Abstract
Nitrate serves as a potent signal to control gene expression in plants and algae, but little is known about the signaling role of nitrite, the direct product of nitrate reduction. Analysis of several nitrate-induced genes showed that nitrite increases mRNA levels as rapidly as nitrate in nitrogen-starved Arabidopsis (Arabidopsis thaliana) roots. Both nitrite and nitrate induction are apparent at concentrations as low as 100 nm. The response at low nitrite concentrations was not due to contaminating nitrate, which was present at <1% of the nitrite concentration. High levels of ammonium (20 mm) in the growth medium suppressed induction of several genes by nitrate, but had varied effects on the nitrite response. Transcriptome analysis using 250 or 5 μm nitrate or nitrite showed that over one-half of the nitrate-induced genes, which included genes involved in nitrate and ammonium assimilation, energy production, and carbon and nitrogen metabolism responded equivalently to nitrite; however, the nitrite response was more robust and there were many genes that responded specifically to nitrite. Thus, nitrite can serve as a signal as well as if not better than nitrate.
Nitrate acts as a signal and a nutrient that supports and regulates plant growth, development, and metabolism (Crawford, 1995; Stitt, 1999; Crawford and Forde, 2002; Forde, 2002). Early studies on nitrate signaling demonstrated that nitrate induces de novo synthesis of nitrate reductase (NR), the first enzyme in the nitrate assimilation pathway (Zielke and Filner, 1971; Somers et al., 1983; Remmler and Campbell, 1986). Subsequent work demonstrated that nitrate induces other genes in the nitrate assimilation pathway, namely, nitrate transporters (NRTs) and nitrite reductase (NiR), as well as genes involved in energy metabolism especially in the pentose phosphate pathway (Wang et al., 2000; for review, see Redinbaugh and Campbell, 1991; Stitt, 1999). Genomic analyses have now provided a comprehensive dataset of over 1,000 nitrate-responsive genes in Arabidopsis (Arabidopsis thaliana; Wang et al., 2003, 2004; Scheible et al., 2004; Gutierrez et al., 2007). These transcriptome studies have confirmed that nitrate assimilation and aspects of energy metabolism are nitrate responsive and have added glycolysis, trehalose-6-P metabolism, amino acid and nucleotide metabolism, and regulation (including kinases and transcription factors) as processes that respond to nitrate within 2 to 3 h of treatment.
As a nutrient, nitrate is converted first to nitrite, then to ammonium, and then to amino acids by the nitrate assimilation pathway. As such, it is possible that the downstream metabolites of nitrate could play a role in the nitrate response by serving as signaling compounds. For example, a specific gene that is induced 2 to 3 h after nitrate treatment may be responding to changes in the concentration of nitrite or ammonium instead of, or in addition to, nitrate. To investigate this possibility, a NR-null mutant lacking NR activity (i.e. produces no nitrite from NR) was constructed in Arabidopsis (Wang et al., 2004). A 2-h nitrate treatment was performed and the responses of both shoot and root transcriptomes were analyzed. It was discovered that, of the 1,596 genes that responded to nitrate in wild-type plants under these conditions, 595 genes also responded similarly in the NR-null mutant. These 595 genes did not require nitrate reduction to be induced or depressed and thus can respond directly to nitrate. Another 492 genes (out of the original 1,596 genes) responded only in the wild type and not in the mutant and thus required nitrate reduction to be induced or depressed. This class of genes may respond to downstream metabolites. To test this idea, we examined the effect of nitrite on the Arabidopsis root transcriptome.
Nitrite is thought to be a transient intermediate in the nitrate assimilation pathway, being produced from nitrate by NR and then being rapidly reduced to ammonium by NiR. Nitrite is considered a toxic metabolite because if it is allowed to accumulate, it can have deleterious effects on the plant. Nitrite concentrations in illuminated leaves from wild-type plants have been measured at 10 μm (Rockel et al., 2002). In contrast, NiR-deficient plants, which accumulate high nitrite levels, display chlorosis and dramatically reduced growth in tobacco (Nicotiana tabacum; Vaucheret et al., 1992) and rapid death in barley (Hordeum vulgare; Duncanson et al., 1993) if grown with nitrate. Such plants also produce high levels of nitric oxide (NO; Morot-Gaudry-Talarmain et al., 2002) because nitrite serves as a substrate for the formation of NO (Yamasaki et al., 1999; Rockel et al., 2002; Lea et al., 2004; Meyer et al., 2005; Planchet et al., 2005). It should be noted that nitrite can have a beneficial effect because it protects maize (Zea mays) roots from anoxia by reducing cytoplasmic acidosis (Libourel et al., 2006).
Very little is known about the signaling effects of nitrite. Being a precursor to NO, nitrite could serve as an indirect signal. For example, nitrite-dependent NO synthesis has been implicated in abscisic acid-induced stomatal closure (Desikan et al., 2004; Bright et al., 2006). However, not much is known about direct signaling by nitrite. Classic studies have shown that nitrite treatments can affect different steps in the nitrate assimilation pathway, but it was not clear whether nitrite was the actual signal. In barley roots, nitrite could inhibit nitrate uptake, but so could ammonium (King et al., 1993). In leaves of barley seedlings, nitrite increased NiR activity, but only after the nitrite was oxidized to nitrate, indicating that nitrate and not nitrite was the inducing signal (Aslam and Huffaker, 1989). In Chlamydomonas, several articles reported both positive and negative effects of nitrite on Nia1 promoter activity (Loppes et al., 1999; Llamas et al., 2002), but these were shown to be due to nitrate contamination in the nitrite solutions or to the inhibition of two of the nitrate-nitrite transport systems (Llamas et al., 2002).
More recent work has provided evidence that nitrite might act as a direct signal both to repress and induce gene expression. In Arabidopsis, the nonsymbiotic hemoglobin gene AtGLB1 is strongly induced after 2 h of 5 mm KNO2 treatment, which is similar to the response observed with 5 mm KNO3 (Sakamoto et al., 2004). In Physcomitrella, the NRT gene NRT2;5 is induced by 1 mm nitrite after 4 h, whereas NRT2;1 and NRT2;2 are strongly repressed (Tsujimoto et al., 2007). In Arabidopsis roots, 0.5 to 1.0 mm KNO2 repressed NRT1.1 and NIA1 mRNA levels after 6 and 24 h of treatment (Loque et al., 2003). These results motivated us to perform a systematic investigation of the role of nitrite as a signal that regulates gene expression.
RESULTS
Nitrite Induces Several Test Nitrate-Responsive Genes
To test the ability of nitrite to act as a signal, Arabidopsis plants were grown in liquid cultures with ammonium as the sole nitrogen source for 9 d. Seedlings were then deprived of nitrogen for 24 h before treating with nitrite. RNA was isolated from roots and then examined by real-time quantitative (Q)-PCR from at least three biological replicates. For the control, roots were treated with the same concentration of KCl. NiR (At2g15620) mRNA levels were examined first because NiR is one of the most nitrate-responsive genes in Arabidopsis (Wang et al., 2003). A nitrite concentration of 250 μm was initially tested because we have used this concentration for many of our previous nitrate transcriptome studies (Wang et al., 2003).
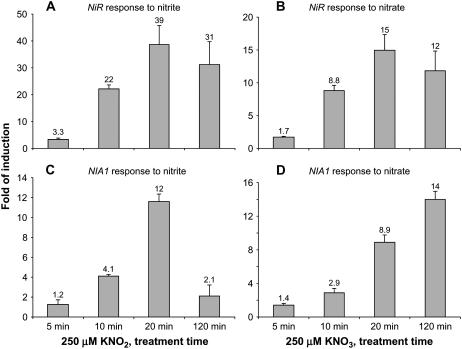
A time course analysis revealed that 250 μm nitrite induced a detectable 3-fold increase in NiR mRNA after only 5 min and a strong increase by 20 min (Fig. 1A). Further incubation (for a total of 2 h) resulted in a small decrease in median NiR mRNA levels relative to 20 min. In comparison, 250 μm nitrate treatment resulted in similar kinetics of induction (Fig. 1B). These results indicate that nitrite treatments induce NiR with similar kinetics to that of nitrate.
Figure 1.
Q-PCR analysis of NR (NIA1) and NiR mRNA in response to nitrite and nitrate. Arabidopsis seedlings were grown in aseptic hydroponics for 9 d in complete medium with 2.5 mm (NH4)2 succinate as the sole nitrogen source, then transferred for 1 d to fresh medium with no nitrogen. On day 10, seedlings were treated with either 250 μm KNO2 (A and C) or 250 μm KNO3 (B and D) for the indicated times. Root RNA was analyzed by real-time Q-PCR to determine relative mRNA levels for NIA1 and NiR as described in “Materials and Methods.” Averages from three biological replicates with ses are shown as fold of induction with KCl treatment serving as the control.
To determine whether nitrite can induce another nitrate-responsive gene, mRNA levels for the NR gene NIA1 (At1g77760) were examined using the same RNA samples described above. The kinetics of induction for NIA1 was slightly delayed compared to NiR; a 4-fold increase in mRNA levels in response to nitrite was first observed at 10 min. There was a substantial increase by 20 min, which decreased dramatically after 2 h (Fig. 1C). In comparison, nitrate treatment resulted in similar kinetics of induction for the first 20 min; however, NIA1 mRNA levels continued to increase after 2 h of nitrate treatment (Fig. 1D). These results indicate that nitrite induces NIA1 with similar initial kinetics observed with nitrate, but at 2 h the response markedly differed.
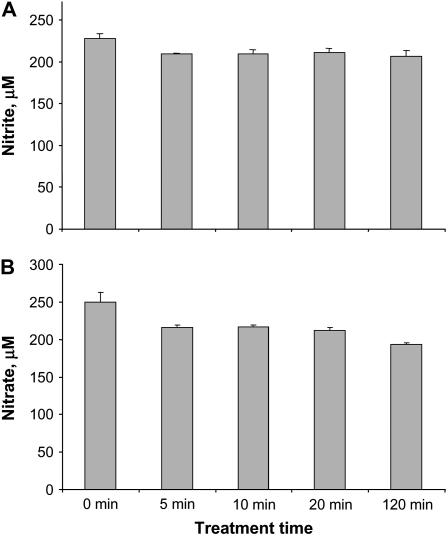
An important parameter in these experiments is the concentration of the signal during the time course of the experiments. Nitrite and nitrate levels were measured in the medium, and both showed only a small decrease during the 2-h treatment (Fig. 2). Thus, the amount of signal remains high during the full course of these experiments.
Figure 2.
Nitrate and nitrite concentrations in medium. Culture medium from experiments described in Figure 1 was sampled and assayed for nitrite (A) and nitrate (B) concentrations as described in “Materials and Methods.” Values are averages of three biological replicates with ses.
The nitrite responses were tested for three additional nitrate-responsive genes to compare with those of NiR and NIA1. Phosphoglycerate mutase (At1g78050), which functions in glycolysis and is highly induced by nitrate, shows high induction at 20 min with both nitrite and nitrate, and declines dramatically at 2 h with both nitrite and nitrate (Supplemental Fig. S1). Glc-6-P dehydrogenase (At1g24280), which is a highly nitrate-responsive gene in the pentose phosphate pathway, shows a similar response to phosphoglycerate mutase, except that the decline at 2 h with nitrite is less dramatic (Supplemental Fig. S2). Last, the NRT gene NRT1.1 (At1g12110) is induced most strongly at 20 min and shows almost no decline after 2 h with both nitrate and nitrite (Supplemental Fig. S3). Thus, all these genes showed similar induction kinetics for nitrate and nitrite for the first 20 min but differed in their extent of decline (relative to the 20-min time point) after 2 h.
Concentration Dependence of the Nitrite and Nitrate Response
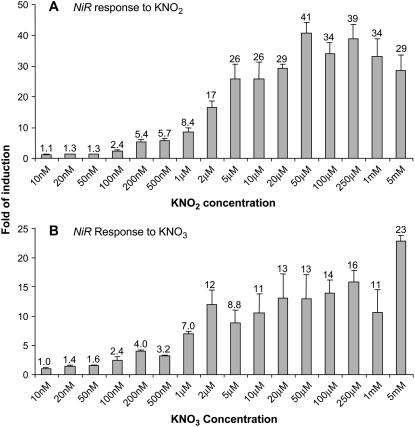
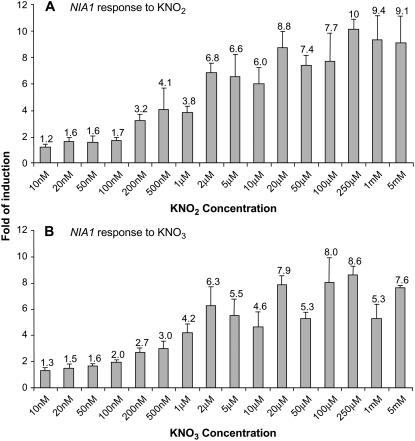
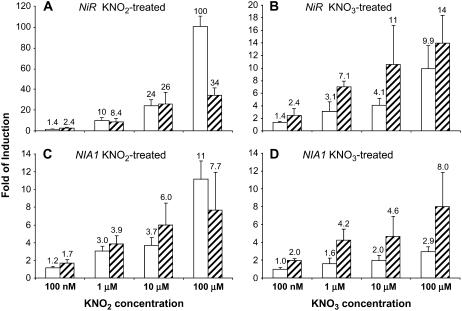
To determine the sensitivity of NiR and NIA1 to nitrite and nitrate, concentration dependence was measured. The same growth and treatment conditions of 9 d with ammonium, 1 d nitrogen deprivation, and 20 min of treatment as described above were used. Concentrations from 10 nm to 5 mm were tested and root mRNA was analyzed by Q-PCR. The lowest nitrite concentration showing a 2-fold or greater increase in NiR mRNA levels was 100 nm (Fig. 3A). The response to nitrate was similar with the first 2-fold or greater increase observed at 100 nm (Fig. 3B). NIA1 behaved similarly with the first increase in NIA1 mRNA levels occurring at 100 or 200 nm for both nitrite (Fig. 4A) and nitrate (Fig. 4B).
Figure 3.
Concentration dependence of the nitrate and nitrite response for NiR. Seedlings were grown for 10 d as described in Figure 1 and then treated for 20 min with KNO2 (A) and KNO3 (B) at concentrations between 10 nm and 5 mm as indicated. Roots were harvested for total RNA preparation and relative NiR mRNA levels were determined by Q-PCR. Averages of three biological replicates with ses are shown as fold of induction with KCl treatment serving as the control.
Figure 4.
Concentration dependence of the nitrate and nitrite response for NIA1. Seedlings were grown for 10 d as described in Figure 1 and then treated for 20 min with KNO2 (A) and KNO3 (B) at concentrations between 10 nm and 5 mm as indicated. Roots were harvested for total RNA preparation and relative NIA1 expression was determined by Q-PCR. Averages of three biological replicates with ses are shown as fold of induction with KCl treatment serving as the control.
Transcriptome Response to 250 μm Nitrite and Nitrate
Because the kinetics and concentration dependence of the nitrate and nitrite responses were similar for NiR and NIA1, we examined the nitrite response at a genomic level and compared it with the transcriptome response to nitrate. For these experiments, seedlings were treated as described above with 9 d of ammonium, 1 d of nitrogen deprivation, and 20 min of 250 μm nitrite or nitrate treatment (the control was the same treatment with chloride instead of nitrate or nitrite). Root mRNA was analyzed from two biological replicates using the Affymetrix ATH1 Genome Arrays. Datasets were filtered by selecting only those genes that were significantly induced or depressed in both biological replicates as determined by Affymetrix MicroArray Suite 5.0 software (i.e. had I or D call values). In addition, only those genes that showed significant mRNA levels (i.e. given a P call and had signal values of 100 or more) in both replicates for either the treated condition (for induced genes) or in the control treatment (for depressed genes) were selected. The lists of genes that satisfied these criteria are provided in Supplemental Tables S1 and S2. Complete datasets showing average response ratios, gene annotation (using The Arabidopsis Information Resource [TAIR] 7), call, and signal values for all genes in the ATH1 array are given in Supplemental Tables S3 and S4.
Microarray data revealed that the nitrite response was more robust than the nitrate response in terms of the number of genes showing significant changes in mRNA levels in roots. Nitrite induced 384 genes and depressed 501 genes for a total of 885 responsive genes under these conditions (Supplemental Table S1). In comparison, nitrate induced 276 genes and depressed 344 genes for a total of 620 genes (Supplemental Table S2). The average (mean) fold-induction and sd were 3.04 (±3.76) for nitrite and 3.66 (±4.65) for nitrate.
To identify which pathways and processes were most responsive to nitrite in these experiments, the set of nitrite-responsive genes were analyzed by the BioMaps program at Virtual Plant (www.vitrualplant.org). BioMaps identifies functional groups that are most overrepresented in a given set of genes using either the classification system from the Munich Information Center for Protein Sequences (MIPS; http://mips.gsf.de) or the gene ontology assignments from TAIR (www.arabidopsis.org). The MIPS functional groups most overrepresented among the nitrite-induced genes were energy, pentose phosphate pathway, carbon metabolism, nitrogen and sulfur metabolism, metabolism (i.e. general metabolism), assimilation of ammonium, and metabolism of energy reserves (Table I).
Table I.
BioMaps analysis of 250 μm nitrite-induced genes
Frequencies are percent of genes that are classified in a given MIPS functional group. Observed frequency refers to genes in the nitrite-induced set. Expected frequency refers to genes in the entire genome. Groups are ranked by their P values, which were determined by comparing the observed with the expected frequencies for that functional group using the hypergeometric distribution method.
| Functional Group | Observed Frequency | Expected Frequency | P Value |
|---|---|---|---|
| Energy | 6.0% | 1.5% | 1.2 × 10−6 |
| Pentose phosphate pathway | 1.6% | 0.1% | 3.8 × 10−5 |
| Carbon and carbohydrate metabolism | 7.3% | 2.8% | 2.1 × 10−4 |
| Nitrogen and sulfur metabolism | 2.1% | 0.3% | 5.6 × 10−4 |
| Metabolism | 12.2% | 6.5% | 1.1 × 10−3 |
| Assimilation of ammonium | 1.6% | 0.2% | 1.6 × 10−3 |
| Metabolism of energy reserves | 1.3% | 0.2% | 7.4 × 10−3 |
| Total no. of genes | 384 | 26,444 |
When the BioMaps profile for the nitrite-induced genes was compared with the BioMaps profile for the nitrate-induced genes (Table II), extensive overlap was found. Six of the seven groups were identical (energy, pentose phosphate pathway, carbon metabolism, nitrogen and sulfur metabolism, metabolism, and assimilation of ammonium). Thus, among the induced genes, there is extensive overlap between the sets of pathways and processes that are most impacted by nitrate and nitrite.
Table II.
BioMaps analysis of 250 μm nitrate-induced genes
Frequencies are percent of genes that are classified in that MIPS functional group. Groups are ranked by their P values, which were determined by comparing the observed with the expected frequencies for that functional group using the hypergeometric distribution method.
| Functional Group | Observed Frequency | Expected Frequency | P Value |
|---|---|---|---|
| Energy | 7.6% | 1.5% | 5.6 × 10−8 |
| Nitrogen and sulfur metabolism | 3.3% | 0.3% | 2.8 × 10−6 |
| Pentose phosphate pathway | 2.2% | 0.1% | 2.1 × 10−4 |
| Assimilation of ammonium | 2.5% | 0.2% | 1.0 × 10−5 |
| Glycolysis and gluconeogenesis | 3.3% | 0.6% | 1.3 × 10−3 |
| Carbon and carbohydrate metabolism | 7.3% | 2.8% | 4.1 × 10−3 |
| Metabolism | 12.7% | 6.5% | 4.6 × 10−3 |
| Total no. of genes | 275 | 26,444 |
Such similarity among functional groups was not observed for the depressed genes. No overlap in functional groups between nitrate- and nitrite-depressed genes was found. In fact, very few functional groups were overrepresented among the depressed genes. For the nitrite-depressed genes, only one group (secondary metabolism) showing a weak P value of 1.4 × 10−3 was found using MIPS. If the gene ontology assignments from TAIR were used instead of MIPS, the categories of peptide transport and oligopeptide transport were found with significant P values of <1 × 10−5. None of these groups were found among the nitrate-depressed genes; however, the group cell wall was identified with a highly significant P value of 1.7 × 10−10 using MIPS.
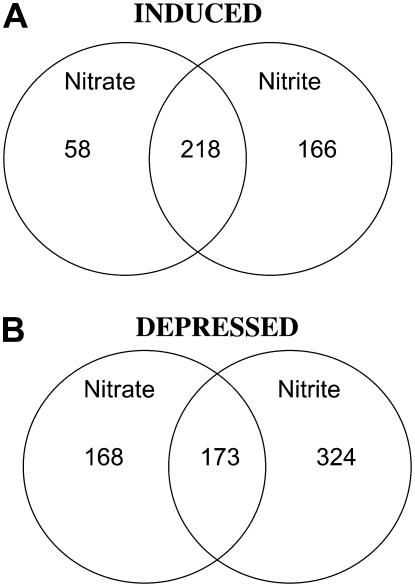
To examine the overlap between the nitrate and nitrite response more carefully, pair-wise gene comparisons were made between the two lists. A Venn diagram (Fig. 5) was produced showing how many genes were induced or depressed by both nitrate and nitrite using the filtering criteria described above. Almost 80% of the nitrate-induced genes (218) can also be induced by nitrite. Among the nitrate-depressed genes, slightly >50% can also be depressed by nitrite. Thus, there is substantial overlap between the sets of nitrate and nitrite-responsive genes.
Figure 5.
Summary of 250 μm microarray data. Venn diagrams show number of genes that respond to 250 μm nitrate and/or nitrite in Arabidopsis roots after 20 min. Nitrate and nitrite treatments and RNA preparation were performed as described in “Materials and Methods.” Microarray analysis was performed using the Affymetrix ATH1 GeneChip array. Only genes that had call values of I (induced) or D (depressed) in both replicates and had a P call value and signal values of 100 or more in both replicates in at least one of the treatment conditions were included. In addition, genes with no TAIR ID number (six genes total) were not included. Numbers in the intersection refer to genes respond to both nitrate and nitrite (induced [A] or depressed [B]); numbers outside the intersections refer to genes that responded to nitrite or nitrate alone using the filtering criteria described above.
We also identified genes that responded specifically to nitrite and not nitrate. For this analysis, the filtering criteria were made more stringent than that for Figure 5 in that only genes showing no corresponding induction or depression by nitrate in either replicate were included. Of the 885 nitrite-responsive genes, 110 genes were found not to be similarly nitrate responsive (Supplemental Table S5). BioMaps analysis showed no significant overrepresentation in any functional group for this group of genes. Thus, no pathway or process can be found that might be specifically affected by 250 μm nitrite and not nitrate; however, genes could be identified whose response was nitrite specific.
Alternative Mechanisms for the Nitrite Response
There are several possible mechanisms whereby nitrite could increase or depress mRNA levels. Our experiments indicate that nitrite itself is serving as a signal, but we checked several other possibilities. Because our plants were nitrogen-deprived, the addition of nitrite provides nitrogen to the plants. The speed of the response observed in our experiments suggests that it is unlikely that reprovision of nitrogen is the cause of the nitrite response; however, we checked this possibility by treating plants with another inorganic nitrogen source (ammonium) at 250 μm for 20 min. No increases in NiR or NIA1 mRNA levels were detected (Supplemental Fig. S4; data not shown). Another possible mechanism is that nitrite is converted to NO, which elicits the response. NO production was assessed by staining roots of wild-type plants with the NO-reactive dye 4-amino-5-methylamino-2′,7′-difluorofluorescein diacetate. A 20-min treatment with 250 μm nitrite showed no increase in 4-amino-5-methylamino-2′,7′-difluorofluorescein diacetate fluorescence (Supplemental Fig. S5). We only observed an increase in fluorescence above 1 mm nitrite (Supplemental Fig. S6). We also tested the NR-null mutant for its response to nitrite. NR can convert nitrite to NO (Yamasaki et al., 1999; Rockel et al., 2002). We found little difference in NiR induction by nitrite in the NR-null mutant compared to wild-type plants (Supplemental Fig. S4).
Last, we checked the possibility that our nitrite solutions were contaminated with nitrate and that the nitrite response is in fact due to nitrate. Nitrite solutions were tested by HPLC analysis and found to have nitrate at levels slightly <1% of the nitrite concentration. Because the concentration dependence for nitrate and nitrite were so similar (both showed responses at concentrations as low as 100 nm), it is very improbable that 1% nitrate contamination (e.g. at levels of <10 nm in a 1.0 μm nitrite solution) could account for the nitrite response at low nitrite concentrations. Thus, we find no evidence that the nitrite response is mediated by NO or ammonium or by nitrate at low nitrite concentrations.
Transcriptome Response to 5 μm Nitrite and Nitrate
Because of the low levels of nitrate found in the nitrite solutions, a concentration of 250 μm nitrite would have slightly <2.5 μm nitrate. This concentration of nitrate can induce NiR and NIA1 (Figs. 3 and 4). To rule out any influence of nitrate in the nitrite response for the transcriptome experiments, microarray analyses were performed on roots treated with 5 μm nitrite, which has <50 nm nitrate, a concentration where no response was observed for NiR and NIA1 (Figs. 3 and 4). RNA samples used for the 5 μm nitrite and nitrate data points in Figures 3 and 4 were analyzed using the ATH1 chips as described above (datasets were filtered by selecting only those genes that were significantly induced or depressed in both biological replicates [i.e. had I or D call values]) and had significant mRNA levels (i.e. given a P call and had signal values of 100 or more) in both replicates for either the treated condition (for induced genes) or the control treatment (for depressed genes). The lists of genes that satisfied these criteria are provided in Supplemental Tables S6 and S7. Complete datasets showing average response ratios, gene annotation (using TAIR 7), call, and signal values for all genes in the ATH1 array are given in Supplemental Tables S8 and S9.
The results from these analyses showed that, based on the number of genes, the 5 μm nitrite response was much stronger than the 5 μm nitrate response using the filtering criteria described above (Table III). There were over 3-fold more nitrite-induced than nitrate-induced genes and over 25-fold more nitrite-depressed than nitrate-depressed genes. Compared to the 250 μm response, the 5 μm nitrite response was comparable (790 versus 885 genes), whereas the 5 μm nitrate response was much lower (144 versus 620 genes).
Table III.
Number of genes that respond to nitrite or nitrate
| Conditions | Nitrite | Nitrate |
|---|---|---|
| 5 μm induced | 444 | 131 |
| 5 μm depressed | 346 | 13 |
| 250 μm induced | 384 | 276 |
| 250 μm depressed | 501 | 344 |
A BioMaps analysis of the 5 μm nitrite-induced genes showed that seven of the overrepresented functional groups are the same as those observed for the nitrate-induced genes (energy, metabolism, nitrogen and sulfur metabolism, assimilation of ammonium, glycolysis and gluconeogenesis, carbon and carbohydrate metabolism, and pentose phosphate pathway; Tables II and IV). In addition, there are four processes related to ion transport that are also overrepresented in the nitrite-induced genes. No overrepresented functional groups were identified among the nitrite-depressed genes.
Table IV.
BioMaps analysis of 5 μm nitrite-induced genes
Frequencies are percent of genes that are classified in that MIPS functional group. Groups are ranked by their P values, which were determined by comparing the observed with the expected frequencies for that functional group using the hypergeometric distribution method.
| Functional Group | Observed Frequency | Expected Frequency | P Value |
|---|---|---|---|
| Assimilation of ammonium | 2.9% | 0.2% | 7.2 × 10−12 |
| Nitrogen and sulfur metabolism | 3.2% | 0.3% | 1.1 × 10−9 |
| Energy | 6.6% | 1.5% | 2.4 × 10−9 |
| Metabolism | 15.2% | 6.5% | 4.7 × 10−9 |
| Amino acid metabolism | 4.5% | 0.9% | 2.5 × 10−7 |
| Ion transport | 3.4% | 0.5% | 9.4 × 10−7 |
| Glycolysis and gluconeogenesis | 3.4% | 0.6% | 2.9 × 10−6 |
| Transported compounds | 4.5% | 1.1% | 7.8 × 10−6 |
| Anion transport | 2.0% | 0.2% | 1.0 × 10−5 |
| Carbon and carbohydrate metabolism | 7.5% | 2.8% | 2.2 × 10−5 |
| Pentose phosphate pathway | 1.4% | 0.1% | 1.1 × 10−4 |
| Transport facilitation | 4.8% | 1.6% | 5.4 × 10−4 |
| Total no. of genes | 441 | 26,444 |
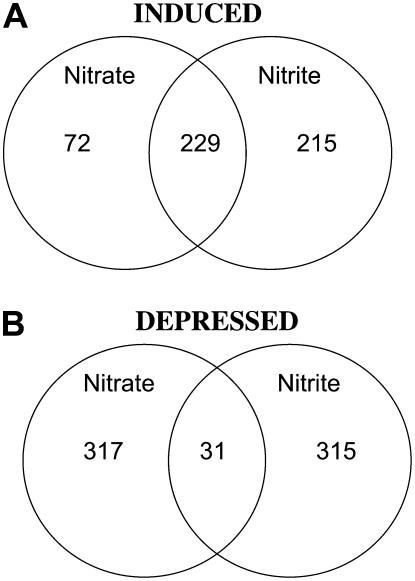
Pair-wise comparisons were performed between the nitrate and the 5 μm nitrite-responsive genes (Supplemental Table S10). Because so few genes responded to 5 μm nitrate, all genes that responded to either 5 or 250 μm nitrate in both replicates were included in the comparison. Most of the nitrate-induced genes (229% or 76%) were also induced by nitrite (Fig. 6), which was also found for the 250 μm nitrite response described above (Fig. 5). However, among the depressed genes, there was almost no overlap (<10%; Fig. 6). If the nitrite-specific genes were examined using BioMaps, only the nitrite-specific induced genes showed any functional groups that were overrepresented and these groups were ion transport (P value of 3.6 × 10−4) and metabolism (P value of 2.0 × 10−3). Among the ion transporters were the NRT gene NRT2.5 and the ammonium transporter gene AMT1.3. This would suggest that these genes may have a nitrite-specific function (e.g. perhaps NRT2.5 is involved in nitrite uptake). However, these genes were not induced at 250 μm nitrite; thus, their relationship to nitrite is not clear.
Figure 6.
Summary of 5 μm microarray data. Venn diagrams show number of genes that respond to 5 μm nitrite and/or 5 or 250 μm nitrate in Arabidopsis roots after 20 min as described in Figure 5 legend. Numbers in the intersection refer to genes responding to both nitrite and at least one of the nitrate treatments (induced [A] or depressed [B]); numbers outside the intersections refer to genes that did not fit this profile.
Effect of High Ammonium on the Nitrite and Nitrate Response
Ammonium has been shown to affect the expression of several hundred genes in Arabidopsis roots (Fizames et al., 2004). High ammonium levels (20 mm or greater) have been used as a nitrogen source for several nitrate transcriptome studies (Wang et al., 2000). We tested the effect of high ammonium levels on the nitrite and nitrate responses of NiR and NIA1 to determine whether they had any effect on these responses. In the experiments described above, plants were initially provided with 5 mm ammonium and then starved for nitrogen for 24 h from days 9 to 10. In the new experiments, plants were grown as above, except the initial medium contained 20 mm ammonium [10 mm (NH4)2 succinate]. The medium was replaced at 9 d with medium containing 20 mm ammonium; therefore, plants were exposed to high ammonium throughout their growth. At 10 d, plants were treated with various concentrations of nitrite or nitrate; root RNA was isolated and analyzed by Q-PCR (Fig. 7). The data show that the 100 nm to 10 μm nitrite induction ratios for NiR and NIA1 were slightly suppressed in most cases by the high ammonium (blue bars) relative to the nitrogen-deficient (red bars) medium. However, at 100 μm nitrite, the response was unexpectedly increased by the high ammonium. For nitrate, the data show that high ammonium strongly decreased the median induction ratios at all nitrate concentrations tested. It also decreased the overall mRNA levels of NiR 3-fold and of NIA1 26-fold (data not shown). Of special note is that high ammonium obscured the nitrate response at very low concentrations of nitrate because the level of nitrate required to observe a 2-fold induction or greater was increased from 100 nm to 10 μm for NIA1 and from 100 nm to 1 μm for NiR.
Figure 7.
Effect of high ammonium on NiR and NIA1 response. Seedlings were grown in medium with 2.5 mm (NH4)2 succinate as the nitrogen source for 9 d, then transferred to fresh medium with no nitrogen for 1 d (nitrogen-starved, cross-hatched bars) or with 10 mm (NH4)2 succinate for 9 d, then transferred to 10 mm (NH4)2 succinate for 1 d (high ammonium, white bars). Seedlings were then treated for 20 min with KNO2 (A and C) and KNO3 (B and D) at the indicated concentrations. Root RNA was analyzed by Q-PCR to determine relative NiR (A and B) and NIA1 (C and D) mRNA levels. Results are averages of three biological replicates with ses as indicated. Data are shown as fold of induction compared with KCl treatment.
DISCUSSION
Our results demonstrate that nitrite can serve as a signal that rapidly induces and depresses mRNA levels within minutes of application to roots of Arabidopsis plants. Nitrite concentrations as low as 100 nm can elicit a response of 2-fold or more for NiR. Experiments that examined possible involvement of ammonium or NO (metabolites of nitrite) in the nitrite response gave negative results. These findings suggest that nitrite is working directly as a signal to regulate gene expression.
An important issue in these experiments is possible contamination of the nitrite solutions with nitrate. Because there was extensive overlap between the nitrate and nitrite responses (in terms of pathways and genes affected), this issue is of some concern. Direct measurements using HPLC showed that the nitrite solutions had slightly <1% contamination with nitrate. Thus, responses to nitrite solutions at 10 μm or less should have little to no interference from nitrate (e.g. the 5 μm nitrite transcriptome experiment) given that little or no response was observed at concentrations of nitrate <100 nm. For nitrite solutions >10 μm, nitrate could contribute to the response (e.g. the 250 μm transcriptome experiment). However, even at higher nitrite concentrations, much of the nitrite response cannot be accounted for by nitrate because there are clear nitrite-specific responses (e.g. >100 genes showed a nitrite, but not nitrate, response at 250 μm).
These findings raise the question: Is there a physiological role for the regulation of genes by nitrite? Normally, nitrite is restricted to cells in which it is made and is present at low micromolar levels compared to high (millimolar) nitrate levels; however, when roots experience hypoxia or anoxia, nitrite levels increase to levels where nitrite is secreted from roots (Botrel et al., 1996; Allegre et al., 2004; Libourel et al., 2006). It has been proposed that nitrite helps protect plants from anoxia by reducing cytoplasmic acidosis (Libourel et al., 2006) and supporting anaerobic metabolism (Stoimenova et al., 2003, 2007; Igamberdiev et al., 2005). From our bioinformatics analysis and from inspection of the gene lists, we found no evidence for nitrite-specific effects on processes involved in anaerobic metabolism (having found only ion transport and general metabolism in the BioMaps analysis), but it is worth further study to test the idea that nitrite may regulate anoxic responses.
A previous publication had indicated that nitrite represses two nitrate-induced genes. Specifically, 0.5 to 1.0 mm KNO2 repressed NRT1.1 and NIA1 mRNA levels after 6 and 24 h of treatment in Arabidopsis roots (Loque et al., 2003). At first glance, these results might appear to contradict our findings; however, these experiments examined treatments for much longer time frames (6–24 h versus 20 min in our experiments). Interestingly, in our experiments, NIA1 shows strong depression after 2 h of nitrite treatment so that it is certainly possible that even longer treatments would result in even lower NIA1 mRNA levels.
There were several surprises from our studies. First was the finding that nitrite could serve as such a potent signal. Several recent reports provided evidence that nitrite could function as a signal in plants, but there was no indication or expectation that it could serve as a transcriptome signal rapidly affecting the mRNA levels of almost 900 genes at micromolar concentrations. In fact, our data indicate that nitrite is a more potent signal than nitrate after 20 min of treatment. This was most apparent in the 5 μm transcriptome experiment, where 3- and 25-fold more genes were induced and depressed, respectively, by nitrite than nitrate (Table III). This effect could be due to stronger affinity of the sensing system to nitrite compared to nitrate or to faster uptake of nitrite. It is also possible that the small amount of nitrate in the nitrite solutions could be potentiating or synergistically enhancing the nitrite response.
Second, the overlap between the nitrate and nitrite response was quite extensive. This was most apparent for the induced genes where approximately three-fourths of the nitrate-induced genes were also induced by nitrite. Also, almost all of the pathways and processes induced by nitrate are also induced by nitrite. This extensive overlap, as well as the strong similarity among the concentration dependence and initial kinetics for both nitrate and nitrite, leads us to propose that the nitrate-sensing system in Arabidopsis roots recognizes nitrite as well as, if not better, than nitrate.
Last, the concentration dependence of the observed nitrate response (Fig. 4) indicates that sensitivity to nitrate is almost 2 orders of magnitude greater than previously thought. Previous publications had reported responses to nitrate at concentrations above 10 μm (Tischner et al., 1993), whereas our results show a response threshold around 100 nm as long as plants are starved for nitrogen for 24 h and not treated with high ammonium (20 mm). Thus, nitrate (and nitrite) serves as signals at nanomolar concentrations in nitrogen-starved Arabidopsis roots.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) plants were of Columbia ecotype unless otherwise indicated. The NR-null mutant was made by crossing a nia1 insertion mutant (SALK_088070) with a nia2 insertion mutant (SALK_088070; Alonso et al., 2003) supplied by the Arabidopsis Resources Center (www.arabidopsis.org).
Growth and Treatment Conditions
Arabidopsis seedlings were grown under hydroponic conditions as previously described (Wang et al., 2003). Briefly, approximately 100 seedlings were grown at 25°C with continuous light supported on nylon mesh so that roots were submerged in liquid medium and shoots were above the medium. Seedlings were germinated, then grown in 25 mL of liquid medium containing 2.5 mm (NH4)2 succinate as the nitrogen source, 0.5% (w/v) Suc, and other essential macro- and micronutrients for 9 d, then shifted to 100 mL of fresh medium lacking (NH4)2 succinate for 24 h (note that the initial Cl− concentration was over 2 mm). At day 10, treatments were initiated by adding 250 μL solution of KNO2 or KNO3 (treatment) or KCl (control) to the culture to reach the specified concentrations in the medium. Plants were grown for the specified amount of time and then roots were harvested for RNA extraction.
For nitrate and nitrite treatments in the presence of high ammonium, plants were grown similarly as described above, except that the initial (NH4)2 succinate concentration was 10 mm. After 9 d of incubation, seedlings were transferred to 100 mL of fresh medium containing 10 mm (NH4)2 succinate.
Nitrate and Nitrite Assay
Nitrate was assayed by reversed-phase ion-pair HPLC as described by Zuo et al. (2006) with a Luna 5 μm C18(2) 100A 250 × 2.0 mm column from Phenomenex.
Nitrite concentration in hydroponics medium was determined by the Griess assay. A 0.5-mL sample was mixed with 0.5 mL of 1% sulfanilamide in 3 n HCl; then, with 0.5 mL of 0.02% N-(1-naphthyl) ethylene-diamine dihydrochloride in water. The solution was incubated at room temperature for 15 min and then centrifuged at 12 K rpm to remove any precipitate. The OD540 was then determined. Standard curve was made with NaNO2 in water at concentrations between 2 and 50 μm.
RNA Preparation
Total RNA was prepared from roots using a total RNA miniprep kit (BioPioneer) and quantified with a Genesis 6 spectrophotometer.
Real-Time Q-PCR
Real-time PCR was performed using a LightCycler system from Roche Diagnostics. Template cDNA samples were prepared using the SuperScript first-strand synthesis system kit (Invitrogen) for real-time PCR with 1.5 μg of total RNA in a reaction volume of 10 μL. The cDNA synthesis reaction mixture was diluted 10 times before being used for PCR. Primers for PCR reactions were designed to have a melting temperature of about 60°C to 65°C and to give a PCR product between 175 to 300 bp. The oligo primers used were as follows: NIA1 (forward primer, 5′-ATCGTCAAAGAAACCGAAGTC; and reverse primer, 5′-ACGGAGCATGGATGAGTT); NiR (forward primer, 5′-CCGGTAGCCAGTTCTGCG; and reverse primer, 5′-CCTATTCGTCCCCCGACGT); Clathrin (At4g24550, as the reference for Q-PCR; forward primer, 5′-ATACGCGCTGAGTTCCC and reverse primer, 5′-CTGACTGGCCCTGCTT).
The LightCycler FastStart DNA Master SYBR Green I Q-PCR kit (Roche Diagnostics) was used for PCR reactions. Each PCR reaction contained 2 μL of cDNA (diluted 10× of first-strand cDNA synthesis reactions) and 0.5 μm of each primer. The initial denaturing time was 10 min, followed by 45 PCR cycles consisting of 94°C for 0 s, 63°C for 5 s, and 72°C for 10 s. A melting curve was run after the PCR cycles. Quantification was performed with LightCycler relative quantification software 1.0.
Target Preparation and Processing for GeneChip Analysis
Procedures for target preparation and processing for GeneChip Analysis were as previously described (Wang et al., 2003).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. PGM mRNA response.
Supplemental Figure S2. G6PDH mRNA response.
Supplemental Figure S3. NRT1.1 mRNA response.
Supplemental Figure S4. NiR mRNA response to ammonium.
Supplemental Figure S5. NO accumulation at low nitrite.
Supplemental Figure S6. NO accumulation at high nitrite.
Supplemental Table S1. Nitrite (250 μm)-responsive genes.
Supplemental Table S2. Nitrate (250 μm)-responsive genes.
Supplemental Table S3. Nitrite (250 μm) response, complete list.
Supplemental Table S4. Nitrate (250 μm) response, complete list.
Supplemental Table S5. Nitrite-specific genes.
Supplemental Table S6. Nitrite (5 μm)-responsive genes.
Supplemental Table S7. Nitrate (5 μm)-responsive genes.
Supplemental Table S8. Nitrite (5 μm) response, complete list.
Supplemental Table S9. Nitrate (5 μm) response, complete list.
Supplemental Table S10. Comparison of nitrate and nitrite genes.
Supplementary Material
Acknowledgments
We wish to thank Mamoru Okamoto for valuable technical advice.
This work was supported by the National Science Foundation (grant no. IOB–0519985).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Nigel Crawford (ncrawford@ucsd.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Allegre A, Silvestre J, Morard P, Kallerhoff J, Pinelli E (2004) Nitrate reductase regulation in tomato roots by exogenous nitrate: a possible role in tolerance to long-term root anoxia. J Exp Bot 55 2625–2634 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Aslam M, Huffaker RC (1989) Role of nitrate and nitrite in the induction of nitrite reductase in leaves of barley seedlings. Plant Physiol 91 1152–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botrel A, Magne C, Kaiser WM (1996) Nitrate reduction, nitrite reduction and ammonium assimilation in barley roots in response to anoxia. Plant Physiol Biochem 34 645–652 [Google Scholar]
- Bright J, Desikan R, Hancock JT, Weir IS, Neill SJ (2006) ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant J 45 113–122 [DOI] [PubMed] [Google Scholar]
- Crawford NM (1995) Nitrate: nutrient and signal for plant growth. Plant Cell 7 859–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NM, Forde BG (2002) Molecular and developmental biology of inorganic nitrogen nutrition. In E Meyerowitz, C Somerville, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0011, www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Desikan R, Cheung MK, Bright J, Henson D, Hancock JT, Neill SJ (2004) ABA, hydrogen peroxide and nitric oxide signalling in stomatal guard cells. J Exp Bot 55 205–212 [DOI] [PubMed] [Google Scholar]
- Duncanson E, Gilkes AF, Kirk DW, Sherman A, Wray JL (1993) nir1, a conditional-lethal mutation in barley causing a defect in nitrite reduction. Mol Gen Genet 236 275–282 [DOI] [PubMed] [Google Scholar]
- Fizames C, Munos S, Cazettes C, Nacry P, Boucherez J, Gaymard F, Piquemal D, Delorme V, Commes T, Doumas P, et al (2004) The Arabidopsis root transcriptome by serial analysis of gene expression: gene identification using the genome sequence. Plant Physiol 134 67–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde BG (2002) Local and long-range signaling pathways regulating plant responses to nitrate. Annu Rev Plant Biol 53 203–224 [DOI] [PubMed] [Google Scholar]
- Gutierrez RA, Gifford ML, Poultney C, Wang R, Shasha DE, Coruzzi GM, Crawford NM (2007) Insights into the genomic nitrate response using genetics and the Sungear Software System. J Exp Bot 58 2359–2367 [DOI] [PubMed] [Google Scholar]
- Igamberdiev AU, Baron K, Manac'h-Little N, Stoimenova M, Hill RD (2005) The haemoglobin/nitric oxide cycle: involvement in flooding stress and effects on hormone signalling. Ann Bot (Lond) 96 557–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King BJ, Siddiqi MY, Ruth TJ, Warner RL, Glass ADM (1993) Feedback regulation of nitrate influx in barley roots by nitrate, nitrite and ammonium. Plant Physiol 102 1279–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea US, Ten Hoopen F, Provan F, Kaiser WM, Meyer C, Lillo C (2004) Mutation of the regulatory phosphorylation site of tobacco nitrate reductase results in high nitrite excretion and NO emission from leaf and root tissue. Planta 219 59–65 [DOI] [PubMed] [Google Scholar]
- Libourel IGL, van Bodegom PM, Fricker MD, Ratcliffe RG (2006) Nitrite reduces cytoplasmic acidosis under anoxia. Plant Physiol 142 1710–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llamas A, Igeno MI, Galvan A, Fernandez E (2002) Nitrate signalling on the nitrate reductase gene promoter depends directly on the activity of the nitrate transport systems in Chlamydomonas. Plant J 30 261–271 [DOI] [PubMed] [Google Scholar]
- Loppes R, Radoux M, Ohresser MCP, Matagne RF (1999) Transcriptional regulation of the Nia1 gene encoding nitrate reductase in Chlamydomonas reinhardtii: effects of various environmental factors on the expression of a reporter gene under the control of the Nia1 promoter. Plant Mol Biol 41 701–711 [DOI] [PubMed] [Google Scholar]
- Loque D, Tillard P, Gojon A, Lepetit M (2003) Gene expression of the NO3− transporter NRT1.1 and the nitrate reductase NIA1 is repressed in Arabidopsis roots by NO2−, the product of NO3− reduction. Plant Physiol 132 958–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C, Lea US, Provan F, Kaiser WM, Lillo C (2005) Is nitrate reductase a major player in the plant NO (nitric oxide) game? Photosynth Res 83 181–189 [DOI] [PubMed] [Google Scholar]
- Morot-Gaudry-Talarmain Y, Rockel P, Moureaux T, Quillere I, Leydecker MT, Kaiser WM, Morot-Gaudry JF (2002) Nitrite accumulation and nitric oxide emission in relation to cellular signaling in nitrite reductase antisense tobacco. Planta 215 708–715 [DOI] [PubMed] [Google Scholar]
- Planchet E, Jagadis Gupta K, Sonoda M, Kaiser WM (2005) Nitric oxide emission from tobacco leaves and cell suspensions: rate limiting factors and evidence for the involvement of mitochondrial electron transport. Plant J 41 732–743 [DOI] [PubMed] [Google Scholar]
- Redinbaugh MG, Campbell WH (1991) Higher plant responses to environmental nitrate. Physiol Plant 82 640–650 [Google Scholar]
- Remmler JL, Campbell WH (1986) Regulation of corn leaf nitrate reductase. II. Synthesis and turnover of the enzyme's activity and protein. Plant Physiol 80 442–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockel P, Strube F, Rockel A, Wildt J, Kaiser WM (2002) Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J Exp Bot 53 103–110 [PubMed] [Google Scholar]
- Sakamoto A, Sakurao S, Fukunaga K, Matsubara T, Ueda-Hashimoto M, Tsukamoto S, Takahashi M, Morikawa H (2004) Three distinct Arabidopsis hemoglobins exhibit peroxidase-like activity and differentially mediate nitrite-dependent protein nitration. FEBS Lett 572 27–32 [DOI] [PubMed] [Google Scholar]
- Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136 2483–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DA, Kuo TM, Kleinhofs A, Warner BL, Oaks A (1983) Synthesis and degradation of barley nitrate reductase. Plant Physiol 72 949–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M (1999) Nitrate regulation of metabolism and growth. Curr Opin Plant Biol 2 178–186 [DOI] [PubMed] [Google Scholar]
- Stoimenova M, Igamberdiev AU, Gupta KJ, Hill RD (2007) Nitrite-driven anaerobic ATP synthesis in barley and rice root mitochondria. Planta 226 465–474 [DOI] [PubMed] [Google Scholar]
- Stoimenova M, Libourel IGL, Ratcliffe RG, Kaiser WM (2003) The role of nitrate reduction in the anoxic metabolism of roots. II. Anoxic metabolism of tobacco roots with or without nitrate reductase activity. Plant Soil 253 155–167 [Google Scholar]
- Tischner R, Waldeck B, Goyal S, Rains WD (1993) Effect of nitrate pulses on the nitrate-uptake rate, synthesis of mRNA coding for nitrate reductase, and nitrate reductase activity in the roots of barley seedlings. Planta 189 533–537 [Google Scholar]
- Tsujimoto R, Yamazaki H, Maeda S, Omata T (2007) Distinct roles of nitrate and nitrite in regulation of expression of the nitrate transport genes in the moss Physcomitrella patens. Plant Cell Physiol 48 484–497 [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Kronenberger J, Lepingle A, Vilaine F, Boutin JP, Caboche M (1992) Inhibition of tobacco nitrite reductase activity by expression of antisense RNA. Plant J 2 559–569 [PubMed] [Google Scholar]
- Wang R, Guegler K, LaBrie ST, Crawford NM (2000) Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 12 1491–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Okamoto M, Xing X, Crawford NM (2003) Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol 132 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Tischner R, Gutierrez RA, Hoffman M, Xing X, Chen M, Coruzzi G, Crawford NM (2004) Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol 136 2512–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki H, Sakihama Y, Takahashi S (1999) An alternative pathway for nitric oxide production in plants: new features of an old enzyme. Trends Plant Sci 4 128–129 [DOI] [PubMed] [Google Scholar]
- Zielke HR, Filner P (1971) Synthesis and turnover of nitrate reductase induced by nitrate in cultured tobacco cells. J Biol Chem 246 1772–1779 [PubMed] [Google Scholar]
- Zuo Y, Wang C, Van T (2006) Simultaneous determination of nitrite and nitrate in dew, rain, snow and lake water samples by ion-pair high-performance liquid chromatography. Talanta 70 281–285 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.