Abstract
Following the discovery of glutamine synthetase/glutamate (Glu) synthase, the physiological roles of Glu dehydrogenase (GDH) in nitrogen metabolism in plants remain obscure and is the subject of considerable controversy. Recently, transgenics were used to overexpress the gene encoding for the β-subunit polypeptide of GDH, resulting in the GDH-isoenzyme 1 deaminating in vivo Glu. In this work, we present transgenic tobacco (Nicotiana tabacum) plants overexpressing the plant gdh gene encoding for the α-subunit polypeptide of GDH. The levels of transcript correlated well with the levels of total GDH protein, the α-subunit polypeptide, and the abundance of GDH-anionic isoenzymes. Assays of transgenic plant extracts revealed high in vitro aminating and low deaminating activities. However, gas chromatography/mass spectrometry analysis of the metabolic fate of 15NH4 or [15N]Glu revealed that GDH-isoenzyme 7 mostly deaminates Glu and also exhibits low ammonium assimilating activity. These and previous results firmly establish the direction of the reactions catalyzed by the anionic and cationic isoenzymes of GDH in vivo under normal growth conditions and reveal a paradox between the in vitro and in vivo enzyme activities.
Nitrogen is taken up by plants mainly as nitrates or ammonium ions. Nitrates are enzymatically reduced to ammonium, which is subsequently used in plant cells for amino acid biosynthesis. Previously, it was widely accepted that Glu dehydrogenase (GDH; EC 1.4.1.2), which catalyzes in vitro the reductive amination of 2-oxoglutarate (2-OG) and the oxidative deamination of Glu, was the enzyme responsible for the assimilation of ammonium. However, with the discovery that ammonia assimilation in plants is mainly catalyzed by the Gln synthetase (GS)/Glu synthase cycle (EC 6.3.1.2./EC 1.4.7.1; Lea and Miflin, 1974), the physiological role(s) of GDH had been obscure.
GDH is a hexamer of α- and β-subunit polypeptides associated in an ordered ratio to form two homohexamers and five hybrids [β6 (isoenzyme 1), β5,α1……β1,α5, α6 (isoenzyme 7); Loulakakis and Roubelakis-Angelakis, 1991; Turano et al., 1996]. gdh cDNAs encoding for the two polypeptides have been isolated from several higher plant species (Purnell et al., 2005). GDH is mostly localized in the mitochondria of the phloem companion cells (Lea and Thurman, 1972; Loulakakis and Roubelakis-Angelakis, 1990a; Paczek et al., 2002) but also in the cytosol of senescing flower receptacles, in senescing shoots, and in epidermal root tip cells (Dubois et al., 2003). Although GDH protein is abundant in plant cells, the physiological role of its seven isoenzymes was for a long time a matter of controversy. Numerous reports suggested that GDH operates in vivo in the direction of ammonium assimilation (Yamaya et al., 1986; Yamaya and Oaks, 1987; Magalhaes et al., 1990; Magalhaes, 1991; Melo-Oliveira et al., 1996), whereas others suggested that the sole role of the enzyme is the deamination of Glu (Robinson et al., 1992; Fox et al., 1995; Stewart et al., 1995; Aubert et al., 2001). Plant GDH has been proposed to be a stress-responsive enzyme (Syntichaki et al., 1996; Restivo, 2004), and consistent with this function, recombinantly produced GDH exhibits considerable thermal stability (Syntichaki et al., 1996). In addition, high intracellular ammonium, provided either externally (Cammaerts and Jacobs, 1985; Srivastava and Singh Rana, 1987; Lea and Ireland, 1999) or as a result of protein hydrolysis (Masclaux et al., 2000; Limami et al., 2002), generally leads to de novo synthesis of the α-subunit, assembly of the anionic isoenzymes, and increased in vitro aminating activity (Loulakakis and Roubelakis-Angelakis, 1992). Furthermore, the gene encoding for the α-subunit is induced in senescing plant organs, accompanied by an increase in immunoreactive protein and aminating in vitro activity (Loulakakis et al., 1994, 2002).
Recent results have started to resolve the in vivo physiological roles of GDH-isoenzymes. Skopelitis et al. (2006) showed that the reactive oxygen species induced by salinity signal the expression of the gene encoding for the α-subunit polypeptide of GDH, the assembly of the anionic GDH isoenzymes, and increased in vivo aminating activity, as evidenced by the incorporation of 15NH4 in Glu and Pro in the presence of Met sulfoximine (MSX), a potent inhibitor of GS. Also, Purnell and Botella (2007), using transgenic tobacco (Nicotiana tabacum) plants overexpressing the tomato (Solanum lycopersicum) gene encoding for the β-subunit polypeptide of GDH (Purnell et al., 2005), found that GDH-isoenzyme 1 deaminates in vivo Glu.
Here, we present results from transgenic tobacco plants overexpressing the Vvgdh-NAD;A gene from grapevine (Vitis vinifera) encoding for the α-subunit polypeptide of GDH. Molecular and biochemical results coupled with gas chromatography/mass spectrometry (GC/MS) and NMR analysis provide very strong evidence that under standard growth conditions, the GDH-isoenzyme 7 primarily acts toward the deamination of Glu and also exhibits low aminating activity.
RESULTS
Vvgdh-NAD;A1 Transcript Levels in Transgenic Plants Corresponded with GDH-Total Protein Levels, GDH α-Subunit Polypeptide Levels, and Abundance of GDH-Anionic Isoenzymes
The two subunit polypeptides of GDH α and β are encoded by two genes, gdh-NAD;A and gdh-NAD;B, respectively. The respective clones have been cloned from several plant species (summarized by Purnell et al., 2005). Tobacco plants were transformed using T-DNA constructs containing the full-length cDNAs coding for the grapevine α-GDH subunit (Vvgdh-NAD;A1) in sense orientation. For every construct, several transgenic lines were produced and screened through the T0, T1, and T2 generations. From the homozygous transformants, several sense-T2 lines were tested. Here, we report results from the lines S2 and S5. Replicate wild-type tobacco ‘Xanthi’ were used as controls. Southern blot and PCR analysis of the lines showed the successful insertion of the transgene(s) (data not shown). More specifically, line S5 contained a single T-DNA insertion and line S2 contained two copies (data not shown).
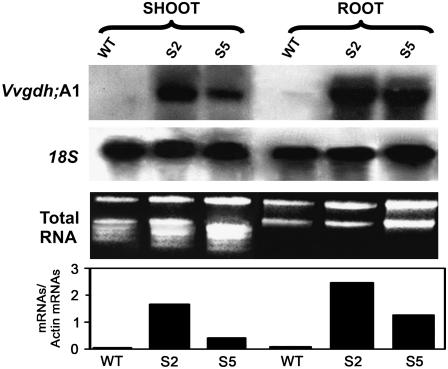
Relative Vvgdh-NAD;A1 transcript abundance was determined in shoots and roots of the transgenic lines and the wild-type plants. Northern-blot analysis revealed that in the overexpressing transgenic lines, the levels of the transgene were significantly higher in both shoots and roots than in the wild type (Fig. 1).
Figure 1.
Transcript abundance of Vvgdh-NAD;A1 in root and shoot of 30-d-old transgenic tobacco plants grown in half-strength Murashige and Skoog solution. Shoots and roots of wild-type (WT) tobacco and Vvgdh-NAD;A1 transgenic lines S2 and S5.
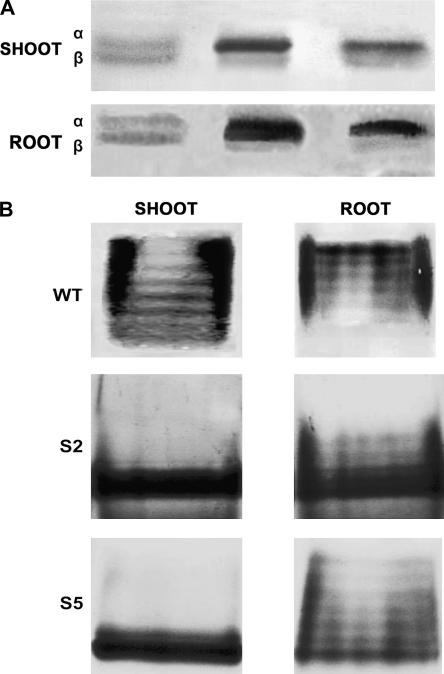
The relative abundance of GDH protein was assessed by western-blot analysis using a GDH antibody (Loulakakis and Roubelakis-Angelakis, 1990b). The α-subunit levels of immunoreactive GDH protein in the transgenic lines (Fig. 2A) corresponded well with Vvgdh-NAD;A1 transcript levels (Fig. 1). Shoots of lines S2 and S5 contained 5.5- and 2.5-fold more GDH α-subunits than the wild-type control, while in roots, it was 8.0- and 3.5-fold higher than in the wild type, respectively (Fig. 2A), as densitometric analysis showed.
Figure 2.
Abundance of α- and β-GDH polypeptide and isoenzyme distribution of gdh-NAD;A1 in the shoots and roots of 30-d-old wild-type tobacco plants (WT) and transgenic lines grown in half-strength MS solution. A, α- and β-immunoreactive subunit polypeptides of GDH in wild-type tobacco and Vvgdh-NAD;A1 transgenic lines S2 and S5. B, Isoenzyme profiles of GDH in wild-type tobacco and Vvgdh-NAD;A1 transgenic lines S2 and S5.
Expression of Vvgdh-NAD;A1 resulted in a marked increase in the levels of the α-subunit polypeptide (Fig. 2A), as would be expected from previous phylogenetic analysis (Purnell et al., 2005). The relative abundance of the GDH α-subunit polypeptide in the transgenics was in good agreement with the levels of the respective transcripts (Fig. 1) and with the amount of total GDH protein (Fig. 2A). In the shoots of lines S2 and S5, the abundance of α-GDH subunit was 6.0- and 2.5-fold higher than in wild-type controls. These results confirmed that Vvgdh-NAD;A1 encodes for the α-GDH subunit polypeptide. Overexpression of the α-subunit polypeptide resulted in increased abundance of the anionic isoenzymes, especially the homohexameric isoenzyme 7 (Fig. 2B).
These results firmly showed that the Vvgdh-NAD;A gene of GDH in fact encodes for the α-GDH subunit polypeptide and that this polypeptide participates in the assembly of the anionic GDH isoenzymes, confirming previous results (Loulakakis and Roubelakis-Angelakis, 1991; Turano et al., 1996).
The in Vitro Aminating and Deaminating Activities Observed in Transgenic Lines Did Not Correspond with the in Vivo Activities
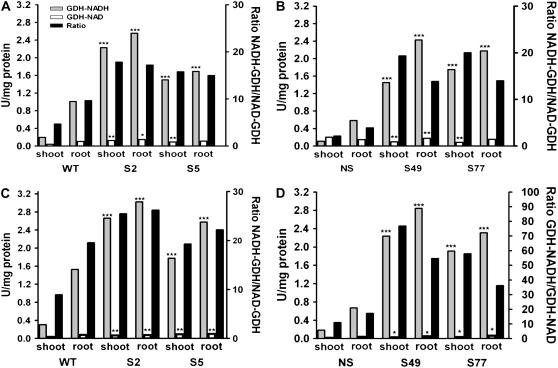
Following the molecular characterization of the α-GDH transgenics, the next step was their biochemical characterization. For comparative purposes, in addition to the α-GDH transgenics, we also analyzed the β-GDH transgenics overexpressing the tomato Slgdh-NAD;B2 and the respective isogenic control null segregant (NS; Purnell et al., 2005). The in vitro aminating activities were very high in shoots and roots of the overexpressing transgenic lines, whereas the measured deaminating activities were significantly lower (P < 0.01; Fig. 3, A and B). In Vvgdh-NAD;A1 overexpressing lines, the aminating activity was increased 10.9-fold (S2 line) and 7.0-fold (S5 line) in shoots and 2.5-fold (S2 line) and 1.7-fold (S5 line) in roots compared to the wild-type control (P < 0.01; Fig. 3A). The average ratio of aminating to deaminating in vitro activities in the Vvgdh-NAD;A1 transgenics was 14 in the shoots and 16.5 in the roots. In Slgdh-NAD;B2 overexpressers, the respective increases in shoots were 17.4-fold (S49 line) and 16.1-fold (S77 line) and 4.2-fold (S49 line) and 3.7-fold (S77 line) in roots compared to the NS control (all P < 0.01; Fig. 3B). In the Slgdh-NAD;B2 transgenics, the average ratio of aminating to deaminating activities was 19.5 and 14.0 in shoots and roots, respectively (Fig. 3, A and B).
Figure 3.
In vitro aminating (NADH) and deaminating (NAD) GDH activities in wild-type and Vvgdh-NAD;A1 tobacco transgenic lines S2 and S5 and NS and in Slgdh-NAD;B2 tobacco transgenic lines S49 and S77. A and B, Enzyme activities were determined in semipurified plant extracts. C and D, Enzyme activities were determined in the same plant extracts following dialysis. Statistical analysis of data was performed by one-way ANOVA. Asterisks indicate that means (n = 4) of tobacco transgenics are significantly different from the means of the corresponding wild-type and NS for either roots or shoots at P < 0.05 (*) and P < 0.01 (**).
To account for any possible contribution of malate dehydrogenase to the measured in vitro GDH activities in plant extracts (Fricke and Pahlich, 1992), GDH aminating and deaminating activities were also determined following the removal of malate from the plant extracts by dialysis. Dialyzed plant extracts exhibited higher aminating and lower deaminating activities (Fig. 3, C and D) compared to values before dialysis (P < 0.05; Fig. 3, A and B), resulting in even greater ratios of aminating to deaminating activity ratios. The respective average ratios after dialysis for the Vvgdh-NAD;A1 transgenics were 22.5 for shoots and 25.0 for roots (Fig. 3, C and D).
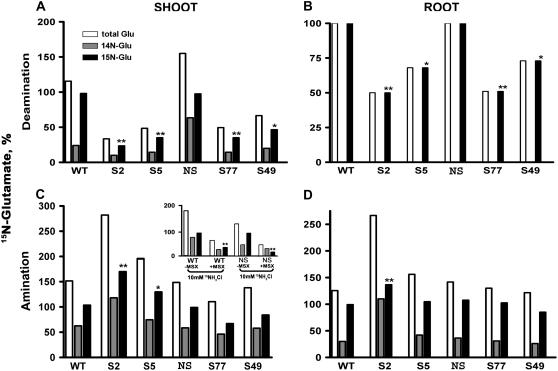
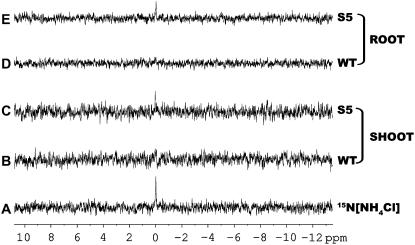
It was of interest to ascertain whether the in vitro GDH enzymatic activities reflected the in vivo GDH reaction direction(s). In doing so, the transgenic plants overexpressing the Vvgdh-NAD;A (this work) and Slgdh-NAD;B2 (Purnell et al., 2005) and the respective wild-type and NS controls, respectively, were supplied [15N]Glu or 15NH4. Given the low GDH-deaminating activities measured in vitro (Fig. 3), the overexpressing lines were expected to exhibit low in vivo rates of Glu deamination. However, both shoots and roots of all Vvgdh-NAD;A1 (S2 and S5) or Slgdh-NAD;B2 (S49 and S77) overexpressers supplied with [15N]Glu for 4 h contained significantly lower levels of residual [15N]Glu compared to the wild-type and NS controls, respectively (P < 0.05; Fig. 4, A and B). More specifically, the shoots of the transgenic lines S2 and S5 contained only 24.0% (P < 0.01) and 37.6% (P < 0.01), respectively, of the total amount of [15N]Glu present in the shoots of wild-type plants. Very similar trends were found in the shoots of the Slgdh-NAD;B2 lines; [15N]Glu levels in S49 (P < 0.05) and S77 (P < 0.01) were 51.1% and 38.2%, respectively, of those in NS controls (P < 0.05; Fig. 4A). Roots showed similar trends (Fig. 4B). [15N]Glu levels were, compared with respective controls, 50.0% and 67.7% in S2 (P < 0.01) and S5 (P < 0.05), 72.9% and 51.0% in S49 (P < 0.05) and S77 (P < 0.01). To further confirm these findings, the accumulation of [15N]NH4 in roots and shoots of wild-type and Vvgdh-NAD;A1 transgenic lines was assessed at 4 h after [15N]Glu treatment by NMR analysis. The transgenic line S5 accumulated more [15N]NH4 in both root and shoot compared to that of wild type (Fig. 5), fully confirming the findings of GC/MS analysis that the principal role of GDH is the deamination of Glu. Combined, these results clearly demonstrate that in vivo, all GDH isoenzymes deaminate Glu despite the low in vitro deaminating activities measured and confirm the recently published results of Purnell and Botella (2007), who monitored by NMR the fate of [15N]Glu in tobacco plants overexpressing the gene Slgdh-NAD;B2.
Figure 4.
GC/MS analysis to monitor the fate of [15N]Glu (A and B) and 15NH4 (C and D) in wild-type (WT) and Vvgdh-NAD;A1 tobacco transgenic lines S2 and S5 and in NS and Slgdh-NAD;B2 tobacco transgenic lines S49 and S77. A and B, The in vivo deaminating activity was determined as the percent residual [15N]Glu compared to wild-type/NS values. C and D, The in vivo amination GDH activity was determined as the 15NH4 incorporated into [15N]Glu with MSX, and the percent amination in the transgenic lines was calculated with respect to the wild-type/NS values (insert). Statistical analysis of data was performed by one-way ANOVA. Asterisks indicate that means (n = 4) of tobacco transgenics are significantly different from these of the corresponding wild-type and NS for either roots or shoots at P < 0.05 (*) and P < 0.01 (**).
Figure 5.
Catabolism of 15N[Glu] into 15N[NH4] in roots and shoots of wild-type tobacco (WT) and Vvgdh-NAD;A1 transgenic line S5. A, NMR spectrum of 20 mm 15N[NH4Cl] standard. B and C, NMR spectrum of 15N[NH4] accumulation at 4 h after 15N[Glu] treatment in shoots of wild-type tobacco and transgenic line S5. D and E, NMR spectrum of 15N[NH4] accumulation at 4 h after 15N[Glu] treatment in roots of wild-type tobacco and transgenic line S5.
In light of the high GDH-aminating activities measured in vitro (Fig. 3), all overexpressing transgenic lines would be expected to exhibit high in vivo rates of ammonium assimilation in the presence of MSX, a potent inhibitor of GS. To test this hypothesis, plants from all transgenic lines and wild-type and NS plants were supplied 15NH4 in the presence of MSX. Presence of the inhibitor resulted in approximately 90% inhibition of in vitro GS activity (data not shown). The insert of Figure 4C shows the levels of [15N]Glu resulting from the supply of 15NH4 to wild-type and NS tobacco shoots, with or without MSX. In the presence of MSX, the levels of [15N]Glu in the shoots of the wild-type and NS controls were 32.3% and 18.4%, respectively, of the amount without MSX. This labeled Glu could be the product of the residual GS activity, together with any GDH aminating activity. If the [15N]Glu originated exclusively from the residual GS activity, then the amount of [15N]Glu in the presence of MSX in both groups of transgenic lines would not differ significantly from the respective controls. This was the case for shoots and roots of the Slgdh-NAD-B2 overexpressing lines S49 and S77, in which [15N]Glu levels in the presence of MSX were not statistically different from the NS control (P < 0.05). In shoots and roots of the Vvgdh-NAD;A1 overexpressing line S2, in the presence of MSX, there was 34% and 27% more [15N]Glu than in the wild-type control (both P < 0.05; Fig. 4, C and D). In shoots of Vvgdh-NAD;A1 overexpressing line S5, the difference was smaller but it was still statistically significant (P < 0.05), which could be due to the relatively lower levels of labeled ammonium ions. These results support that GDH-isoenzyme 7 strongly deaminates Glu (Fig. 4, A and B) but also exerts low aminating activity (Fig. 4, C and D) under normal growth conditions. In addition, it is shown that the in vitro aminating/deaminating GDH enzymatic activities do not reflect the in vivo activities and explain the existing paradox and conflict in the literature.
DISCUSSION
Although the GDH homohexamers have nearly identical kinetic properties in vitro (Loulakakis and Roubelakis-Angelakis, 1996), they may catalyze opposite reactions in vivo and hence have different physiological roles. Clarification of the physiological role(s) of GDH isoenzymes is of great interest for biologists, because GDH occupies a biochemically critical position at the junction between carbon (2-OG) and nitrogen (Glu) metabolism and participates in the balancing of the cellular levels of three major components: the ammonium ions, 2-OG, and Glu. Until recently, there had been much controversy over the physiological function(s) of GDH isoenzymes in plants (Miflin and Habash, 2002; Stitt et al., 2002; Dubois et al., 2003), as there has been over ectomycorrhizal fungi (Morel et al., 2006), although they have been the focus of many projects and were at the center of an ongoing scientific discussion for three decades. The controversial results were largely due to several constrains: (1) in most studies, the in vitro enzymatic activities had been considered as indices of the in vivo function of GDH; (2) the in vitro activity of GDH is readily reversible; (3) the on-gel activity of the seven GDH isoenzymes is assessed using Glu as substrate for activity staining; (4) GDH is expressed in the form of seven GDH isoenzymes consisting of two subunit polypeptides at different ratios (Loulakakis and Roubelakis-Angelakis, 1991; Turano et al., 1996) encoded by different genes (Loulakakis and Roubelakis-Angelakis, 1990b, 1991; Restivo, 2004; Purnell et al., 2005); and (5) lack of suitable mutants and transgenic plants with altered genes encoding for the two subunit polypeptides of GDH.
Under physiological growth conditions, if GDH operates in the aminating direction, it may assimilate excessive ammonium ions in concert with GS/Glu synthase cycle reactions and/or assimilate some photorespiratory ammonium. Conversely, if GDH operates in the deaminating direction, it may fuel the tricarboxylic acid cycle under conditions of carbon deficit (Rhodes et al., 1989; Robinson et al., 1992; Aubert et al., 2001; Loulakakis and Roubelakis-Angelakis, 1991; Miflin and Habash, 2002; Dubois et al., 2003). Until recently, transgenic plants were transformed using bacterial or fungi gdh genes (Ameziane et al., 2000; Kisaka and Kida, 2003). The transformed tobacco lines overexpressing the bacterial gdh gene encoding for the α-subunit of GDH showed increased tolerance to water stress, herbicides, and toxic levels of ammonia and higher biomass production (Ameziane et al., 2000; Mungur et al., 2005). However, it was not known if these effects were brought about by the improved metabolic efficiency of ammonia assimilation or other adaptive mechanisms, because no detailed molecular or biochemical/enzymatic analyses were performed. Also, fruits from transgenic tomato plants carrying a gene for NADP-GDH (gdh;A) from Aspergillus nidulans contained 2- to 3-fold more free amino acids and 2-fold more Glu (Kisaka and Kida, 2003). An Arabidopsis (Arabidopsis thaliana) mutant lacking the α-subunit displayed retarded growth on media containing high concentration of nitrates and ammonium (Melo-Oliveira et al., 1996). In potato (Solanum tuberosum) tubers, GDH (of unknown isoenzyme distribution) was bidirectional in vivo, and the net reaction was strongly in the direction of Glu deamination (Aubert et al., 2001).
In 2006, Fontaine et al. reported the development of transgenic tobacco plants with decreased levels of the α-GDH subunit and identified an Arabidopsis mutant similarly affected. Also, very recently, NMR analysis of the transgenic tobacco plants overexpressing the tomato gene encoding for the β-GDH subunit (Purnell et al., 2005), grown under standard physiological conditions, clarified that the GDH isoenzyme 1 catalyzes in vivo the deamination of Glu (Purnell and Botella, 2007). In this work, transgenic tobacco plants overexpressing the gene encoding for the α-GDH subunit (this work) and β-GDH subunit (Purnell et al., 2005) administered with 15NH4 and [15N]Glu and grown under standard physiological conditions strongly deaminated Glu (Fig. 4A). Only the tobacco transgenic plants overexpressing the gene encoding for the α-GDH subunit exhibited low aminating activity, which at least in one line was significantly different from the wild-type plants in both shoots and roots (Fig. 4, C and D). That the deamination activity was stronger is supported by the low residual [15N]Glu after 24 h in the presence of MSX, which inhibits further use of [15N]Glu for synthesis of Gln by GS (data not shown) and in the presence of aminoacetic acid, which inhibits transamination of [15N]Glu (data not shown). Furthermore, earlier studies have shown that treatment with MSX does not inhibit GDH activity (Bechtold et al., 1998; Ameziane et al., 2000; Skopelitis et al., 2006).
From the results presented herein, it is clear that: (1) expression of the plant genes gdh-NAD;A is consistent with synthesis of the α-subunit polypeptide of GDH (Figs. 1 and 2); (2) accumulation of the α-subunit polypeptides results in the assembly of the anionic GDH isoenzymes, in agreement with previous results (Loulakakis and Roubelakis-Angelakis, 1991; Turano et al., 1996); (3) the measured values of in vitro high aminating and low deaminating GDH enzymatic activities (Fig. 3) do not reflect the in vivo directions of the enzymatic action of GDH (Fig. 4) and explain the existing paradox and conflict in literature regarding the assessment of the physiological functions of GDH (as shown in Figs. 3 and 4, although high ratios of in vitro aminating to deaminating activities were measured, the reverse was the case in vivo, i.e. very low ratios of aminating to deaminating activities); (4) the only reliable method for the assessment of the in vivo activities of GDH activity is the GC/MS or NMR analysis of the fate of [15N]Glu and 15NH4; and (5) results from these studies using the transgenic tobacco plants overexpressing either of the two genes that encode for the α- and β-subunit polypeptides (Purnell et al., 2005; Purnell and Botella, 2007; this work) clarified that in vivo, all GDH isoenzymes strongly deaminate Glu to NH4 and only GDH-isoenzyme 7 and the anionic ones exhibit low aminating activity toward synthesis of Glu under standard growth conditions (Fig. 4).
In light of these and recently published results (Skopelitis et al., 2006) that showed that under stress (salinity) conditions, the generated reactive oxygen species signal the expression of gdh-NAD;A, resulting in synthesis of GDH-isoenzyme 7, high in vivo aminating activity, leading to synthesis of Glu that in turn is directed toward Pro synthesis, and the available transgenic lines (Purnell et al., 2005; Fontaine et al., 2006; this work) will further provide a firm platform to answer pending questions about the physiological role(s) of GDH in plant carbon/nitrogen metabolism and to elucidate the developmental, trophic, and environmental cues regulating the expression of GDH genes.
CONCLUSION
These results support that GDH-isoenzyme 7 strongly deaminates Glu (Fig. 4, A and B) but also exerts low aminating activity (Fig. 4, C and D) under normal growth conditions. Therefore, adding together these results and those of Purnell and Botella (2007), it is now safe to conclude the under normal conditions, GDH-isoenzyme 1 and the cationic isoenzymes exclusively deaminate Glu, whereas GDH-isoenzyme 7 and the anionic isoenzymes exhibit bidirectional activities, high deaminating, and low aminating. Furthermore, it is clearly shown that the in vitro aminating/deaminating GDH enzymatic activities do not reflect the in vivo activities and explain the existing paradox and conflict in the literature.
MATERIALS AND METHODS
Plant Material
Tobacco (Nicotiana tabacum) L. ‘Xanthi’ plants (wild type) for Vvgdh-NAD;A1 and ‘Ti68’ and ‘NS’ for Slgdh-NAD;B2 transgenics were grown as described (Purnell et al., 2005).
Gene Constructs and Plant Transformation
Gene constructs, plant transformation, and screening of the transformants was performed as described earlier (Purnell et al., 2005) except for the Vvgdh-NAD;A1 transgenics, for which pART7/pART27 plant transformation vectors (Gleave, 1992) were used for both gene overexpression and suppression. For overexpression, the full-length cDNA clone Vvgdh-NAD;A1 from grapevine (Vitis vinifera; formerly gdhA; Loulakakis and Roubelakis-Angelakis, 1991) was ligated in sense orientation to the 35S cauliflower mosaic virus promoter in the vector pART7. For gene suppression, specific primers (sense primer 5′-CTTCTAGAGAGATCAAGGTGGAGTGCACGATTC-3′, antisense primer 5′-GGTACCCAAGATCCAACATTGCCAAAACCC-3′), with appropriate restriction sites, were used to PCR amplify a 560-bp fragment from the Vvgdh-NAD;A1 clone. This fragment was cloned into the vector pT3T7 and then recloned into two different sites of the plasmid pBluscript SK in opposite orientation separated by an intron sequence. The hairpin cassette was subcloned into the vector pART7 and then transferred to the vector pART27. For transformation, Agrobacterium tumefaciens strain LBA4404 was used. In all cases, T2 generation plants were used for further work.
Protein Extraction, Enzyme Assays, and Electrophoresis
Total proteins were extracted from plant tissues as described previously (Loulakakis and Roubelakis-Angelakis, 1990a; Siminis et al., 1994; Primikirios and Roubelakis-Angelakis, 1999). GDH activity was determined in both aminating and deaminating directions. The standard amination reaction mixture contained 100 mm Tris-HCl, pH 8.0, 20 mm a-ketoglutarate, 200 mm NH4Cl, 1 mm CaCl2, 0.2 mm NAD(P)H, enzyme solution, and deionized water to a final volume of 1 cm3. The standard deamination reaction mixture contained 100 mm Tris-HCl, pH 9.3, 100 mm l-Glu, 1 mm NAD(P)+, 0.5 mm CaCl2, enzyme solution, and deionized water to a final volume of 1 cm3. All assays were performed at 30°C. The absorption change at 340 nm was measured using a Perkin-Elmer UV/VIS spectrophotometer. One unit of GDH activity was defined as the reduction or oxidation of 1 μmol of coenzyme [NAD(P)+, NAD(P)H, respectively] min−1 at 30°C. Analytical and preparative SDS-PAGE of protein extracts (Loulakakis and Roubelakis-Angelakis, 1990b) and transfer of electrophoretically resolved proteins onto nitrocellulose filters (0.2-μm pore size, Schleicher and Schuell; Loulakakis and Roubelakis-Angelakis, 1991) were carried out as described. GDH isoenzymes were localized in nondenaturating polyacrylamide gels according to Loulakakis and Roubelakis-Angelakis (1991).
To account for any possible contribution of malate dehydrogenase to the measured in vitro GDH activities, crude extract was precipitated with ammonium sulfate, and the 35% to 70% fraction was collected. The pellet was dissolved in 200 mm Tris-HCl, pH 7.4, containing 14 mm mercaptoethanol and dialyzed (Loulakakis and Roubelakis-Angelakis, 1992).
Northern-Blot Analysis
Extraction of total RNA from plant tissues and northern blotting were performed as described (Loulakakis and Roubelakis-Angelakis, 1992; Loulakakis et al., 1996). The full-length cDNA Vvgdha-NAD;A1 (Loulakakis et al., 1996) and Slgdh-NAD;B2 (Purnell et al., 1997) were used as probes.
GC/MS Analysis of [15N]Glu in Tobacco Plants Grown in the Presence of 15NH4 or [15N]Glu
Plants (30 d postemergence) were kept in distilled water for 12 h and transferred to Murashige and Skoog (1962) medium containing 20 mm 15NH4Cl for 48 h or 10 mm 15[N]Glu for 4 h. For GS inhibition, plants were sprayed with 1.5 mm MSX solution of the potential GS inhibitor and roots were immersed in the same solution for 10 min. Two hours after the treatment with MSX, when GS activity was reduced by approximately 90%, plants were transferred to the [15N]-enriched growth medium. Plants were harvested and kept at −80°C. Tissue was extracted with 0.01 n HCl and 100 μL of the extract was transferred to a vial and dried at 90°C under nitrogen. Subsequently, 100 μL of acetonitrile and 100 μL of MTBSTFA were added to the residue. The mixture was sonicated for 30 s, heated at 70°C for 30 min, and filtered. 1-Chlorohexadecane was added as internal standard and 1 μL of the resulting solution was administered to GC/MS for amino acid determination (Sobolevsky et al., 2003) using a Hewlett-Packard 5971A mass-selective detector with the appropriate data system. A Hewlett-Packard model 5890 gas chromatograph, equipped with a Grob-type split-splitless injector, was directly coupled with the fused-silica capillary column (HP-5 MS with 0.25-mm film, 30 m × 0.25 mm i.d.) to the ion source. Helium was used as the carrier gas with a back pressure of 0.8 atm. The injector temperature was 280°C and the oven temperature program started at 70°C for 2 min and increased at a rate of 5°C/min up to 290°C, where it remained for 10 min. The ions used for 15N enrichment calculations for Glu were 432 and 433.
NMR Spectroscopy Analysis of [15N]Ammonium in Tobacco Plants Grown in the Presence of [15N]Glu
Thirty-day-old tobacco plants, treated as described in GC/MS analysis, were used in this experiment. A total of 300 mg of plant tissues (roots and shoots) from wild-type and S5 line treated with 15[N]Glu were sampled at 4 h to assess [15N]ammonium. Root and shoot samples from three individual plants were pooled and extracted with 5 volumes ice-cold 100% methanol and reduced to dryness under vacuum. Subsequently, the pellets were resuspended in 450 mL of a 0.2 m HCl/KOH buffer, pH 2.0 (Weast, 1974), and centrifuged (14,000g, 4°C, 20 min) and placed in 5-mm tubes for NMR analysis.
NMR spectra were recorded on a Bruker AMX500 spectrometer operating at 50.67 MHz for the 15N nucleus, at 26°C, with a 5-mm inverse broadband probe. All chemical shifts (δ) were referenced to the NH4Cl signal at δ = 0 ppm.
15N spectra were acquired using polarization transfer with a refocused INEPT pulse sequence (Burum and Ernst, 1980) using 16 K complex data points with a spectral width of 6.1 kHz and a repetition time of 3.5 s. The value of the 1JN-H evolutionary delay was set to 74 Hz and the refocusing delay was set to 1/8 1JN-H. 1H broadband decoupling (WALTZ16) was applied only during acquisition of the flame ionization detector. Typically, 512 transients were acquired with a total data acquisition time of 30 min. Flame ionization detectors were Fourier transformed following exponential multiplication with a line-broadening factor of 0.5 Hz for 15N spectra.
Preparation of Figures
The in situ-stained polyacrylamide gels were photographed using a Kodak DC120 digital camera (Eastman Kodak), western and northern blots were scanned with a HP ScanJet 6100 scanner (Hewlett-Packard), and densitometry was performed with Phoretix 1D version 2003.02 Image Analysis software.
Statistical analysis of data was performed with one-way ANOVA. Asterisks indicate that means (n = 4) of tobacco transgenics are significantly different from the means of the corresponding wild type and NS for either roots or shoots at P < 0.05 (*) and P < 0.01 (**).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number X86924.
Acknowledgments
We thank Dr. M. Purnell and Professor J. Botella for kindly providing the β-transgenics. The authors thank Professor E. Mikros from the School of Health Sciences, Department Pharmaceutical Chemistry for his valuable help in NMR analysis.
This work was supported by the European Social Fund Herakleitos, Pythagoras, and National Resources.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Kalliopi A. Roubelakis-Angelakis (poproube@biology.uoc.gr).
Open Access articles can be viewed online without a subscription.
References
- Ameziane RK, Bernhard RB, Lightfoot D (2000) Expression of the bacterial gdhA gene encoding NADPH glutamate dehydrogenase in tobacco affects plant growth and development. Plant Soil 221 47–57 [Google Scholar]
- Aubert S, Bligny R, Douce R, Ratcliffe RG, Roberts JKM (2001) Contribution of glutamate dehydrogenase to mitochondrial metabolism studied by 13C and 31P nuclear magnetic resonance. J Exp Bot 52 37–45 [PubMed] [Google Scholar]
- Bechtold U, Pahlich E, Lea PJ (1998) Methionine sulphoximine does not inhibit pea and wheat glutamate dehydrogenase. Phytochemistry 49 347–354 [Google Scholar]
- Burum BP, Ernst RR (1980) Net polarization transfer via a J-ordered state for signal enhancement of low-sensitivity nuclei. J Magn Reson 39 163–168 [Google Scholar]
- Cammaerts D, Jacobs M (1985) A study of the role of glutamate dehydrogenase (EC 1.4.1.2) in the nitrogen metabolism of Arabidopsis thaliana. Planta 163 517–526 [DOI] [PubMed] [Google Scholar]
- Dubois F, Tercé-Laforgue T, Gonzalez-Moro MB, Estavillo MB, Sangwan R, Gallais A, Hirel B (2003) Glutamate dehydrogenase in plants: is there a new story for an old enzyme? Plant Physiol Biochem 41 565–576 [Google Scholar]
- Fontaine JX, Saladino F, Agrimonti C, Bedu M, Tercé-Laforgue T, Tétu T, Hirel B, Restivo FM, Dubois F (2006) Control of the synthesis and subcellular targeting of the two GDH genes products in leaves and stems of Nicotiana plumbaginifolia and Arabidopsis thaliana. Plant Cell Physiol 47 410–418 [DOI] [PubMed] [Google Scholar]
- Fox GG, Ratcliffe RG, Robinson SA, Stewart GR (1995) Evidence for deamination by glutamate dehydrogenase in higher plants. Commentary. Can J Bot 73 1112–1115 [Google Scholar]
- Fricke W, Pahlich E (1992) Malate: a possible source of error in the NAD glutamate dehydrogenase. J Exp Bot 43 1515–1518 [Google Scholar]
- Gleave AP (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20 1203–1207 [DOI] [PubMed] [Google Scholar]
- Kisaka H, Kida T (2003) Transgenic tomato plant carrying a gene for NADP-dependent Glu dehydrogenase (gdhA) from Aspergillus nidulans. Plant Sci 164 35–42 [Google Scholar]
- Lea PJ, Ireland RJ (1999) Nitrogen metabolism in higher plants. In BK Singh, ed, Plants Amino Acids. Marcel Dekker, New York, pp 1–47
- Lea PJ, Miflin BJ (1974) An alternative route for nitrogen assimilation in plants. Nature 251 680–685 [DOI] [PubMed] [Google Scholar]
- Lea PJ, Thurman DA (1972) Intracellular location and properties of plant L-glutamate dehydrogenases. J Exp Bot 23 440–449 [Google Scholar]
- Limami AM, Rouillon C, Glevarec G, Gallais A, Hirel B (2002) Genetic and physiological analysis of germination efficiency in maize in relation to nitrogen metabolism reveals the importance of cytosolic glutamine synthetase. Plant Physiol 130 1860–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loulakakis KA, Primikirios NI, Nikolantonakis MA, Roubelakis-Angelakis KA (2002) Immunocharacterization of Vitis vinifera L. ferredoxin-dependent glutamate synthase and its spatial and temporal changes during leaf development. Planta 215 630–638 [DOI] [PubMed] [Google Scholar]
- Loulakakis KA, Roubelakis-Angelakis KA (1990. a) Intracellular localization and properties of NADH-glutamate dehydrogenase from Vitis vinifera L.: purification and characterization of the major isoenzyme. J Exp Bot 41 1223–1230 [Google Scholar]
- Loulakakis KA, Roubelakis-Angelakis KA (1990. b) Immunocharacterization of NADH-glutamate dehydrogenase from Vitis vinifera L. Plant Physiol 94 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loulakakis KA, Roubelakis-Angelakis KA (1991) Plant NAD(H) glutamate dehydrogenase consists of two subunit polypeptides and their participation in the seven isoenzymes occurs in an ordered ratio. Plant Physiol 97 104–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loulakakis KA, Roubelakis-Angelakis KA (1992) Ammonium-induced increase in NADH-glutamate dehydrogenase activity is caused by de novo synthesis of the α-subunit. Planta 187 322–327 [DOI] [PubMed] [Google Scholar]
- Loulakakis KA, Roubelakis-Angelakis KA (1996) The seven NAD(H)-glutamate dehydrogenase isoenzymes exhibit similar anabolic and catabolic activities. Physiol Plant 96 29–35 [Google Scholar]
- Loulakakis KA, Roubelakis-Angelakis KA, Kanellis AK (1994) Regulation of glutamate dehydrogenase and glutamine synthetase in avocado fruit during development and ripening. Plant Physiol 106 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loulakakis KA, Roubelakis-Angelakis KA, Kanellis AK (1996) Isolation of functional RNA from grapevine tissues poor in nucleic acid content. Am J Enol Vitic 47 181–185 [Google Scholar]
- Magalhaes JR (1991) Kinetics of 15NH4 assimilation in tomato plants: evidence for 15NH4 assimilation via GDH in tomato roots. J Plant Nutr 4 1341–1353 [Google Scholar]
- Magalhaes JR, Grace PJ, Rich D, Rhodes D (1990) Kinetics of 15NH4 assimilation in Zea mays. Plant Physiol 94 647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux C, Valadier MH, Brugière N, Morot-Gaudry JF, Hirel B (2000) Characterization of the sink/source transition in tobacco (Nicotiana tabacum L.) shoots in relation to nitrogen management and leaf senescence. Planta 211 510–518 [DOI] [PubMed] [Google Scholar]
- Melo-Oliveira R, Oliveira IC, Coruzzi GM (1996) Arabidopsis mutant analysis and gene regulation define a nonredundant role for glutamate dehydrogenase in nitrogen assimilation. Proc Natl Acad Sci USA 93 4718–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miflin BJ, Habash D (2002) The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. J Exp Bot 53 979–987 [DOI] [PubMed] [Google Scholar]
- Morel M, Buée M, Chalot M, Brun A (2006) NADP-dependent glutamate dehydrogenase: a dispensable function in ectomycorrhizal fungi. New Phytol 169 179–189 [DOI] [PubMed] [Google Scholar]
- Mungur R, Glass ADM, Goodenow DB, Lightfoot DA (2005) Metabolite fingerprinting in transgenic Nicotiana tabacum altered by the Escherichia coli glutamate dehydrogenase gene. J Biomed Biotechnol 2 198–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige I, Skoog F (1962) Revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Paczek V, Dubois F, Sangwan R, Morot-Gaudry JF, Roubelakis-Angelakis KA, Hirel B (2002) Cellular and subcellular localization of glutamine synthetase and glutamate dehydrogenase in grapes gives new insights on the regulation of C and N metabolism. Planta 216 245–254 [DOI] [PubMed] [Google Scholar]
- Primikirios NI, Roubelakis-Angelakis KA (1999) Characterization and expression of arginine decarboxylase in Vitis vinifera L. Planta 208 574–582 [DOI] [PubMed] [Google Scholar]
- Purnell MP, Botella JR (2007) Tobacco isoenzyme 1 of NAD(H)-dependent glutamate dehydrogenase catabolizes glutamate in vivo. Plant Physiol 143 530–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purnell MP, Skopelitis DS, Roubelakis-Angelakis KA, Botella JR (2005) Modulation of glutamate dehydrogenase activity in transgenic tobacco reveals cryptic regulation of gene expression and protein level. Planta 222 167–180 [DOI] [PubMed] [Google Scholar]
- Purnell MP, Stewart GR, Botella JR (1997) Cloning and characterisation of a glutamate dehydrogenase cDNA from tomato (Lycopersicon esculentum L). Gene 186 249–254 [DOI] [PubMed] [Google Scholar]
- Restivo FM (2004) Molecular cloning of glutamate dehydrogenase genes of Nicotiana plumaginifolia: structure analysis and regulation of their expression by physiological and stress conditions. Plant Sci 166 971–982 [Google Scholar]
- Rhodes D, Brunk OG, Magalhaes JR (1989) Assimilation of ammonium by glutamate dehydrogenase? In TE Poulten, JT Romeo, EE Conn, eds, Recent Advances in Phytochemistry. Plenum Press, New York, pp 191–226
- Robinson SA, Stewart GR, Phillips P (1992) Regulation of glutamate dehydrogenase activity in relation to carbon limitation and protein catabolism in carrot cell suspension cultures. Plant Physiol 98 1190–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siminis CI, Kanellis AK, Roubelakis-Angelakis KA (1994) Catalase is differentially expressed in dividing and nondividing protoplasts. Plant Physiol 105 1375–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skopelitis DS, Paranychianakis NV, Paschalidis KA, Pliakonis ED, Yakoumakis D, Delis ID, Kouvarakis A, Papadakis AK, Stephanou E, Roubelakis-Angelakis KA (2006) Abiotic stress generated ROS signal expression of anionic glutamate dehydrogenases to form glutamate for proline synthesis. Plant Cell 18 2767–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky TG, Revelsky AI, Miller B, Oriedo V, Chernetsova ES, Revelsky IA (2003) Comparison of silylation and esterification/acylation procedures in GC-MS analysis of amino acids. J Sep Sci 26 1474–1478 [Google Scholar]
- Srivastava HS, Singh-Rana P (1987) Role and regulation of glutamate dehydrogenase activity in higher plants. Phytochemistry 26 597–610 [Google Scholar]
- Stewart GR, Shatilov VR, Turnbull MH, Robinson SA, Goodall R (1995) Evidence that glutamate dehydrogenase plays a role in oxidative deamination of glutamate in seedlings of Zea mays. Aust J Plant Physiol 22 805–809 [Google Scholar]
- Stitt M, Muller C, Matt P, Gibon Y, Carillo P, Morcuende R, Scheible WR, Krapp A (2002) Steps towards an integrated view of nitrogen metabolism. J Exp Bot 53 959–970 [DOI] [PubMed] [Google Scholar]
- Syntichaki KM, Loulakakis KA, Roubelakis-Angelakis KA (1996) The amino acid sequence similarity of plant glutamate dehydrogenase with the extremophilic archaeal enzyme conforms to its stress related function. Gene 168 87–92 [DOI] [PubMed] [Google Scholar]
- Turano FJ, Dashner R, Upadhayaya A, Caldwell CR (1996) Purification of mitochondrial glutamate dehydrogenase from dark-grown soybean seedlings. Plant Physiol 112 1357–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weast RC (1974) Handbook of Chemistry and Physics. CRC Press, Cleveland
- Yamaya T, Oaks A (1987) Synthesis of glutamate by mitochondria-an anaplerotic function for glutamate dehydrogenase. Physiol Plant 70 749–756 [Google Scholar]
- Yamaya T, Oaks A, Rhodes D, Matsumoto H (1986) Synthesis of [15N]glutamate from [15N]H4+ and [15N]glycine by mitochondria isolated from pea and corn roots. Plant Physiol 81 754–757 [DOI] [PMC free article] [PubMed] [Google Scholar]