. J. Gen. Physiol. 130:335–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez, R., B.C. Schroeder, A. Accardi, T.J. Jentsch, and M. Pusch. 2003. Conservation of chloride channel structure revealed by an inhibitor binding site in ClC-1. Neuron. 38:47–59. [DOI] [PubMed] [Google Scholar]
- Faraldo-Gómez, J.D., and B. Roux. 2004. Electrostatics of ion stabilization in a ClC chloride channel homologue from Escherichia coli. J. Mol. Biol. 339:981–1000. [DOI] [PubMed] [Google Scholar]
- Gervasio, F.L., M. Parnello, M. Ceccarellil, and M.L. Klein. 2006. Exploring the gating mechanism in the ClC chloride channel via metadynamics. J. Mol. Biol. 361:390–398. [DOI] [PubMed] [Google Scholar]
- Lin, C.W., and T.Y. Chen. 2000. Cysteine modification of a putative pore residue in ClC-0:implication for the pore stoichiometry of ClC chloride channels. J. Gen. Physiol. 116:535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C.W., and T.-Y. Chen. 2003. Probing the pore of ClC-0 by substituted cysteine accessibility method using methane thiosulfonate reagents. J. Gen. Physiol. 122:147–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y.-W., C.-W. Lin, and T.-Y. Chen. 1999. Elimination of the slow gating of ClC-0 chloride channel by a point mutation. J. Gen. Physiol. 114:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludewig, U., M. Pusch, and T.J. Jentsch. 1996. Two physically distinct pores in the dimeric ClC-0 chloride channel. Nature. 383:340–343. [DOI] [PubMed] [Google Scholar]
- Ludewig, U., T.J. Jentsch, and M. Pusch. 1997. Analysis of a protein region involved in permeation and gating of the voltage-gated Torpedo chloride channel ClC-0. J. Physiol. 498:691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduke, M., C. Williams, and C. Miller. 1998. Formation of CLC-0 chloride channels from separated transmembrane and cytoplasmic domains. Biochemistry. 37:1315–1321. [DOI] [PubMed] [Google Scholar]
- Middleton, R.E., D.J. Pheasant, and C. Miller. 1996. Homodimeric architecture of a ClC-type chloride ion channel. Nature. 383:337–383. [DOI] [PubMed] [Google Scholar]
- Miloshevsky, G.V., and P.C. Jordan. 2004. Anion pathway and potential energy profiles along curvilinear bacterial ClC Cl− pores: electrostatic effects of charged residues. Biophys. J. 86:825–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusch, M., U. Ludewig, A. Rehfeldt, and T.J. Jentsch. 1995. Gating of the voltage-dependent chloride channel ClC-0 by the permeant ion. Nature. 373:527–531. [DOI] [PubMed] [Google Scholar]
- Traverso, S., G. Zifarelli, R. Aiello, and M. Pusch. 2006. Proton sensing of ClC-0 mutant E166D. J. Gen. Physiol. 127:51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, J., R. Taylor, C. Chothia, and M. Gerstein. 1999. The packing density in proteins: standard radii and volumes. J. Mol. Biol. 290:253–266. [DOI] [PubMed] [Google Scholar]
- Yin, J., Z. Kuang, U. Mahankali, and T.L. Beck. 2004. Ion transit pathways and gating in ClC chloride channels. Proteins. 57:414–421. [DOI] [PubMed] [Google Scholar]
- Zhang, X.D., Y. Li, W.P. Lu, and T.Y. Chen. 2006. Roles of K149, G352, H401 in the channel functions of ClC-0: testing the predictions from theoretical calculations. J. Gen. Physiol. 127:435–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.