Abstract
ClC-0 is a chloride channel whose gating is sensitive to both voltage and chloride. Based on analysis of gating kinetics using single-channel recordings, a five-state model was proposed to describe the dependence of ClC-0 fast-gate opening on voltage and external chloride (Chen, T.-Y., and C. Miller. 1996. J. Gen. Physiol. 108:237–250). We aimed to use this five-state model as a starting point for understanding the structural changes that occur during gating. Using macroscopic patch recordings, we were able to reproduce the effects of voltage and chloride that were reported by Chen and Miller and to fit our opening rate constant data to the five-state model. Upon further analysis of both our data and those of Chen and Miller, we learned that in contrast to their conclusions, (a) the features in the data are not adequate to rule out a simpler four-state model, and (b) the chloride-binding step is voltage dependent. In order to be able to evaluate the effects of mutants on gating (described in the companion paper, see Engh et al. on p. 351 of this issue), we developed a method for determining the error on gating model parameters, and evaluated the sources of this error. To begin to mesh the kinetic model(s) with the known CLC structures, a model of ClC-0 was generated computationally based on the X-ray crystal structure of the prokaryotic homolog ClC-ec1. Analysis of pore electrostatics in this homology model suggests that at least two of the conclusions derived from the gating kinetics analysis are consistent with the known CLC structures: (1) chloride binding is necessary for channel opening, and (2) chloride binding to any of the three known chloride-binding sites must be voltage dependent.
INTRODUCTION
Members of the CLC family orchestrate the movement of chloride necessary for proper neuronal, muscular, cardiovascular, and epithelial function (Jentsch et al., 2002, 2005; Uchida and Sasaki, 2005; Sile et al., 2006). Defects in these proteins are directly responsible for human diseases of muscle, kidney, bone, and brain (Lloyd et al., 1996; Simon et al., 1997; Cleiren et al., 2001; Kornak et al., 2001; Pusch, 2002; Haug et al., 2003; Naesens et al., 2004). ClC-1, which is highly expressed in mammalian skeletal muscle, is responsible for restoring the resting potential of the cell after muscle contraction (Lehmann-Horn and Jurkat-Rott, 1999; Colding-Jorgensen, 2005). Hence mutations in this channel can lead to myotonia, a condition where voluntary contraction of muscles is only slowly reversible (Lehmann-Horn and Jurkat-Rott, 1999; Colding-Jorgensen, 2005; Jurkat-Rott and Lehmann-Horn, 2005). Many of the myotonia-inducing mutations in ClC-1 cause shifts in voltage-dependent gating (Pusch, 2002). Understanding the voltage dependence of CLC gating is thus of great physiological relevance.
ClC-0, found in Torpedo electric rays, was the first CLC to be discovered and characterized (White and Miller, 1979; Miller and Richard, 1990; Bauer et al., 1991), and has ∼50% sequence identity to ClC-1. Since ClC-0 has a single-channel conductance higher than any other CLC yet studied (∼10 pS), it is the most thoroughly studied CLC and serves as the family prototype (Maduke et al., 1999; Estevez and Jentsch, 2002; Pusch, 2004; Chen, 2005; Jentsch et al., 2005; Dhani and Bear, 2006; Dutzler, 2006). The functional form of ClC-0 is a dimer in which each subunit forms its own pore (Ludewig et al., 1996; Middleton et al., 1996). One gating mechanism, termed “slow” gating, closes both pores simultaneously. A second gating mechanism, termed “fast” gating, governs the independent opening and closing of each pore. Both slow and fast gating are sensitive to transmembrane voltage, chloride, and pH (Miller, 1982; Hanke and Miller, 1983; Pusch et al., 1995, 1999; Chen and Miller, 1996; Chen and Chen, 2001; Pusch, 2004; Traverso et al., 2006), and have unprecedented mechanisms in which the permeant ion plays a key role. Slow gating is energetically coupled to the transmembrane chloride gradient (Richard and Miller, 1990); fast gating voltage dependence arises from the movement of the permeant ion through the transmembrane field (Pusch et al., 1995; Chen and Miller, 1996). Hence gating, permeation, and chloride binding are all tightly coupled in ClC-0.
An interesting feature of the fast gate of ClC-0, revealed by the voltage dependence of its opening rate constant, is that it can be activated by either hyperpolarization or depolarization. These two gating pathways can be distinguished further since only the depolarization-activated pathway is sensitive to external chloride concentration.
Analysis of single-channel data led Chen and Miller (1996) to propose a five-state model (Scheme 2) for fast-gate opening that explains the effects of voltage and chloride on the opening rate constant. We aimed to gain insight into what structural changes occur during the steps in this model by examining a series of mutants with altered gating and determining how specific steps in the model are affected (Engh et al., 2007). As a first step toward this goal, we used macroscopic recordings to investigate the effects of voltage and external chloride on the gating kinetics of wild-type ClC-0. While our data display all the same features as those published by Chen and Miller (1996), the use of a different method (global fitting) to fit the data to the theoretical models leads to a substantially different interpretation. By performing a thorough error analysis of both our data and those of Chen and Miller (1996), we show the limitations of fits to the five-state model, and provide an estimate of the uncertainty of these results. We conclude that a simpler four-state model is sufficient to explain the data and that the chloride-binding step is depolarization activated, not voltage independent as previously proposed.
MATERIALS AND METHODS
Channel Expression
We used a ClC-0 construct in a plasmid derived from the pBluescript vector (Stratagene) (Jentsch et al., 1990; Maduke et al., 1998), which contained the point mutation C212S. This mutation removes voltage-dependent slow-gate inactivation and has no other measurable effect on ClC-0 function (Lin et al., 1999). Plasmids were linearized with FspI (New England Biolabs), cleaned using the DNA Clean and Concentrator-5 (Zymo Research), and transcribed in vitro using the mMessage mMachine T3 RNA-polymerase transcription kit (Ambion). RNA was dissolved in RNAase-free water (Invitrogen) containing 0.77 U/μL SUPERase-In (Ambion).
Excised Patch Recording
Defolliculated Xenopus oocytes were injected with 27.5 nl RNA at ∼1 mg/ml and incubated at 16°C for 2–5 d before recording. Data were then collected from excised inside-out patches using Axopatch 200B and pClamp software. Before patching, the vitelline membrane was removed manually with the oocyte bathed in internal (bath) solution (in mM: 110 NMDG, 110 HCl, 5 MgCl2, 10 HEPES, 1 EGTA, brought to pH 7.3 using NaOH). Electrical contact between the recording chamber and the ground electrode was made via agarose bridges. Recording electrodes were pulled from 100-μl calibrated pipettes (VWR), polished to 0.2–1.5 MΩ, and filled with external solution. All external (pipette) solutions contained 1 mM EGTA, 10 mM HEPES, and were brought to pH 7.3 using NaOH. For the seven external chloride concentrations used (in mM: 5, 15, 30, 65, 110, 310, 610), the concentration of the other components are shown in Table I. All solutions were sterilized using 0.2-μm filters.
TABLE I.
External (Pipette) Solutions
| [Cl] (mM)
|
5
|
15
|
30
|
65
|
110
|
310
|
610
|
|---|---|---|---|---|---|---|---|
| Components (mM) | |||||||
| NMDG | 100 | 100 | 100 | 100 | 100 | 300 | 600 |
| Glutamate | 95 | 85 | 70 | 35 | 0 | 0 | 0 |
| HCl | 5 | 15 | 30 | 65 | 100 | 300 | 600 |
| MgCl2 | 0 | 0 | 0 | 0 | 5 | 5 | 5 |
| MgSO4 | 5 | 5 | 5 | 5 | 0 | 0 | 0 |
Each column represents a separate pipette solution. The top row lists the final chloride concentration for each solution, and the ensuing rows show the concentration of each of the components of the solution.
Measuring Junction Potentials
Having different solutions on either side of the patch causes significant junction potentials. To accurately correct for these junction potentials we measured them independently. We compared the junction potentials we measured with those calculated using the JPcalc feature of Clampex 8.1 software (Fig. S1, available at http://www.jgp.org/cgi/content/full/jgp.200709759/DC1). When the difference between internal and external solution is extreme (e.g., for external solutions with 5, 300, or 600 mM chloride; internal has 120 mM chloride), there is a significant difference between the measured junction potential and the calculated junction potential. These differences are reduced if activities of the most abundant ions are entered into JPcalc instead of concentrations. We corrected the command voltages using measured junction potentials.
Determining Open Probability
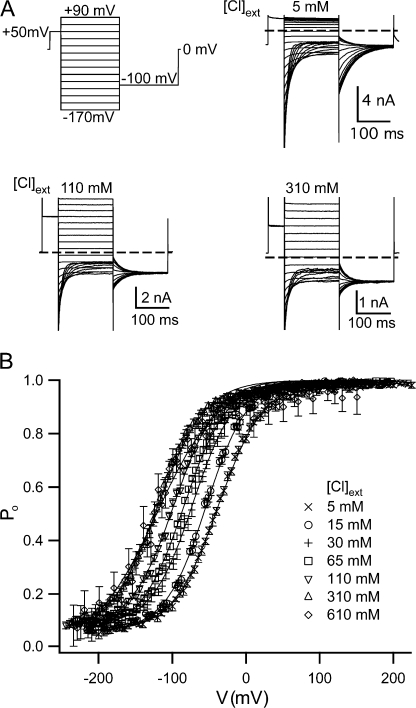
We used a voltage protocol designed to obtain the apparent open probability (Po) as a function of voltage. (This protocol was also used to determine the opening and closing rate constants, as described in the legend of Fig. 2.) In this protocol, illustrated in Fig. 1, a 50-ms prepulse to a positive voltage (at least +50 mV) is followed by a 170-ms test pulse, starting at least +40 mV above the prepulse voltage and decrementing by 20 mV, and then a 170-ms tail pulse to −100 mV. The prepulse voltage and the range of the test-pulse voltages were adjusted so that the Po could be seen to plateau at either end of the voltage range used.
Figure 2.
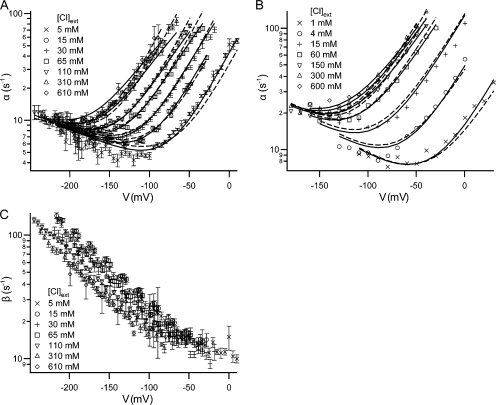
Effect of voltage and external chloride on the kinetics of fast gating. (A and C) opening (α) and closing (β) rate constants for each test voltage. For test pulses (see examples in Fig. 1 A) where current decay could be seen, the decay was fit to a single exponential to derive the decay constant (1/τ). This was used in conjunction with the open probability (Po) to calculate α and β (Chen and Chen, 2001): α = P o/τ ; β = (1/τ) − α. For each condition (voltage (V), external chloride activity ([Cl]ext), we recorded from at least five patches and derived the rate constant for each patch. For the 610 mM chloride condition, we recorded two to five patches for each voltage. For a given condition, the rate constants from all patches were averaged; the error bars show the SEM. (B) For comparison, opening rate constant data were taken from the graphs published by Chen and Miller (1996) and replotted. (A and B) Each opening rate constant dataset was globally fit to the five-state model (Scheme 2, Eqs. S5–S8 in the online supplemental material) either holding z c constant (dashed lines) or letting all six parameters find their best-fit values (solid lines). In the case where z c was held constant, the values of z c (0.09 in [A] and 0.08 in [B]) were chosen based on the analysis using the Method 1 and the four-state model, where only a subset of voltages were included (see analysis in the supplemental material and parameter values in Fig. 3). The parameters derived from all of these fits are shown in Fig. 5.
Figure 1.
Effects of external chloride on gating. (A) Voltage pulse protocol (top left) and current responses in 5, 110, and 310 mM external chloride. (B) Tail-current analysis was used to determine the apparent open probability (Po) as a function of voltage (V) (see Materials and methods) (n = 5–26 for external chloride concentrations <600 mM, and n = 2–5 for 610 mM). These plots were fit as described in Materials and methods (solid lines), and gating parameters were derived (Table II).
Since the −100-mV tail pulse provides a common driving force to compare the relative number of open channels in each preceding test pulse, we used the current at the beginning of the tail pulse (the instantaneous tail current) to calculate the steady-state open probability reached during each test pulse. The instantaneous tail current, I, was determined by fitting each tail-current decay to a single exponential, and the apparent open probability was calculated as P o = I/I max, where I max is the maximum instantaneous tail current. The gating parameters P min (the minimum apparent open probability), z (the effective gating charge), and V o (the midpoint of the voltage-activation curve) were calculated by fitting the apparent open probability (solid lines) to an equation of the form Po = P min + (1 − P min)/{1 + exp[−zF(V − V o)/RT]}, and are listed in Table II.
TABLE II.
Wild-Type Voltage-dependent Gating Parameters at Various External Chloride Concentrations
| [Cl]ext | V o | z | P min | n |
|---|---|---|---|---|
| mM | mV | |||
| 5 | −37 ± 1.4 | 0.87 ± 0.03 | 0.070 ± 0.011 | 13 |
| 15 | −55 ± 1.4 | 0.93 ± 0.02 | 0.064 ± 0.009 | 7 |
| 30 | −69 ± 2.0 | 0.90 ± 0.03 | 0.075 ± 0.011 | 10 |
| 65 | −83 ± 1.1 | 0.91 ± 0.02 | 0.077 ± 0.013 | 11 |
| 110 | −95 ± 1.1 | 0.90 ± 0.01 | 0.070 ± 0.008 | 26 |
| 310 | −119 ± 1.1 | 0.85 ± 0.03 | 0.078 ± 0.014 | 16 |
| 610 | −121 ± 4.8 | 0.76 ± 0.04 | 0.051 ± 0.020 | 5 |
Plots of apparent open probability (P o) versus voltage were fit as described in Materials and methods to derive the voltage at the midpoint in the voltage-activation curve (V o), apparent gating charge (z), and minimum open probability (P min). The mean ± SEM is given, and n indicates the number of patches used.
Determining Opening and Closing Rate Constants
The approach we used to determine opening (α) and closing (β) rate constants is described in the legend of Fig. 2. This method becomes inaccurate under the following circumstances: (a) when the current decay is so fast that it is difficult to separate from the capacitive transient, making it difficult to accurately determine 1/τ, (i.e., at very negative voltages and/or low external chloride); (b) when P o is small and therefore difficult to accurately determine (i.e., at very negative voltages); (c) when the amplitude of the current decay is small so it is difficult to determine 1/τ accurately (i.e., at higher voltages); (d) when there is little current passing through the patch, such that the signal-to-leak ratio is small and neither 1/τ nor P o can be accurately determined.
To avoid inaccuracies arising from the circumstances mentioned above, we investigated any patches that gave α values that appeared to be outliers. We excluded data from patches where the raw data had any obvious problems, such as (a) the prepulse current continually drifted down between sweeps, (b) the test-pulse current at high voltages drifted down radically over the course of the pulse, and (c) the tail currents from all the sweeps did not approach the same final value. These three measures resulted in the exclusion of <6% of the 256 records taken (for each patch we took one to four records, with each record testing a different set of voltages). We also excluded data from individual sweeps where (a) there were large and biased residuals for the single exponential fit to the test-pulse or tail-pulse decay, (b) noise in the record was so large that the instantaneous tail current was less than zero, resulting in a P o less than zero, or (c) the resulting α value failed a Dixon test for outliers. We used the Dixon tests and 95% confidence cutoff values as described in Outliers in Statistical Data, tests N7–N13 (Barnett and Lewis, 1994). These three measures resulted in the exclusion of <2% of the 2185 sweeps recorded. Finally, after averaging the α values for each condition (voltage, external chloride concentration), we excluded conditions where there were extremely large error values (mean/SEM > 0.4), since the data taken at these conditions are likely to be too variable to be trusted. This last measure resulted in the exclusion of <3% of the 246 conditions tested.
The use of macroscopic recordings to derive α values is based on two important assumptions. First, this method assumes that the maximum P o is equal to 1, which has been shown using single-channel recordings over a wide range of external chloride concentrations (4–600 mM; Chen and Miller, 1996). Second, this method assumes that there are only two conductance levels (open and closed) for each pore in the channel. Single-channel recordings have shown that this is the case for wild type (Miller, 1982; Hanke and Miller, 1983; Richard and Miller, 1990; Ludewig et al., 1996; Middleton et al., 1996; Accardi and Pusch, 2003) and K149L (Zhang et al., 2006). These assumptions seem reasonable because the wild-type α values we measured using macroscopic patch recordings show the same dependence on voltage and external chloride as those obtained from single-channel recordings (as reported by Chen and Miller, 1996).
Global Fitting
We used multivariate fitting procedures to fit the opening rate constant as a function of voltage and external chloride activity to equations for either the four-state or the five-state model. To do these fits, we used the Levenberg-Marquardt algorithm as implemented in Igor (Wavemetrics). To improve fitting in Igor, we fit log(α) rather than α, weighted the fits by 1/SEM, and wrote user-defined functions such that the parameters sought were all of the same order of magnitude. We subsequently transformed the parameters to their actual order of magnitude.
Determining the Activity of Chloride in our Solutions
The analyses and calculations shown were done using chloride activities; similar results were obtained using chloride concentrations. To calculate the chloride activity, we had to consider the effects of other ions present in the solution. The major ions in our solutions were NMDG, chloride, and in the cases of low chloride concentration, glutamate. Unfortunately, the activity coefficient for NMDG-Cl has not been measured. Since the difference between the activity coefficient of potassium chloride and choline chloride is small over the relevant range of concentrations (Fig. S2), we assume that the difference between the activity coefficient of NMDG-Cl and choline chloride is small. Since NMDG is closer in size to choline than to potassium, we used the activity coefficient of choline chloride to calculate the chloride activity for each of our solutions (activity = activity coefficient * concentration).
Sequence Alignment and Structural Modeling
To construct a three-dimensional model of the atomic structure of wild-type ClC-0, we followed the homology modeling procedure described below, based on the structure of the prokaryotic proton/chloride exchanger ClC-ec1 (Dutzler et al., 2003). The underlying assumption of this approach is that the true structure of ClC-0 (whether wild type or mutant) does not depart substantially from that of its homolog, especially in regions of high sequence conservation. We consider this a reasonable assumption based on results showing the reliability of the ClC-ec1 structure as a guide to ClC-0 (Estevez et al., 2003; Lin and Chen, 2003; Engh and Maduke, 2005).
First, to identify the optimal sequence alignment between ClC-0 and ClC-ec1 (Fig. S3), we obtained a consensus prediction from (a) a multiple-sequence alignment including all known members of the CLC family, performed with Clustal-W (Thompson et al., 1994), and (b) a profile-to-profile alignment, performed with HMAP (Petrey et al., 2003). The latter method includes both primary sequence information, derived from a multiple-sequence alignment of the output of a PSI-BLAST search, as well as secondary-structure information, derived from the known structure of the ClC-ec1 and from a consensus prediction for ClC-0 (using JNET, PSIPRED, and PHD).
Based on the resulting alignment and the structure of the ClC-ec1 dimer (PDB ID 1OTS), an initial model of wild-type ClC-0 was built using the molecular modeling software CHARMM (Brooks et al., 1983), preserving the three-dimensional structure of the backbone of the prokaryotic homolog, and excluding insertions of more than five amino acids. The conformation of conserved or sterically similar side chains (e.g., Glu and Gln) was also preserved, in addition to the chloride ions bound to ClC-ec1 in the S int and S cen sites; otherwise, the prediction algorithm SCWRL (Canutescu et al., 2003) was employed to determine the most plausible side chain conformations. Subsequently, a stepwise scheme was employed to energy minimize the atomic model, where structural constraints based on the degree of conservation were gradually released. These energy minimizations also were performed with CHARMM, using the CHARMM22 all-atom force field (MacKerell et al., 1998), a 14-Å cutoff for the nonbonded interactions, and a distance-dependent dielectric constant.
For comparison, we also built a model inspired by the structure of the E148Q mutant of ClC-ec1 (Dutzler et al., 2003) using an analogous procedure to that described above. In this structure an additional chloride ion is bound to the S ext site; in the wild-type structure, this site is occupied by the side chain of E148. In our ClC-0 homology model inspired by the E148Q structure, the S ext site may be occupied by chloride, while the side chain of E166 (the E148 equivalent) is protonated and rotated around the χ1 dihedral angle. After energy minimization of this model, E166 forms a hydrogen bond with C212.
Electrostatic Binding Energies and Transmembrane Potential
The electrostatic contribution to the free energy of binding of each chloride ion i to the remainder of the protein–ion complex p was computed according to the expression:
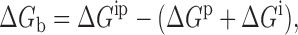 |
(1) |
where ΔG ip, ΔG p, and ΔG i represent the electrostatic energies of the full complex, the complex of protein and chloride ions other than i, and the isolated ion i in solution, respectively. For each of these systems, the electrostatic energy is defined as:
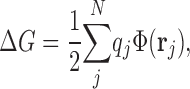 |
(2) |
where qj and r j denote the charge and location of each of the N atoms in the protein–ion complex and Φ(r j) is the electrostatic potential evaluated at those positions. To compute ΔG ip(r), ΔG p(r), and ΔG i(r), we employed the Poisson-Boltzmann equation in its weak-field, linear approximation:
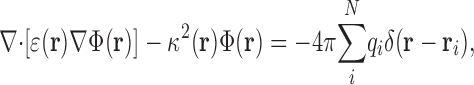 |
(3) |
where the position-dependent quantities ɛ(r) and κ(r) are the dielectric constant and the Debye-Huckel ionic screening factor, respectively. The dielectric distribution ɛ(r) was partitioned into different regions: the region delimited by the solvent-accessible surface of the protein–ion complex (constructed with a 1.4-Å probe radius) was assigned a dielectric constant ɛp equal to 4, 8, or 20 in independent calculations; the membrane region, represented by a continuous slab of overlapping, uncharged pseudo-atoms in which the protein–ion complex is embedded, was assigned a dielectric constant ɛ m = 2; and the remainder of the system was assigned a water-like dielectric constant ɛ w = 80. Similarly, the screening factor κ(r) due to the ionic solution (in this case of monovalent ions, e.g., NaCl, with concentration C = 150 mM) was partitioned as
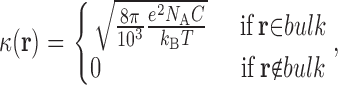 |
(4) |
where N A and k B are Avogadro's and Boltzmann's constants, respectively, and T the temperature; here, the “bulk” region comprises the portion of the system that is accessible to the ions in solution, i.e., the region outside the solvent-accessible surface of protein and membrane, minus a 2-Å-wide Stern layer.
Following Roux (1997) and Nonner et. al. (2004), we also used the Poisson-Boltzmann framework to analyze the spatial dependence of the electrostatic potential Φtm(r) resulting from an externally applied transmembrane voltage V tm, via the expression:
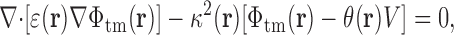 |
(5) |
where θ(r) is a step function equal to one on one side of the membrane, and equal to zero on the other side.
To solve the Poisson-Boltzmann equation, we used the grid-based, finite-difference solver PBEQ (Im et al., 1998) implemented in version C32A2 of the CHARMM molecular modeling software (Brooks et al., 1983), with a modification that allows for the use of pseudo-atoms with an arbitrary dielectric constant. Atomic charges were obtained from the CHARMM22 force field (MacKerell et al., 1998), alongside the optimized atomic radii derived by Nina et al. (1997).
Grid focusing and translational averaging were implemented in all calculations of the electrostatic binding energy in order to minimize discretization errors without significantly increasing the computational cost. The dimensions of the initial grid were 116 × 74 × 80 Å, with a grid-point spacing of 1.0 Å. Boundary conditions were interpolated from this calculation and used for a second grid, of dimensions 104 × 62 × 78 Å with a grid-point spacing equal to 0.5 Å. The focusing procedure was repeated for eight different locations of the protein–membrane system relative to the center of the three-dimensional grids (shifted 0–0.25 Å in each direction); the ΔG b values reported hereafter are averages over these independent calculations, as well as over the two protein subunits. For the calculation of Φtm(r), the electrostatic potential was computed along a putative permeation pathway across the channel, for a transmembrane voltage V tm = 200 mV and a grid-point spacing of 0.5 Å. For each position along this pathway, the values of V/V tm reported are averages of Φtm over nine sites shifted on the plane of the membrane by ±1 Å, as well as over both protein subunits. Standard deviations of the averages of ΔG b and V/V tm are shown as error bars in Fig. 6.
Figure 6.
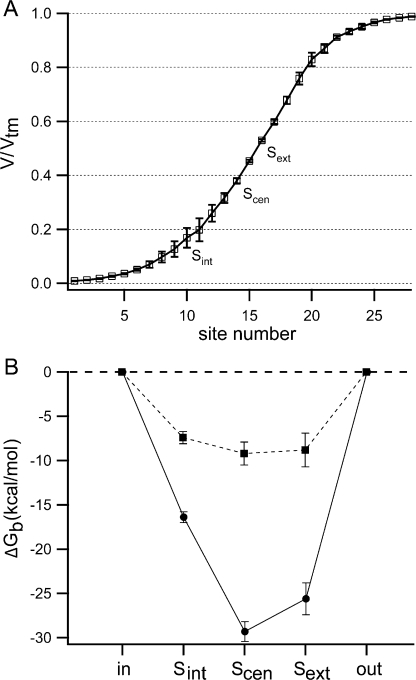
Chloride binding and movement in the pore. (A) The ClC-0 homology models created as described in Materials and methods were used to calculate the electrostatic potential at various positions in the pore (V/V tm) under an externally applied transmembrane voltage of −200 mV. Shown here is the fraction of the transmembrane voltage along a hypothetical permeation pathway, with S int, S cen, S ext at sites 10, 14, and 16, respectively. (B) The ClC-0 homology model was used to calculate the electrostatic contribution to the free energy of chloride binding (ΔG b) to each site in the pore (starting from the cytoplasmic side: S int, S cen, S ext) with either zero (circles) or two (squares) chlorides already bound in the pore. The dielectric constant used for the protein was 4. In both A and B, lines connect data points for ease in viewing, and error bars represent an estimate of the discretization error arising from the grid-based Poisson-Boltzmann calculations (see Materials and methods).
Online Supplemental Material
The supplemental material is available online at http://www.jgp.org/cgi/content/full/jgp.200709759/DC1. Fig. S1 shows the measured and calculated junction potentials. Fig. S2 shows the activity coefficients for chloride in the patching solutions used. Fig. S3 shows the alignment of ClC-0 and ClC-ec1 used to create the ClC-0 homology model. The supplemental material also includes a large part of the kinetic analysis, including a detailed description of Method 1 (Fig. S4), a comparison of the results obtained using Method 1 and Method 2 (Fig. S5), and a section on determining the interdependency of parameters in the gating model (Fig. S6). Also included are the results of calculations analyzing the effects of chloride occupancy on chloride binding affinity, in which the electrostatic contribution to chloride binding (ΔG b) to various sites in the pore is determined under various occupancy conditions (Fig. S7).
RESULTS
The Effect of Voltage and Chloride on Fast-Gate Open Probability
To assess the effects of voltage (V) and external chloride concentration ([Cl]ext) on fast gating, we measured currents through macroscopic excised inside-out patches from Xenopus oocytes expressing ClC-0. To do this we used the pulse protocol described in Materials and methods and illustrated in Fig. 1 A (top left). Examples of the current response to this protocol at different external chloride concentrations (Fig. 1 A) illustrate that over this wide range of chloride concentration, tail currents can be used to derive the apparent open probability (Fig. 1 B). As has been previously reported (Pusch et al., 1995; Chen and Miller, 1996), we found that increasing external chloride concentration activates the fast gate, causing a leftward shift in the plot of the open probability as a function of voltage.
The Effect of Voltage and Chloride on Fast-Gate Kinetics
The results from our macroscopic recordings of wild-type ClC-0 show similar features to those obtained by Chen and Miller using single-channel recordings: (a) the opening rate constant (α) as a function of voltage gives a nearly V-shaped curve similar to a chevron plot (Chan and Dill, 1998), where for the righthand arm of chevron (voltages less negative than ∼−175 mV, depending on external chloride concentration), opening is dominated by a depolarization-activated process; and for the lefthand arm of the chevron (voltages more negative than ∼−175 mV), opening is dominated by a hyperpolarization-activated process (Fig. 2, A and B); (b) only the righthand arm of the curve is shifted by external chloride (Fig. 2, A and B); (c) the closing rate constant shows a log-linear dependence on voltage and thus appears to be governed by only one process (Fig. 2 C). Fast-gate closing is activated by hyperpolarization and is slightly sensitive to changes in external chloride concentration.
Methods of Fitting the Data
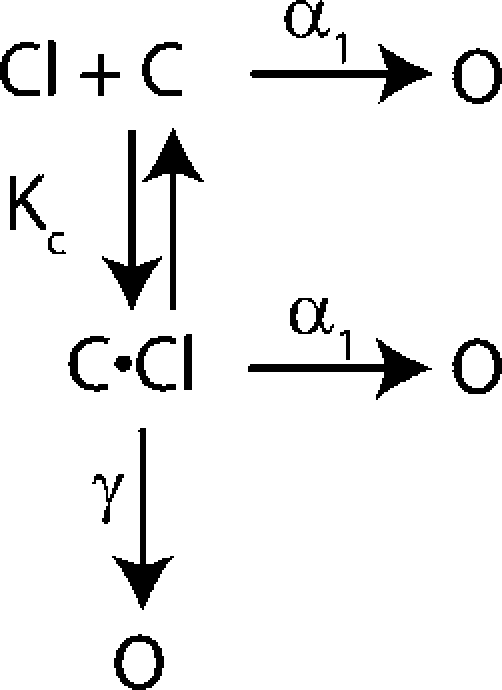
Chen and Miller (1996) proposed both a four-state model (Scheme 1) and five-state model (Scheme 2) to describe the dependence of fast-gate opening on voltage and chloride. These two models are similar in that they both include a chloride-bound (C⋅Cl) and a chloride-free (C) closed state, and an open state (O).
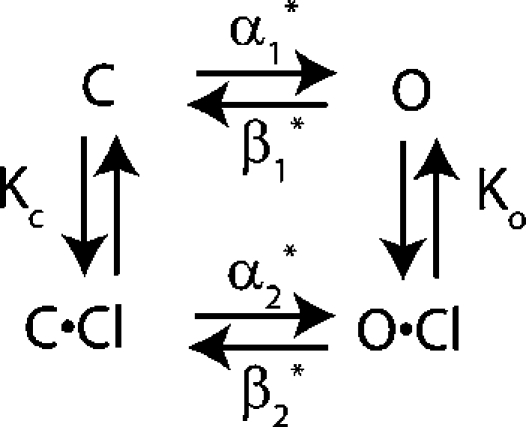 |
(SCHEME 1) |
 |
(SCHEME 2) |
In both models the channel can open from either closed state. The difference between these models is not the states that the channel can visit but the connectivity between states. In the five-state model the channel can open after the chloride-binding step via a hyperpolarization-activated or a depolarization-activated pathway. In contrast, in the four-state model the channel must open through a depolarization-activated pathway once the chloride-binding event has occurred. Mathematically, however, these two models are very similar (see online supplemental material, available at http://www.jgp.org/cgi/content/full/jgp.200709759/DC1) and both have six free parameters. We fit our data to both the four- and five-state gating models and in the process of doing so, discovered that the method of fitting the data greatly affects the results and the estimation of errors. To better compare our results to those of Chen and Miller, we reanalyzed their data as well as ours using several methods, as described below.
Chen and Miller's Method of Fitting (Method 1)
As described in detail in the supplemental material, Chen and Miller used a two-step method (Method 1) to fit the opening rate constant to the four- and five-state models, as a function of voltage and chloride. We used Method 1 to fit our data and Chen and Miller's to both the four- and five-state models. To best compare these two datasets we needed to compare the parameter values resulting from the fits to the kinetic models. We found that the range of voltages included in the fit significantly affected the resulting parameter values (Fig. 3). This suggests that the error on the parameter values is large and that the datasets cannot be compared without accurate error estimates. Unfortunately, since Method 1 is a two-step process, it is difficult to estimate the error for the parameter values determined using this method.
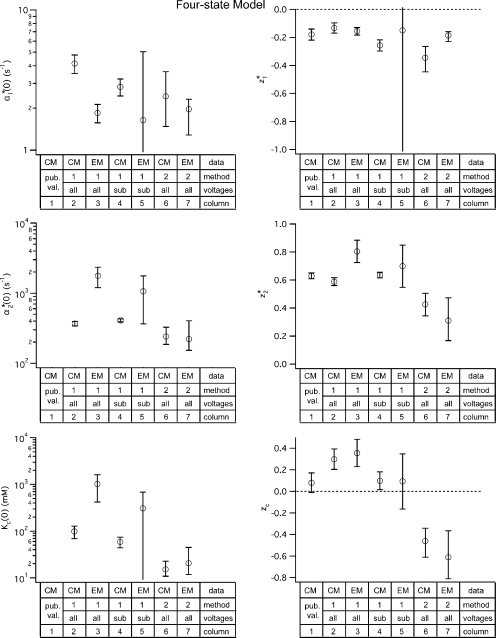
Figure 3.
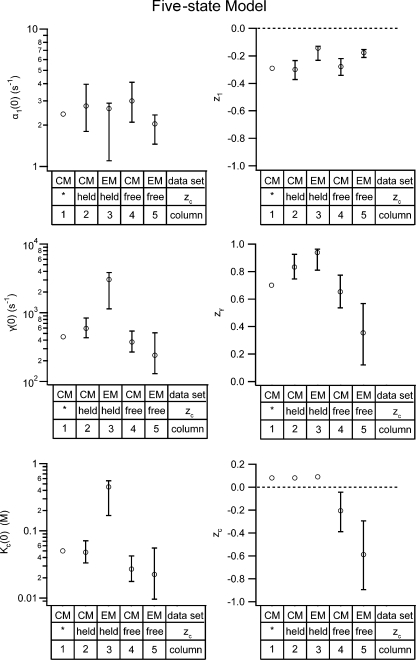
Parameter values for the four-state gating model. Values for the four-state model parameters were derived for both the dataset published by Chen and Miller in 1996 (CM) and for the dataset we obtained using macroscopic recordings (EM). These parameter values were derived using two different fitting methods: (1) Method 1, used by Chen and Miller, shown in Figure S4; and (2) Method 2, globally fitting the opening rate constant as a function of voltage and external chloride (α(V, [Cl]ext)), allowing all six parameters to be free, shown as solid lines in Fig. S5, C and F. Method 1 was performed including either all the voltages (all) or some subset of the voltages (sub). For Method 1, the error bars show the standard deviation calculated by the fitting algorithm (for fits shown in Fig. S4, B and D); for Method 2, the error bars show the 95% confidence limits determined as described in the text. For comparison, the first column shows the parameter values (z 1 *, z 2 *, and z c) published by Chen and Miller (pub. val.); values for α1 *(0), α2 *(0), and K c(0) were not reported.
Fitting the Data Using Global Fits (Method 2)
Because of the difficulty in estimating errors when using Method 1, we used a different method of fitting the data that allowed us to estimate the error in the parameter values calculated. This enabled us to determine whether our dataset yields significantly different results from that of Chen and Miller. In this approach, Method 2, the data were globally fit, allowing all six parameters to be free. When Method 2 is used, both Chen and Miller's dataset (solid lines in Fig. 2 B and Fig. S5 C) and our dataset (solid lines in Fig. 2 A and Fig. S5 F) fit well to both the four- and five-state models.
Four-State versus Five-State model
Since Chen and Miller (1996) rejected the four-state model because it did not fit their data well, we were surprised that when using Method 2, both our data and those of Chen and Miller fit well to the four-state model. The reason for this discrepancy is that when Method 1 is used (as in Chen and Miller, 1996), the four-state model parameters derived predict that within the range of the data (a) there will be an intersection point for all the curves of α versus V (where each curve represents a different chloride concentration); and (b) once the chloride binding is saturated, α will vary log-linearly with voltage (instead of having a chevron shape). However, when Method 2 is used to analyze the data, the resulting parameters are such that the intersection point occurs outside the range of the data, and α is no longer predicted to vary log-linearly with voltage at saturating chloride concentrations. Therefore at this point there is no strong evidence to suggest whether the four-state or the five-state model is more accurate. Although both models have six free parameters, and therefore mathematically are of equal complexity, the four-state model is conceptually simpler. Since it is important to compare our results for wild-type ClC-0 to those published by Chen and Miller, we will continue to include analysis using the five-state model since that was the model Chen and Miller chose to use for their final analysis. In the Discussion, however, we will only refer to the four-state model and its parameters.
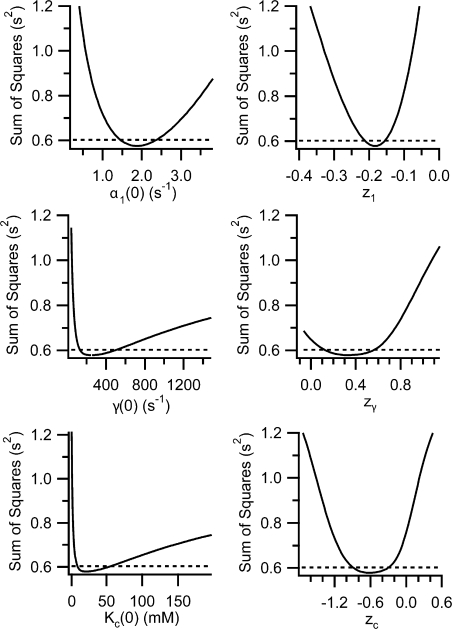
Error Analysis on Global Fits
Since Method 2 is a one-step fitting process where all the data from a particular dataset are fit at once, we were able to readily estimate the errors in parameter values calculated by this global fitting method. To estimate these errors, we used a procedure to test how changing a given parameter value affects the goodness of fit to the five-state model. To estimate how much each parameter value could vary and still yield a good fit to the data, we used the following procedure: (a) the parameter of interest was held at a test value and α(V, [Cl]ext) was globally fit with the other five parameters allowed to find their best-fit values; (b) the best-fit values of the other five parameters were stored and the sum of squares (SS), a measure of the goodness of fit, was computed as
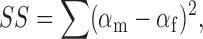 |
(6) |
where for every α(V, [Cl]ext) point, αm is the measured α value, and αf is the α value calculated by the fit; (c) steps (a) and (b) were repeated over a large range of test values. For each of the six parameters in the model, steps (a)–(c) were performed for a wide range of test values. Plotting the sum of squares (SS) as a function of each parameter (Fig. 4) results in minima near the best-fit value (for the fit where all parameters were free) (compare values in Fig. 5, column 5, to parameter values at the SS minima in Fig. 4). The plots in Fig. 4 show how markedly the goodness of fit depends on each parameter, allow us to be sure that we sampled a large enough range of parameter values, and show that there are no other minima. We also used the plots in Fig. 4 to estimate confidence intervals for each parameter that was determined by globally fitting α(V, [Cl]ext). To obtain 95% confidence bars, we determined the sum of squares at 95% confidence:
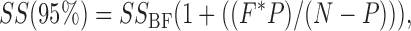 |
(7) |
where SS BF is the sum of squares for the best fit when all parameters are free, P is the number of free parameters in the model, N is the number of data points being fit, and F is the critical value of the F distribution for a p-value of 0.05 (Motulsky and Christopoulos, 2004). For each parameter determined by global fitting, 95% confidence intervals were obtained as the two parameter values that result in a sum of squares equal to the sum of squares at 95% confidence (see the intersection of the two lines in plots shown in Fig. 4). These 95% confidence intervals are shown as error bars in Figs. 3 and 5, and are much larger than the standard error values calculated by the Igor fitting algorithm.
Figure 4.
Error analysis on fits to the five-state model. To determine the certainty of the best-fit values found when globally fitting α(V, [Cl]ext) to the five-state model (Method 2), we used the procedure described in the text. Each of the plots shows how varying one of the six parameters affects the goodness of fit to our data as given by the sum of squares (solid lines). The sum of squares at 95% confidence is shown as a dashed line, and the parameter values at the intersections of the solid line and dashed line represent the 95% confidence limits, shown as error bars in Fig. 5. A similar procedure was used to calculate the 95% confidence limits on the four-state model parameter values, which are shown as error bars in Fig. 3, columns 6 and 7 of each graph.
Figure 5.
Parameter values for the five-state gating model. Values for the five-state model parameters were derived for both the dataset published by Chen and Miller in 1996 (CM) and for the dataset we obtained using macroscopic recordings (EM). In both cases, parameter values were derived by globally fitting the opening rate constant as a function of voltage and external chloride (α(V, [Cl]ext)) (Method 2) (fits shown in Fig. 2, A and B), either with all six parameters free (free) or while holding z c at 0.08 (held) (see the supplemental material for explanation, and Figs. 3 and S4, subset of voltages). The error bars show the 95% confidence limits determined as described in the text. For comparison, the first column shows the parameter values published by Chen and Miller (*).
With these error bars it is easy to see that when Method 2 is used, there is no significant difference between the parameter values that result for our dataset and that of Chen and Miller, for both the four-state (Fig. 3, compare the last two columns in each chart) or five-state models (Fig. 5, compare the last two columns in each chart). Thus, despite the differences in data collection techniques, internal chloride concentrations used, and voltage range analyzed, the parameters for ClC-0 opening are not significantly different.
The Value of zc
The error bars in Figs. 3 and 5 illustrate that some of the parameter values calculated using Method 2 are significantly different from those previously published (for each graph compare the first column to the last two). The difference that is conceptually important is that in our analysis of both our data and those of Chen and Miller, using Method 2 and either the four- or five-state models (Fig. 3 and 5, respectively), we found that z c is significantly different from zero, between −0.9 and −0.1. This demonstrates that chloride binding during fast-gate opening (the K c step) is voltage dependent, contrary to the conclusions of Chen and Miller (1996). Since z c represents the voltage dependence of K c, the equilibrium constant for chloride dissociation, the z value for chloride association, z a, is greater than zero (z a = −z c). This suggests that the K c step involves the binding of chloride from the extracellular milieu to site(s) within the transmembrane electric field.
Since this is an important result, we wanted to evaluate the probability that the chloride-binding step is not voltage dependent. To evaluate this possibility, we tested to see how holding z c at a value near to zero would affect the other results. We used Method 2 and globally fit both our data and those of Chen and Miller to the five-state model while holding z c constant at 0.08, the value published by Chen and Miller (1996). This approach resulted in the fits shown as dashed lines in Fig. 2 (A and B), and the parameter values in Fig. 5 (column 2 and column 3 in each graph). Since holding z c to 0.08 gives reasonable-looking fits, but significantly affects some of the parameter values, we needed to know if this alternate method of fitting could be more valid.
To this end, we used an F-test to compare the two methods for fitting the data to the five-state model: the method where only five parameters are free (z c is held) and the method where all six parameters are free. The F-test takes into account the difference in goodness of fit and the difference in degrees of freedom. In this test, described by Motulsky and Christopolous (2004), an F ratio is calculated:
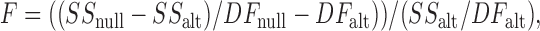 |
(8) |
where SS is the sum of squares for the best fit to the data, and DF is the degrees of freedom ((number of data points) − (number of free parameters)). In this case, the fit using six free parameters is the more complex “alt” model and the fit using five free parameters is the less complex “null” model. We calculated F ratios of 43 and 121 for Chen and Miller's dataset and ours, respectively. These correspond to p-values of 2 × 10−9 and 3 × 10−23. The p-values indicate the likelihood that the simpler model, with only five free parameters, is the better model to use. This statistic indicates that the method of using a global fit with all six parameters free was a better method to use. Therefore it is unlikely that z c is zero, and from these data we conclude that chloride binding during fast gating is voltage dependent.
The Interdependence of Parameters in the Five-State Model
Since holding z c at 0.08 changed the values calculated for the other parameters in the global fit, we realized that the parameters are likely to be interdependent. This could explain why some of the error bars on the gating parameters are so large (last two columns in each graph of Figs. 3 and 5). Interdependence between parameters means that there are not enough features in the relationship between the opening rate constant as function of voltage and external chloride to allow all six parameters in the gating model(s) to be uniquely determined. Therefore, there are a number of fits that are all equally good.
As described in the supplemental material, we used the data from the error analysis to determine whether any of the parameters in the four- or five-state models are interdependent. These results are summarized in Table III. Although some of the pairs of parameters are expected to be interdependent (e.g., for the five-state model α1(0) and z 1; γ(0) and z γ; K c(0) and z c, see Eqs. S5–S8 in the supplemental material), there are several sets of interdependent parameters that were unexpected. Namely, interdependencies were also found for the five-state model parameter pairs γ(0)-K c(0), z γ-K c(0), γ(0)-z c, and z γ-z c, and the four-state model parameter pairs α2 *(0)-K c(0), z 2 *-K c(0), α2 *(0)-z c, and z 2 *-z c. This was true for both our data and those of Chen and Miller (unpublished data). This strongly suggests that these parameters cannot be independently determined when globally fitting the α(V, [Cl]ext) data to the five-state model, and that the large error bars on the calculated best-fit parameter values are at least partially due to these interdependencies. The results of this analysis are similar when the four-state model is used.
TABLE III.
Interdependence of Parameters
| Four-state model | |||||
|---|---|---|---|---|---|
| z1 * | α2 * | z2 * | Kc(0) | z c | |
| α1 * | + | – | – | – | – |
| z1 * | – | – | – | – | |
| α2 * | ++ | ++ | + | ||
| z2 * | ++ | ++ | |||
| Kc(0) | + | ||||
| Five-state model | |||||
| z1 | γ | zγ | Kc(0) | z c | |
| α1 | ++ | – | – | – | – |
| z1 | – | – | – | – | |
| γ | + | ++ | + | ||
| zγ | + | ++ | |||
| Kc(0) | + |
Contour plots such as those shown in Fig. S6 were used to judge the interdependence of pairs of parameters when α(V, [Cl]ext) is globally fit (using Method 2) to the four- and five-state models. + indicates interdependence, ++ indicates strong interdependence, – indicates no interdependence.
Chloride Binding during the Kc Step
We wondered whether our finding that K c is voltage dependent, in combination with information from the X-ray crystal structures of ClC-ec1, could tell us about what chloride movements occur during the K c step. To address this question, we used information from the X-ray crystal structures of ClC-ec1 to try to learn more about the electrostatics of chloride binding in the pore. First, we used the structures of ClC-ec1 (Dutzler et al., 2003) in conjunction with sequence alignments (see Materials and methods and Fig. S3) to construct two homology models for ClC-0. One homology model was based on the structure of wild-type ClC-ec1, which has been proposed to resemble the ClC-0 closed state; the other homology model was based on the structure of the E148Q ClC-ec1 mutant, which has been proposed to resemble a ClC-0 conducting state. The difference between these two X-ray crystal structures is that in the wild-type structure the S ext chloride-binding site is occupied by the side chain of E148, and in the E148Q mutant structure the 148 side chain is rotated out and a chloride is bound in S ext. Both of these homology models were made with minimal adjustments to the ClC-ec1 structures (see Materials and methods). Thus, the overall shape of the pore was preserved, as were the three chloride-binding sites detected in that structure, namely S int, S cen, and S ext (moving from the cytoplasm to the extracellular space). We then performed an analysis of the electrostatics of chloride binding to the homology model, using the Poisson-Boltzmann framework.
Our analysis of the homology models suggests that chloride binding to all three sites is voltage dependent. This is illustrated in Fig. 6 A, which shows the fraction of transmembrane potential (V/V tm) a chloride ion has to traverse to reach each of the three binding sites along a putative permeation pathway. (Results from the two homology models are barely distinguishable.) The V/V tm plot shows that a chloride ion coming from the extracellular milieu would have to traverse a significant portion of the transmembrane potential to reach S ext, S cen, or S int. This means that chloride binding to any of these sites, relative to the bulk, would be voltage dependent. Since K c is voltage dependent, having a z c between −0.1 and −0.9, it is likely to involve chloride binding to one of these three sites.
Electrostatic Contribution to the Free Energy of Chloride Binding
Previous experimental work has shown that both conductance and gating show anomalous mole fraction effects in CLC channels (Pusch et al., 1995; Rychkov et al., 1998). Since this suggests that chloride–chloride repulsion in the pore plays a significant role in ClC-0 function, we wanted to characterize the energetic coupling between chloride ions in the pore and evaluate how this coupling could affect fast gating. Using the ClC-0 homology model inspired by the E148Q ClC-ec1 structure, we calculated the electrostatic contribution to the free energy of chloride binding (ΔG b) to S int, S cen, S ext (Eq. 1). Fig. 6 B shows that for chloride binding to any of the three sites, the chloride binding affinity is less when there are two chlorides already in the pore (squares) than when there are no chlorides already bound (circles). The results of this electrostatic analysis are therefore consistent with the anomalous mole fraction data and previous in silico analyses (Cohen and Schulten, 2004; Gervasio et al., 2006). As described in the supplemental material, in terms of electrostatics, the effect of having the E148 side chain (E166 in ClC-0) rotated into the S ext site appears to be comparable to that of having a chloride bound in this site. Hence the calculated ΔG b values for S cen and S int are similar for the homology model based on the wild-type ClC-ec1 structure and the homology model inspired by the E148Q mutant when it has a chloride bound in S ext. As discussed below, comparison of ΔG b for different sites and under different conditions helps to evaluate models for chloride movement during fast gating.
DISCUSSION
Here we have shown that using macroscopic patch recordings we can reproduce the effects of voltage and external chloride on the open probability and gating kinetics reported by Chen and Miller (1996) based on their single-channel recordings. We have also shown that the method of analysis greatly affects the gating parameter values derived from these data. We propose an alternate methodology (Method 2) that involves globally fitting the data with all parameters free. This approach allows for (a) a quantitative determination of the error in the calculated parameter values, which is essential for comparing different datasets, and (b) an assessment of the interdependence of the parameters in the gating model. The interdependence between the parameters probably makes a large contribution to the error in the parameter values determined. Our analysis using Method 2 shows that (a) our data and those of Chen and Miller yield similar gating parameter values; (b) the four- and five-state models fit the data equally well; and (c) the chloride-binding step that leads to channel opening is voltage dependent. This last finding changes our conceptualization of the gating process, as described in the next section.
A Model for Chloride Movement during Gating
Our ultimate goal is to understand the structural changes that occur during fast gating in ClC-0. As a first step toward a structural model for gating, we aimed to use previously published results and our new findings to develop a model for how chloride moves during depolarization-activated fast-gate opening: C→C⋅Cl→O. Given that there are at least three chloride-binding sites in ClC-0, there are many possibilities for how chloride could move during depolarization-activated fast-gate opening.
K c Is Likely to Involve Binding to S ext, S cen, or S int
Our homology model analysis suggests that the chloride-binding sites S ext, S cen, and S int in ClC-0 are all significantly into the transmembrane field (Fig. 6 A). (It is important to note that the transmembrane voltage profile is primarily sensitive to the extent to which the pore can be permeated by high-dielectric solvent. Therefore, for any reasonable model based on the ClC-ec1 structures, it is improbable that any of the binding sites mentioned above would be equipotential with the bulk environment.) However, it is conceivable that the extracellular vestibule is wider in ClC-0 than in ClC-ec1. We wondered whether the extracellular vestibule of ClC-0 could be so wide that S ext is not within the transmembrane field, such that chloride from the extracellular milieu binding to this site would not be voltage dependent. To examine the impact that widening the extracellular vestibule may have on the transmembrane profile, we built a ClC-0 homology model lacking helix L, a helix that lines the extracellular vestibule. In spite of the much greater solvent accessibility of this region of the protein that results from the deletion of this helix, the transmembrane voltage at the chloride-binding sites was found to be essentially unchanged relative to our original model (unpublished data). Since K c is voltage dependent, it is therefore reasonable to conclude based on this analysis that the K c step is likely to involve binding to S ext, S cen, or S int.
There remains the question of whether K c could involve chloride binding to some yet undiscovered site extracellular to S ext. Since z c is between −0.1 and −0.9, such a site would have to be at least 10% into the transmembrane electric field, which corresponds to a position ∼6–7 Å into the pore. One possibility is the S ext* site proposed by Faraldo-Gómez and Roux (2004), which is in the vicinity of H401, or alternatively, the sites proposed by Yin et al. (2004) and Bostick and Berkowitz (2004), near K165. These proposals are worth noting in view of electrophysiological data showing that K165 mutations affect both permeation and fast gating (Lin and Chen, 2000), and that H401 mutations affect permeation (Zhang et al., 2006). However, mutation at H401 does not affect gating, and the effects of mutations at K165 and H401 could occur through allostery. All these predictions of a fourth chloride-binding site are based on computational analyses of the ClC-ec1 structures, and no structural data is as yet available that confirms the existence of a such a site, including the more recent crystallographic studies (Lobet and Dutzler, 2006; Accardi et al., 2006). Thus it is unclear whether H401 and K165 line an additional extracellular chloride-binding site or whether the mutations exert their effects via some other mechanism. Therefore in our models for chloride movement during gating, we have only included the three chloride-binding sites observed in the X-ray crystal structures.
Chloride Movement during the α2 * Step
Analysis of ClC-0 gating kinetics indicates that the step following chloride binding, the α2 * step (referring now only to the four-state model), is voltage dependent. Given the previously observed anomalous mole fraction effects on V o (the voltage at the midpoint of the voltage-activation curve) (Pusch et al., 1995), this voltage dependence has been thought to be due to the movement of chloride through the pore. The calculated V/V tm profile (Fig. 6 A) suggests that a chloride ion has to traverse a significant portion of the transmembrane field to travel between binding sites in the pore, and therefore all steps in gating that require chloride movements between sites are going to be voltage dependent. This leads us to conclude that the α2 * step could involve chloride movement between any of the sites in the pore. Our error analysis shows that the voltage dependence of these steps cannot be determined with great accuracy: z 2 * is between 0.2 and 0.5. Our homology model analysis predicts that chloride would have to traverse ∼20% of the transmembrane field to move between neighboring binding sites in the pore. Therefore, if the actual z value for the α2 * step is >0.2, then this step would have to involve more than just a single chloride moving between neighboring binding sites in the pore. It could involve movements of more than one bound chloride, or could involve the movement of one chloride through more than one site. Finally, it could involve movement of charged protein elements through the transmembrane field, or as recently proposed, movement of protons (Miller, 2006; Traverso et al., 2006).
S ext Gets Filled during Gating
Work by Dutzler et al. (2003) helps narrow down the number of likely models for chloride movement. Dutzler et al. (2003) found that the S ext site is occupied by a glutamate side chain (E148) in the wild-type ClC-ec1 crystal structure, and becomes occupied by chloride when this glutamate has been mutated to alanine or glutamine. Mutation of the equivalent glutamate side chain in ClC-0 (E166) results in channels that are almost always open, which indicates that this residue may act as a physical gate. This leads to the hypothesis that the outward movement of E166 precedes chloride binding to S ext, and that having a chloride in this site prevents E166 from moving inward, thus keeping the gate open. This suggests that S ext is filled during fast-gate opening, meaning that the open state (O) has chloride bound to S ext, and the first closed state (C) does not.
It should be noted that another model for how E166 moves during gating has been proposed by Bisset et al. (2005). According to their calculations, the more conductive state (the open state) has the side chain of E166 facing down into the pore, and the less conductive state (the closed state) has the side chain facing out into the extracellular milieu. In this model, the E166 side chain does not prevent chloride binding to S ext.
Hypothetical Models for Chloride Movement
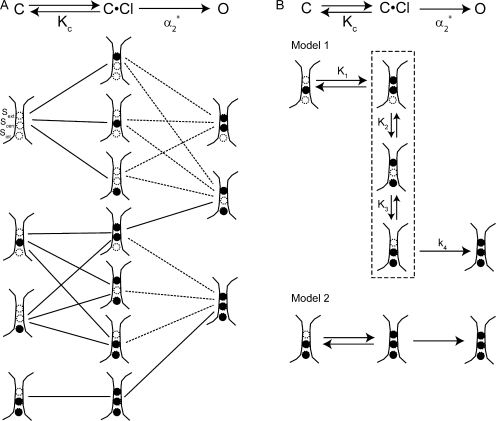
We used the evidence discussed so far to generate models for chloride movement during depolarization-activated gating. After considering a wide range of possibilities, we arrived at a set of more likely models (Fig. 7 A) based on the following criteria: (a) only S ext, S cen, and S int are considered; (b) the K c step involves chloride from the extracellular milieu binding to one of these sites; (c) the α2 * step can involve depolarization-activated chloride movement (solid lines) or binding (dashed lines) (but does not have to; it could involve depolarization-activated conformational changes that do not affect chloride occupancy); (d) S ext becomes occupied during the course of gating, and must be occupied for the channel to remain open. The lines connecting states indicate likely transitions, given the simplifying assumption that only one chloride can bind per step. It is important to note that the open state (O) depicted does not represent all the possible chloride occupancy states explored once chloride has started to conduct through the pore, but rather the minimum chloride occupancy required for the channel to remain open.
Figure 7.
Possible models for chloride movement during depolarization-activated fast gating. Evidence from previously published papers and our kinetic and homology model analysis was used to narrow down the possible models for chloride movement during depolarization-activated fast-gate opening to those shown (see text). The channel begins in the closed state C, binds chloride to reach the closed state C·Cl, and then goes through another depolarization-activated transition to reach the open state O. This is depicted at the top of both A and B. K c is the apparent equilibrium constant for chloride dissociation, and α2 * is the rate constant for the second step, C·Cl→O. In A, depicted below each of these states (C, C·Cl, and O) are their possible chloride occupancy states, with empty circles depicting vacant chloride binding sites, and filled circles depicting filled chloride binding sites. Lines between these chloride-occupancy states indicate transitions corresponding to either C→C·Cl or C·Cl→O. Dashed lines indicate C·Cl→O transitions that involve chloride binding to the pore. Evidence from our analysis of gating kinetics and the ClC-0 homology models further narrowed the possibilities to those shown in B. In Model 1, the dashed line outlines the microstates that are all part of C·Cl. K 1, K 2, and K 3 are microscopic equilibrium constants, and k 4 is a microscopic rate constant. In Model 2, the second step has to involve some conformational change that opens the channel but does not change chloride occupancy.
The Two Most Likely Models
Analyses of ClC-ec1 structures suggest that the likely models shown in Fig. 7 A can be narrowed down further. These analyses come in two forms. First, Lobet and Dutzler (2006) obtained and analyzed ClC-ec1 crystal structures under a variety of anion concentrations in order to estimate the apparent binding affinity of chloride for each of the three sites observed. Second, we estimated the electrostatic contribution to the chloride binding affinity for each of the three sites for all the possible occupancy states, (Fig. 6 B; Fig. S7). Both these analyses suggest the following. (a) ClC-0 has a sufficiently high affinity for chloride such that the state where no chloride is bound is unlikely under our experimental conditions (5–610 mM extracellular chloride, 120 mM intracellular chloride). If the chloride-free state (the top C state in Fig. 7 A) is eliminated from the list of likely possibilities, then by extension we eliminate the C·Cl and O states connected only to this C state. (b) Chloride appears to have a higher affinity for S cen than S int, such that the second C state (from the top) in Fig. 7 A is more likely than the third, and the α2 *-step transition that involves movement of chloride from S cen to S int is unlikely.
Therefore, based on the available information, we can winnow down the possible models in Fig. 7 A to the two most likely options. These two options are shown in Fig. 7 B: Model 1, (top) C has one chloride bound in S cen, C·Cl has two chlorides bound, and O has three chlorides bound; Model 2, (bottom) C has two chlorides bound in S cen and S int, C·Cl has three chlorides bound, and O also has three chlorides bound. In Model 1, we have modified the first option so that each step is simplified and only involves either the binding of chloride to S ext or the downward movement of chloride between adjacent binding sites in the pore. The C·Cl state consists of three microstates in equilibrium (outlined by a dashed line).
These two models are fundamentally different from one another. In Model 1, each of the steps can be simply the chloride binding or chloride movement shown. Although additional protein structural rearrangements are allowed, this model does not require any additional conformational changes other than that the E166 side chain rotate out of S ext to allow the binding of chloride to this site. The only requirement for opening is that all sites are occupied with chloride. In contrast, both steps in Model 2 require protein structural movements: the K c step involves the rotation of E166 and the binding of chloride to S ext; the α2 * step has to involve some structural change that does not change chloride occupancy but makes conduction through the channel easier. For example, this transition could involve the lowering of barriers to chloride movement between the chloride-binding sites. In Model 2 there are two requirements for opening: (1) that all three sites are occupied with chloride and (2) that some additional protein conformational change has occurred so that the channel can conduct. Additional protein conformational changes in CLC gating have been proposed by others (Accardi and Pusch, 2003; Bell et al., 2006). Consistent with the models recently proposed by Miller (2006) and Traverso et al. (2006), the additional conformational change in Model 2 could involve proton movement through the transmembrane field, and this movement could be partly or wholly responsible for the voltage dependence of the α2 * step. Further studies on the pH dependence of gating would be informative regarding this possibility. Model 2 may also be slightly more likely than Model 1 based on the apparent chloride affinities calculated by Lobet and Dutzler (2006). Since for all three sites the apparent affinities Lobet and Dutzler calculate are in the millimolar range, and we used 120 mM chloride on the cytoplasmic side, it is likely, if these apparent affinities hold for ClC-0, that both S int and S cen were filled before the channel opening that we measured.
Conclusion
In conclusion, we have shown that the method of analyzing the dependence of the opening rate constant on voltage and chloride affects the conclusions drawn about the mechanism of fast gating in ClC-0. We have shown that both the four- and five-state gating models fit the data, and that the accuracy of the calculated gating parameter values is compromised by the interdependence of parameters. Despite the uncertainty in determining gating parameter values, we can conclude that chloride binding during gating is a voltage-dependent process, and must involve a binding site at least 10% into the transmembrane field. In this regard, our homology model analysis shows that any of the three known chloride-binding sites could play a role in gating. By considering results from our kinetic analysis, previously published results, and available structural information, we propose two possible models for chloride movement during depolarization-activated fast gating. Although more information is needed to determine which of these two models is more likely, understanding the assumptions in each of these models provides fodder for future experiments. Both the methods of analysis and error estimation described here and the structural models proposed provide a framework for evaluating how mutations in ClC-0 affect fast gating.
Supplemental Material
Acknowledgments
We thank Gilbert Martinez and Drs. Richard Aldrich, Kimberly Matulef, and Joseph Mindell for comments on the manuscript. We also thank Dr. Benoît Roux for helpful discussions and Dr. Dan Cox for advice on data analysis.
This work has been supported by National Institutes of Health grant R01 GM070773 and by the Mathers Foundation. A. Engh was supported in part by a Predoctoral Fellowship from the American Heart Association.
Olaf S. Andersen served as editor.
References
- Accardi, A., and M. Pusch. 2003. Conformational changes in the pore of ClC-0. J. Gen. Physiol. 122:277–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Accardi, A., S. Lobet, C. Williams, C. Miller, and R. Dutzler. 2006. Synergism between halide binding and proton transport in a CLC-type exhanger. J. Mol. Biol. 362:691–699. [DOI] [PubMed] [Google Scholar]
- Barnett, V., and T. Lewis. 1994. Outliers in Statistical Data. Third ed. John Wiley and Sons, New York. 164.
- Bauer, C.K., K. Steinmeyer, J.R. Schwarz, and T.J. Jentsch. 1991. Completely functional double-barrelled chloride channel expressed from a single Torpedo cDNA. Proc. Natl. Acad. Sci. USA. 88:11052–11056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, S.P., P.K. Curran, S. Choi, and J.A. Mindell. 2006. Site-directed fluorescence studies of a prokaryotic ClC antiporter. Biochemistry. 45:6773–6782. [DOI] [PubMed] [Google Scholar]
- Bisset, D., B. Corry, and S.H. Chung. 2005. The fast gating mechanism in ClC-0 channels. Biophys. J. 89:179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick, D.L., and M.L. Berkowitz. 2004. Exterior site occupancy infers chloride-induced proton gating in a prokaryotic homolog of the ClC chloride channel. Biophys. J. 87:1686–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks, B.R., R.E. Bruccoleri, B.D. Olafson, D.J. States, S. Swaminathan, and M. Karplus. 1983. CHARMM: a program for macromoleculr energy, minimization, and dynamics calculations. J. Comp. Chem. 4:187–217. [Google Scholar]
- Canutescu, A.A., A.A. Shelenkov, and R.L. Dunbrack. 2003. A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 12:2001–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, H.S., and K. Dill. 1998. Protein folding in the landscape perspective: chevron plots and non-arrhenius kinetics. Proteins. 30:2–33. [DOI] [PubMed] [Google Scholar]
- Chen, M.F., and T.-Y. Chen. 2001. Different fast-gate regulation by external Cl− and H+ of the muscle-type ClC chloride channels. J. Gen. Physiol. 118:23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T.-Y. 2005. Structure and function of CLC channels. Annu. Rev. Physiol. 67:809–839. [DOI] [PubMed] [Google Scholar]
- Chen, T.-Y., and C. Miller. 1996. Nonequilibrium gating and voltage dependence of the ClC-0 Cl− channel. J. Gen. Physiol. 108:237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleiren, E., O. Benichou, E. Van Hul, J. Gram, J. Bollerslev, F.R. Singer, K. Beaverson, A. Aledo, M.P. Whyte, T. Yoneyama, et al. 2001. Albers-Schonberg disease (autosomal dominant osteopetrosis, type II) results from mutations in the ClCN7 chloride channel gene. Hum. Mol. Genet. 10:2861–2867. [DOI] [PubMed] [Google Scholar]
- Cohen, J., and K. Schulten. 2004. Mechanism of anionic conduction across ClC. Biophys. J. 86:836–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colding-Jorgensen, E. 2005. Phenotypic variability in myotonia congenita. Muscle Nerve. 32:19–34. [DOI] [PubMed] [Google Scholar]
- Dhani, S.U., and C.E. Bear. 2006. Role of intramolecular and intermolecular interactions in ClC channel and transporter function. Pflugers Arch. 451:708–715. [DOI] [PubMed] [Google Scholar]
- Dutzler, R. 2006. The ClC family of chloride channels and transporters. Curr. Opin. Struct. Biol. 16:439–436. [DOI] [PubMed] [Google Scholar]
- Dutzler, R., E.B. Campbell, and R. Mackinnon. 2003. Gating the selectivity filter in ClC chloride channels. Science. 300:108–112. [DOI] [PubMed] [Google Scholar]
- Engh, A.M., and M. Maduke. 2005. Cysteine accessibility in ClC-0 supports conservation of the ClC intracellular vestibule. J. Gen. Physiol. 125:601–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engh, A.M., J.D. Faraldo-Gómez, and M. Maduke. 2007. The role of a conserved lysine in chloride- and voltage-dependent ClC-0 fast gating. J. Gen. Physiol. 130:351–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez, R., and T.J. Jentsch. 2002. CLC chloride channels: correlating structure with function. Curr. Opin. Struct. Biol. 12:531–539. [DOI] [PubMed] [Google Scholar]
- Estevez, R., B.C. Schroeder, A. Accardi, T.J. Jentsch, and M. Pusch. 2003. Conservation of chloride channel structure revealed by an inhibitor binding site in ClC-1. Neuron. 38:47–59. [DOI] [PubMed] [Google Scholar]
- Faraldo-Gómez, J.D., and B. Roux. 2004. Electrostatics of ion stabilization in a ClC chloride channel homologue from Escherichia coli. J. Mol. Biol. 339:981–1000. [DOI] [PubMed] [Google Scholar]
- Gervasio, F.L., M. Parnello, M. Ceccarellil, and M.L. Klein. 2006. Exploring the gating mechanism in the ClC chloride channel via metadynamics. J. Mol. Biol. 361:390–398. [DOI] [PubMed] [Google Scholar]
- Hanke, W., and C. Miller. 1983. Single chloride channels from Torpedo electroplax. Activation by protons. J. Gen. Physiol. 82:25–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haug, K., M. Warnstedt, A.K. Alekov, T. Sander, A. Ramirez, B. Poser, S. Maljevic, S. Hebeisen, C. Kubisch, J. Rebstock, et al. 2003. Mutations in CLCN2 encoding a voltage-gated chloride channel are associated with idiopathic generalized epilepsies. Nat. Genet. 33:527–532. [DOI] [PubMed] [Google Scholar]
- Im, W., D. Beglov, and B. Roux. 1998. Continuum solvation model: computation of electrostatic forces from numerical solutions to the Poisson-Boltzmann equation. Comput. Phys. Commun. 111:59–75. [Google Scholar]
- Jentsch, T.J., K. Steinmeyer, and G. Schwartz. 1990. Primary structure of Torpedo mamorata chloride channel isolated by expression cloning in Xenopus oocytes. Nature. 348:510–514. [DOI] [PubMed] [Google Scholar]
- Jentsch, T.J., V. Stein, F. Weinreich, and A.A. Zdebik. 2002. Molecular structure and physiological function of chloride channels. Physiol. Rev. 82:503–568. [DOI] [PubMed] [Google Scholar]
- Jentsch, T.J., M. Poet, J.C. Fuhrmann, and A.A. Zdebik. 2005. Physiological functions of CLC Cl− channels gleaned from human genetic disease and mouse models. Annu. Rev. Physiol. 67:779–807. [DOI] [PubMed] [Google Scholar]
- Jurkat-Rott, K., and F. Lehmann-Horn. 2005. Muscle channelopathies and critical points in function and genetic studies. J. Clin. Invest. 115:2000–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornak, U., D. Kasper, M.R. Bosl, E. Kaiser, M. Schweizer, A. Schulz, W. Friedrich, G. Delling, and T.J. Jentsch. 2001. Loss of the ClC-7 chloride channel leads to osteopetrosis in mice and man. Cell. 104:205–215. [DOI] [PubMed] [Google Scholar]
- Lehmann-Horn, F., and K. Jurkat-Rott. 1999. Voltage-gated ion channels and hereditary disease. Physiol. Rev. 79:1317–1372. [DOI] [PubMed] [Google Scholar]
- Lin, C.W., and T.-Y. Chen. 2000. Cysteine modification of a putative pore residue in ClC-0: implication for the pore stoichiometry of ClC chloride channels. J. Gen. Physiol. 116:535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C.W., and T.-Y. Chen. 2003. Probing the pore of ClC-0 by substituted cysteine accessibility method using methane thiosulfonate reagents. J. Gen. Physiol. 122:147–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y.-W., C.-W. Lin, and T.-Y. Chen. 1999. Elimination of the slow gating of ClC-0 chloride channel by a point mutation. J. Gen. Physiol. 114:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, S.E., S.H. Pearce, S.E. Fisher, K. Steinmeyer, B. Schwappach, S.J. Scheinman, B. Harding, A. Bolino, M. Devoto, P. Goodyer, et al. 1996. A common molecular basis for three inherited kidney stone diseases. Nature. 379:445–449. [DOI] [PubMed] [Google Scholar]
- Lobet, S., and R. Dutzler. 2006. Ion-binding properties of the ClC chloride selectivity filter. EMBO J. 25:24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludewig, U., M. Pusch, and T.J. Jentsch. 1996. Two physically distinct pores in the dimeric ClC-0 chloride channel. Nature. 383:340–343. [DOI] [PubMed] [Google Scholar]
- MacKerell, A.D., D. Bashford, M. Bellott, R.L. Dunbrack, J.D. Evanseck, M.J. Field, S. Fischer, J. Gao, H. Guo, S. Ha, et al. 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Physiol.Chem. B. 102:3586–3616. [DOI] [PubMed] [Google Scholar]
- Maduke, M., D.J. Pheasant, and C. Miller. 1999. High-level expression, functional reconstitution, and quaternary structure of a prokaryotic ClC-type chloride channel. J. Gen. Physiol. 114:713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduke, M., C. Williams, and C. Miller. 1998. Formation of CLC-0 chloride channels from separated transmembrane and cytoplasmic domains. Biochemistry. 37:1315–1321. [DOI] [PubMed] [Google Scholar]
- Middleton, R.E., D.J. Pheasant, and C. Miller. 1996. Homodimeric architecture of a ClC-type chloride ion channel. Nature. 383:337–383. [DOI] [PubMed] [Google Scholar]
- Miller, C. 1982. Open-state substructure of single chloride channels from Torpedo electroplax. Philos. Trans. R. Soc. Lond. B Biol. Sci. 299:401–411. [DOI] [PubMed] [Google Scholar]
- Miller, C. 2006. ClC chloride channels viewed through a transporter lens. Nature. 440:484–489. [DOI] [PubMed] [Google Scholar]
- Miller, C., and E.A. Richard. 1990. The voltage-dependent chloride channel of Torpedo electroplax: intimations of molecular structure from quirks of single-channel function. In Chloride Channels and Carriers in Nerve, Muscle, and Glial Cells. F.J. Alvarez-Leefmans and J.M. Russell, editors. Plenum Publishing Corp., New York. 383–405.
- Motulsky, H., and A. Christopoulos. 2004. Fitting Models to Biological Data Using Linear and Nonlinear Regression: A Practical Guide to Curve Fitting. Oxford University Press, Inc., New York. 351 pp.
- Naesens, M., P. Steels, R. Verberckmoes, Y. Vanrenterghem, and D. Kuypers. 2004. Bartter's and Gitelman's syndromes: from gene to clinic. Nephron. Physiol. 96:65–78. [DOI] [PubMed] [Google Scholar]
- Nina, M., D. Beglov, and B. Roux. 1997. Atomic radii for continuum electrostatics calculations based on molecular dynamics free energy simulations. J. Physiol. Chem. B. 101:5239–5248. [Google Scholar]
- Nonner, W., A. Peyser, D. Gillespie, and B. Eisenberg. 2004. Relating microscopic charge movement to macroscopic currents: the Ramo-Shockley theorem applied to ion channels. Biophys. J. 87:3716–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrey, D., Z. Xiang, C.L. Tang, L. Xie, M. Gimpelev, T. Mitros, C.S. Soto, S. Goldsmith-Fischman, A. Kernytsky, A. Schlessinger, et al. 2003. Using multiple structure alignments, fast model building, and energetic analysis in fold recognition and homology modeling. Proteins. 53:430–435. [DOI] [PubMed] [Google Scholar]
- Pusch, M. 2002. Myotonia caused by mutations in the muscle chloride channel gene CLCN1. Hum. Mutat. 19:423–434. [DOI] [PubMed] [Google Scholar]
- Pusch, M. 2004. Structural insights into chloride and proton-mediated gating of CLC chloride channels. Biochemistry. 43:1135–1144. [DOI] [PubMed] [Google Scholar]
- Pusch, M., U. Ludewig, A. Rehfeldt, and T.J. Jentsch. 1995. Gating of the voltage-dependent chloride channel ClC-0 by the permeant ion. Nature. 373:527–531. [DOI] [PubMed] [Google Scholar]
- Pusch, M., S.E. Jordt, V. Stein, and T.J. Jentsch. 1999. Chloride dependence of hyperpolarization-activated chloride channel gates. J. Physiol. 515:341–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard, E.A., and C. Miller. 1990. Steady-state coupling of ion-channel conformations to a transmembrane ion gradient. Science. 247:1208–1210. [DOI] [PubMed] [Google Scholar]
- Roux, B. 1997. Influence of the membrane potential on the free energy of an intrinsic protein. Biophys. J. 73:2980–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rychkov, G.Y., M. Pusch, M.L. Roberts, T.J. Jentsch, and A.H. Bretag. 1998. Permeation and block of the skeletal muscle chloride channel, ClC-1, by foreign anions. J. Gen. Physiol. 111:653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sile, S., C.G. Vanoye, and A.L. George Jr. 2006. Molecular physiology of renal ClC chloride channels/transporters. Curr. Opin. Nephrol. Hypertens. 15:511–516. [DOI] [PubMed] [Google Scholar]
- Simon, D.B., R.S. Bindra, T.A. Mansfield, C. Nelson-Williams, E. Mendonca, R. Stone, S. Schurman, A. Nayir, H. Alpay, A. Bakkaloglu, et al. 1997. Mutations in the chloride channel gene, CLCNKB, cause Bartter's syndrome type III. Nat. Genet. 17:171–178. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., D.G. Higgins, and T.J. Gibson. 1994. Clustal-W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverso, S., G. Zifarelli, R. Aiello, and M. Pusch. 2006. Proton sensing of CLC-0 mutant E166D. J. Gen. Physiol. 127:51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida, S., and S. Sasaki. 2005. Function of chloride channels in the kidney. Annu. Rev. Physiol. 67:759–778. [DOI] [PubMed] [Google Scholar]
- White, M.M., and C. Miller. 1979. A voltage-gated anion channel from the electric organ of Torpedo californica. J. Biol. Chem. 254:10161–10166. [PubMed] [Google Scholar]
- Yin, J., Z. Kuang, U. Mahankali, and T.L. Beck. 2004. Ion transit pathways and gating in ClC chloride channels. Proteins. 57:414–421. [DOI] [PubMed] [Google Scholar]
- Zhang, X.D., Y. Li, W.P. Lu, and T.Y. Chen. 2006. Roles of K149, G352, H401 in the channel functions of ClC-0: testing the predictions from theoretical calculations. J. Gen. Physiol. 127:435–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.