Abstract
The involvement of the PsbI protein in the assembly and repair of the photosystem II (PSII) complex has been studied in the cyanobacterium Synechocystis sp. PCC 6803. Analysis of PSII complexes in the wild-type strain showed that the PsbI protein was present in dimeric and monomeric core complexes, core complexes lacking CP43, and in reaction center complexes containing D1, D2, and cytochrome b-559. In addition, immunoprecipitation experiments and the use of a histidine-tagged derivative of PsbI have revealed the presence in the thylakoid membrane of assembly complexes containing PsbI and either the precursor or mature forms of D1. Analysis of PSII assembly in the psbI deletion mutant and in strains lacking PsbI together with other PSII subunits showed that PsbI was not required for formation of PSII reaction center complexes or core complexes, although levels of unassembled D1 were reduced in its absence. However, loss of PsbI led to a dramatic destabilization of CP43 binding within monomeric and dimeric PSII core complexes. Despite the close structural relationship between D1 and PsbI in the PSII complex, PsbI turned over much slower than D1, whereas high light-induced turnover of D1 was accelerated in the absence of PsbI. Overall, our results suggest that PsbI is an early assembly partner for D1 and that it plays a functional role in stabilizing the binding of CP43 in the PSII holoenzyme.
The PSII complex is the multisubunit membrane protein complex catalyzing oxidation of water and reduction of plastoquinone in the thylakoid membranes of higher plants, algae, and cyanobacteria. The membrane-embedded core complex of PSII consists of the D1 and D2 reaction center (RC) subunits, the inner chlorophyll (Chl)-binding antenna proteins, CP47 and CP43, and a number of smaller polypeptides (for review, see Barber, 2006). D1 and D2 form a heterodimer that binds the cofactors involved in primary charge separation and subsequent electron transfer within PSII, while the main purpose of CP47 and CP43 is to deliver energy to the RC for driving electron transfer and, in the case of CP43, to help ligate the CaMn4 cluster.
The roles of many of the small subunits in PSII are still unclear. In the case of PsbI, it is known to be a component of the simplest PSII unit capable of primary charge separation called D1/D2/cytochrome b-559 (Cyt b-559) or PSII RC complex (Nanba and Satoh, 1987; Webber et al., 1989). PsbI is a highly conserved PSII component showing 71% amino acid identity between the Arabidopsis (Arabidopsis thaliana) and Synechocystis sp. PCC 6803 (Synechocystis 6803) proteins. Recent crystal structures of cyanobacterial PSII (Ferreira et al., 2004; Loll et al., 2005) have confirmed that PsbI spans the membrane once and that it is located on the periphery of the PSII complex in close proximity to CP43 and the first and second transmembrane helices of D1, where it participates in binding a Chl molecule (ChlZD1).
The role of PsbI in the assembly of PSII is unclear. In plastids, the D2 protein with bound Cyt b-559 has been postulated to be the first stable assembly complex, which subsequently binds newly synthesized D1 protein to form the PSII RC complex (Müller and Eichacker, 1999). The D1 protein is synthesized as a precursor (pD1) with a carboxyl-terminal extension that is cleaved after residue Ala-344 (Takahashi et al., 1988; Nixon et al., 1992) by a specific processing endoprotease, CtpA (Anbudurai et al., 1994). In higher plants, the extension usually consists of nine residues and it is removed in a single step. In contrast, the 16-amino acid C-terminal extension of D1 in Synechocystis 6803 is cleaved in two steps (Inagaki et al., 2001). The primary cleavage occurs after Ala-352, resulting in formation of a processing intermediate termed iD1 (Komenda et al., 2007), which in Synechocystis 6803 is mainly associated with RC complexes (Komenda et al., 2004).
In contrast to Cyt b-559, PsbI is not critical for assembly of PSII. A mutant of Synechocystis 6803 lacking the psbI gene was able to grow photoautotrophically and its PSII oxygen evolution activity was 70% to 75% of wild-type levels (Ikeuchi et al., 1995). In Chlamydomonas reinhardtii, inactivation of the psbI gene decreased oxygen evolution activity and D1 content in the resulting photoautotrophic mutant to 10% to 20% of wild-type levels (Künstner et al., 1995). A psbI deletion mutant of tobacco (Nicotiana tabacum) contained reduced amounts of dimeric PSII and PSII light-harvesting complex II supercomplexes. The tobacco mutant also exhibited a modified primary quinone acceptor, QA, an increased sensitivity to high light, and no phosphorylation of the RC proteins, D1 and D2 (Schwenkert et al., 2006). In summary, inactivation of psbI causes a decrease in PSII photochemical activity and content in all organisms studied to date, but the underlying molecular basis for this effect remains unknown.
Here we present a detailed analysis of the role of PsbI during assembly and repair of cyanobacterial PSII. The results showed that although the presence of PsbI is not critical for PSII biogenesis, the protein is important for stable incorporation of CP43 into PSII and possibly for stabilizing newly synthesized D1 protein.
RESULTS
Identification of the PsbI Protein in PSII Complexes and Early Assembly Intermediates
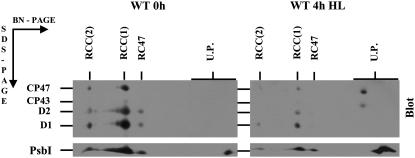
To investigate the role of PsbI in PSII assembly, we first screened for the presence of PsbI in various types of PSII complexes and PSII assembly intermediates. An excellent tool for this purpose is the two-dimensional (2D) separation of membrane proteins consisting of blue native (BN) PAGE in one direction and denaturing PAGE in the second direction (2D BN/SDS-PAGE) followed by immunoblotting. In this way, we probed for the presence of PsbI in PSII complexes of wild type and several mutant strains of Synechocystis 6803 blocked at a particular step in assembly.
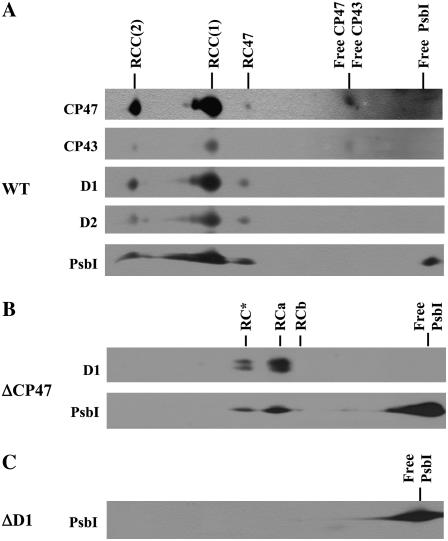
In wild type, the majority of the large PSII protein subunits CP47, D2, and D1 accumulate in monomeric [RCC(1)] and dimeric [RCC(2)] core complexes as well as in a core subcomplex lacking CP43 (RC47; see also Komenda et al., 2004). PsbI was detected in all these complexes and, in addition, was found in the low-molecular-weight region, probably in an unassembled form (Fig. 1A).
Figure 1.
Immunoblots of thylakoid membrane proteins from Synechocystis 6803 wild type (WT; A), psbB deletion mutant ΔCP47 (B), and psbA deletion mutant ΔD1 (C) after their separation by 2D BN/SDS-PAGE. Thylakoid proteins were separated by 2D BN-PAGE, blotted onto PVDF membrane, and immunodecorated using antibodies raised against D1, D2, CP47, CP43, and PsbI. Designation of complexes: RCC(2) and RCC(1), dimeric and monomeric PSII core complexes, respectively; RC47, PSII core complex lacking CP43; RC*, RCa, and RCb, RC complexes. One microgram of Chl was loaded for each sample.
To investigate at what stage PsbI bound to PSII during assembly, we tested for its presence in the PSII complexes found in strain ΔCP47, which is unable to synthesize CP47 and so is unable to assemble PSII beyond formation of a PSII RC complex (Komenda et al., 2004). In this strain, native BN-PAGE resolved three RC complexes with different mobilities. The precise subunit composition of each complex is unclear, but each contained D2, both subunits of Cyt b-559, and either D1 or its processing intermediate, iD1. The most abundant RC complex was RCa, followed by RC*, with a mobility similar to that of RC47, while the smallest RCb complex was present in very low amounts. Immunoblots confirmed the presence of PsbI in both RC* and RCa, although the majority of the protein was detected in the low Mr region as a free protein (Fig. 1B). The low abundance of RCb in the sample analyzed in Figure 1B did not allow a reliable evaluation of the presence of PsbI. However, PsbI could be detected in this particular complex in frozen thylakoids of ΔCP47 stored for a longer time, indicating possible artefactual origin of RCb.
In contrast, in a psbA deletion strain (ΔD1) in which CP47, CP43, and D2 are synthesized but do not assemble into a larger complexes (Komenda et al., 2004), the PsbI protein was found exclusively as a free protein (Fig. 1C).
The overall level of PsbI in the ΔD1 mutant was much lower than in wild type, but after additional inactivation of the slr0228 gene encoding an FtsH protease homolog, the amount of PsbI reached wild-type levels (Supplemental Fig. S1). This observation suggested that unassembled PsbI is removed from the thylakoid membrane through the action of FtsH (slr0228) as observed previously for unassembled D2 and CP47 (Komenda et al., 2006).
Absence of PsbI Destabilizes Binding of CP43 in the PSII Core Complex But Does Not Inhibit Assembly of the PSII RC Complex
To characterize the role of PsbI in the assembly of PSII core and RC complexes, we constructed a psbI deletion mutant (ΔPsbI) in which the psbI gene was replaced by a zeocine resistance cassette. In agreement with earlier data (Ikeuchi et al., 1995), the resulting mutant exhibited a somewhat slower rate of photoautotrophic growth and its PSII photochemical activity, measured as the variable fluorescence yield or the light-saturated rate of oxygen evolution, was lower by 30% to 40% in comparison with wild type (Supplemental Table S1). On the other hand, electron transfer between the plastoquinone electron acceptors, QA and QB, was similar in wild type and mutant (data not shown).
To characterize PSII assembly in wild type and ΔPsbI, we pulse labeled cells with [35S]Met/Cys at 23°C and analyzed the incorporation of radiolabel into PSII subunits following 2D BN/SDS-PAGE to resolve the different PSII complexes. By performing the experiment at 23°C we hoped to slow assembly down so that assembly intermediates could be more easily detected.
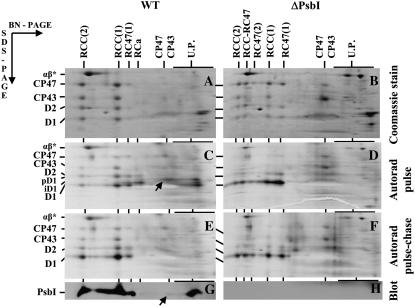
In wild type, 80% to 90% of the D1 subunit was found in the dimeric and monomeric core complexes, RCC(2) and RCC(1), as assessed by the intensity of the Coomassie Blue-stained band, with the remaining 10% to 20% in the RC47 complex (Fig. 2A; Supplemental Table S2). In striking contrast, in ΔPsbI, only 20% and 30% of D1 was present in dimeric and monomeric PSII core complexes, respectively. Instead, a significant level of D1 was found in PSII complexes lacking the full complement of CP43: 20% was detected in the dimeric core complex lacking one copy of CP43 (RCC-RC47), 5% in the dimeric core complex lacking both CP43 copies [RC47(2)], and 25% in the monomeric RC47 (Fig. 2B; Supplemental Table S2). Thus, binding of CP43 was inhibited or destabilized in about 50% of the PSII core complexes in ΔPsbI.
Figure 2.
Pulse-chase analysis of wild type and the psbI deletion strain ΔPsbI. Cells of wild type (A, C, E, and G) and ΔPsbI (B, D, F, and H) were radiolabeled at 500 μmol photons m−2 s−1 and 23°C with a mixture of [35S]Met/Cys for 10 min (pulse) and then CAP (1 mg mL−1) was added and cells incubated at the same temperature at 125 μmol photons m−2 s−1 for another 20 min (chase). Labeled cells were used for isolation of thylakoids, which were analyzed by 2D BN/SDS-PAGE. A and B, Coomassie Blue-stained gels of proteins after the pulse. C and D, Autoradiograms of the same samples with the putative D1-PsbI complex designated by arrow in C. E and F, Autoradiograms of proteins after pulse chase. G and H, Low-Mr region with PsbI detected by immunoblotting (blot) and putative D1-PsbI complex designated by an arrow. Designations of proteins are as described in the legend to Figure 1. RCC-RC47 designates dimeric PSII core complex lacking only one CP43 copy and RC47(2) the dimeric PSII core lacking both CP43 copies. U.P., Unassembled proteins. α- and β-subunits of ATP synthase (designated by αβ*) were used as internal standards during quantification of D1-stained and -labeled bands (see Supplemental Table S2). The arrows in C and G show the complex of PsbI and pD1 in wild type. Six micrograms of Chl was loaded for each sample.
Autoradiograms obtained from the same gels were used to assess the incorporation of radioactive label into the various PSII proteins. For wild type, the D1 protein was preferentially labeled in all PSII complexes, including the RCa complex, as expected because of selective D1 replacement during PSII repair. However, 50% to 60% of labeled D1, including its two incompletely processed forms, pD1 and iD1, was found in the fraction of small complexes and free unassembled proteins (Fig. 2C, U.P.). This unassembled D1 fraction can be detected in wild type only by a short radioactive pulse at a lower temperature (23°C), while at the growth temperature (30°C), the assembly of D1 into PSII complexes is so fast that free D1 forms are not detectable (see Komenda et al., 2006). In the unassembled fraction, pD1 migrated in two bands under native conditions in the first dimension, with the mobility of the larger one (Fig. 2C, arrow) comigrating with a weak band in the PsbI immunoblot (Fig. 2G, arrow). The data suggested the existence of a putative pD1-PsbI complex (see below). When the label was chased for 20 min in the presence of the protein synthesis inhibitor, chloramphenicol (CAP), the labeled D1 protein found in RCa and in the free protein fraction disappeared, and labeling of the mature D1 subunit increased 3.5 times in RCC(1) and 7 times in RCC(2) (Fig. 2E; Supplemental Table S2). This result confirmed that unassembled D1 and the RCa complex were true assembly intermediates and not dead end products.
The D1 protein was also preferentially labeled in the ΔPsbI mutant, but its distribution among the various complexes differed from wild type. After the pulse, 60% of the labeled D1 protein was present in the RC47 complex and no labeled D1 was observed in either RCa or the unassembled protein fraction (Fig. 2D). During the chase, 80% of the labeled D1 disappeared from RC47, but only 20% appeared in PSII complexes larger than RC47 (Fig. 2F; Supplemental Table S2). This result indicated that most of the newly synthesized D1 protein incorporated into RC47 complexes was degraded before the complex could be assembled further into monomeric and dimeric PSII core complexes.
We also constructed a series of double mutants in which the deletion of the psbI gene was performed in strains lacking the large PSII subunits CP43, CP47, or D2. In the strain lacking CP43 (ΔCP43), PSII assembly could not progress beyond formation of the RC47 complex and the strongly radiolabeled D1 protein almost exclusively accumulated in this complex (Supplemental Fig. S2, A and C). A similar pattern was also maintained after additional removal of PsbI, except that there was a slight increase in the amount of labeled D2 in the RC47 complex and in the unassembled protein fraction (Supplemental Fig. S2, B and D). The very similar intensity of radiolabeled D1 band in the RC47 complex indicated similar turnover rates of D1 in both strains.
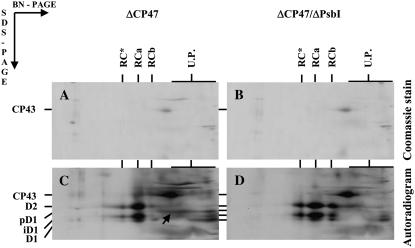
The psbB deletion strain ΔCP47 has already been shown to accumulate small amounts of the RC complexes RC*, RCa, and RCb, and all these complexes contained PsbI (Fig. 1B). To find out whether their formation was dependent on the presence of PsbI, we also constructed and characterized the double mutant ΔCP47/ΔPsbI. On Coomassie Blue-stained gels of radioactively labeled thylakoid proteins separated by 2D BN/SDS-PAGE, only the free unassembled CP43 was visible in both ΔCP47 and ΔCP47/ΔPsbI strains (Fig. 3, A and B). The formation of RC complexes in both strains was confirmed by autoradiography (Fig. 3, C and D), although the amount of RC* and RCb complexes in the PsbI-less mutant was increased at the expense of RCa. The autoradiogram of ΔCP47 also indicated the presence of a putative pD1-PsbI complex (Fig. 3C, arrow) previously found in wild type labeled at 23°C (Fig. 2C). Quantification of D1 by immunoblotting (Supplemental Fig. S3) revealed a slightly lower amount of mature D1 and partially processed iD1 (Komenda et al., 2007) in the double mutant. Overall, these data show that formation of the RC complexes was not dependent on the presence of PsbI.
Figure 3.
Analysis of thylakoid membrane proteins in the psbB deletion mutant ΔCP47 and the double deletion mutant ΔCP47/ΔPsbI. Cells of the Synechocystis 6803 strains were radiolabeled at 500 μmol photons m−2 s−1 and 29°C with [35S]Met/Cys for 30 min and their thylakoid proteins were separated by 2D BN/SDS-PAGE. Designations of proteins are as described in the legend to Figure 1; the arrow indicates the complex of PsbI and pD1. Six micrograms of Chl was loaded for each sample.
In the Absence of D2, the Majority of the D1 Protein Is Associated with PsbI
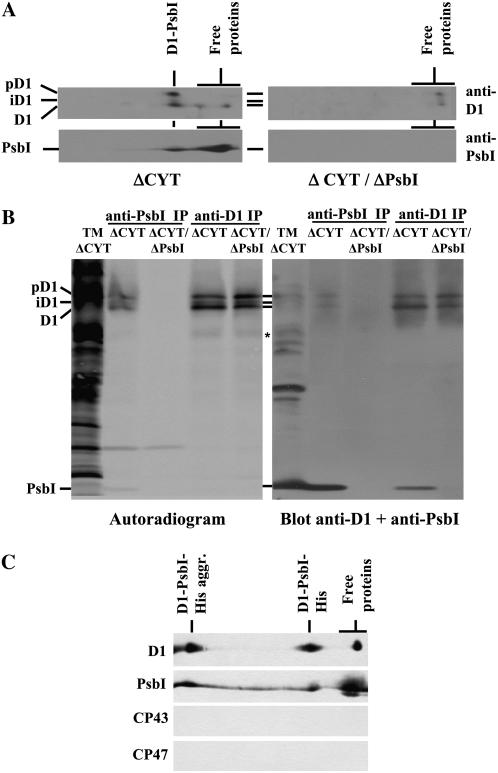
To confirm that PsbI was able to form a complex with D1, experiments were conducted using a psbEFLJ deletion strain, which is unable to synthesize Cyt b-559 and, as a consequence, is also unable to accumulate the D2 protein (Komenda et al., 2004). In this strain (ΔCYT), most of PsbI was present as a free protein, but there was an additional small PsbI complex (Fig. 4A) similar in size to the putative pD1-PsbI complex detected in wild type (Fig. 2G, arrow). Indeed, when the 2D blot of the ΔCYT strain was reprobed with an antibody against D1, each D1 form (i.e. pD1, iD1, and D1) migrated in two bands under native conditions (see also Komenda et al., 2004). About 80% of the protein was present in the larger complex with the same mobility as the small PsbI-containing complex (Fig. 4A). When a strain lacking both the psbEFLJ operon and the psbI gene was analyzed (ΔCYT/ΔPsbI), the larger D1 band was no longer detected and the overall level of D1 decreased to 25% of that found in ΔCYT (see Fig. 4A). The identical D1-PsbI complex was also detected in the ΔCP47 strain (Fig. 3, arrow) but not in the ΔCP47/ΔPsbI double mutant. Overall, these results suggested that in the absence of D2, the majority of D1 was associated with PsbI. This association appeared to stabilize unassembled D1 and enabled its higher accumulation in the thylakoid membrane.
Figure 4.
Identification of a D1-PsbI precomplex. A, Thylakoid membrane proteins (1 μg of Chl) of the ΔCYT and ΔCYT/ΔPsbI strains were separated by 2D BN/SDS-PAGE, transferred onto PVDF membrane, and detected by antibodies against D1 and PsbI. B, Pulse-labeled thylakoid proteins (5 μg of Chl) from the ΔCYT and ΔCYT/ΔPsbI strains were immunoprecipitated using antibodies specific for D1 (anti-D1 IP) or PsbI (anti-PsbI IP) and the immunoprecipitates together with thylakoids from ΔCYT (TM) were analyzed by SDS-PAGE, blotted onto PVDF membrane, autoradiographed (left, autoradiogram), and then probed with antibodies against both PsbI and D1 proteins (right, blot anti-D1 + anti-PsbI). *, A putative 23-kD D1 synthesis intermediate. C, Immunoblots of D1, PsbI, CP43, and CP47 after 2D BN/SDS-PAGE of the protein fraction bound to nickel-affinity column loaded with solubilized thylakoids of the PsbI-His/ΔPsbI/ΔCYT strain. D1-PsbI-His aggr., Aggregates of D1 and PsbI-His.
To strengthen these conclusions, we performed immunoprecipitation experiments using antibodies against PsbI and D1 and radiolabeled thylakoids isolated from the ΔCYT and ΔCYT/ΔPsbI strains. In ΔCYT, both antibodies precipitated PsbI together with all forms of D1 but not the other PSII proteins (Fig. 4B). For the ΔCYT/ΔPsbI strain, we were unable to immunoprecipitate D1 using the antibody against PsbI, while the antibody against D1 precipitated only the D1 protein. These results confirmed the existence of the D1-PsbI complex in the ΔCYT strain. The presence of labeled pD1 in the immunoprecipitate further indicated that binding of PsbI occurred soon after or during synthesis of D1.
The third line of evidence for binding of PsbI to D1 was obtained using a strain lacking the psbEFLJ operon in which the original psbI gene was replaced by a His-tagged copy. Thylakoids from the PsbI-His/ΔPsbI/ΔCYT strain were solubilized with dodecyl-maltoside (DM) and the extract was loaded on the nickel-affinity chromatography column. Bound proteins were analyzed by 2D BN/SDS-PAGE and immunoblotting using antibodies against PsbI, D1, CP43, and CP47. The majority of PsbI-His was present as free protein but a substantial portion was found in a small complex with D1 (Fig. 4C). Neither CP43 nor CP47 were found in the eluate, confirming specific binding of PsbI-His to D1.
PsbI Does Not Undergo Fast Turnover But Its Absence Accelerates Turnover of the D1 Protein
Given that PsbI binds to D1 in the PSII complex (Ferreira et al., 2004; Loll et al., 2005), we were interested to learn whether both PsbI and D1 were turned over during PSII repair or whether PsbI was stable and reused during D1 replacement. Analysis of thylakoids isolated from wild-type cells that had been exposed to high light for 4 h in the presence of the protein synthesis inhibitor lincomycin showed that PsbI is much more stable than D1 (Fig. 5). Under these illumination conditions, conducted in the absence of protein synthesis, the majority of D1 and D2 was degraded, while other proteins like CP47, CP43, and PsbI were released from photodamaged PSII and were detected in the unassembled protein fraction. Pulse-chase experiments (Fig. 6, bottom sections) also indicated that a weakly labeled band corresponding to PsbI remained stable over the period of the chase.
Figure 5.
Immunoblots of thylakoid membrane proteins from the Synechocystis 6803 wild-type strain before and after 4-h exposure to high light. Cells of wild type were exposed to high irradiance (1,000 μmol photons m−2 s−1) for 4 h in the presence of the protein synthesis inhibitor lincomycin (100 μg mL−1). Thylakoid membrane proteins (1 μg of Chl) were separated by 2D BN/SDS-PAGE, transferred onto PVDF membrane, and probed with antibodies against the D1, D2, CP43, CP47, and PsbI proteins. Designations of proteins are as described in the legend to Figure 1. To allow direct comparison of protein bands, thylakoids from control and photoinhibited cells were analyzed on a single gel and blot.
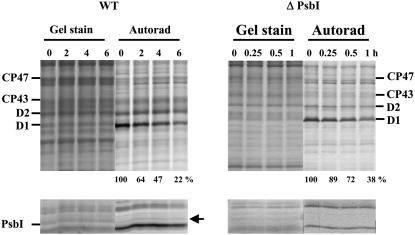
Figure 6.
Degradation of the PSII proteins in the wild-type and ΔPsbI strains under high irradiance monitored by radioactive pulse-chase labeling. Cells of both strains were subjected to 250 μmol photons m−2 s−1 of white light for 20 min in the presence of [35S]Met/Cys. Then the cells were washed, supplemented with unlabeled Met/Cys, and subjected to 500 μmol photons m−2 s−1 of white light for 6 h. Thylakoids were isolated, analyzed by SDS-PAGE, the gel was stained (Gel stain), and the radioactive labeling of the proteins was visualized using a PhosphorImager (Autorad). Quantification of radioactivity in the D1 band was performed by ImageQuant software with samples of each strain equally loaded on Chl basis (2 μg of Chl; see Gel stain) in a single gel. The radioactivity incorporated into the D1 band of each strain during pulse was taken as 100%; numbers show means of three measurements; sd did not exceed 7%. The low-Mr region of the gel is shown in the bottom sections and the stable band of PsbI is designated by an arrow.
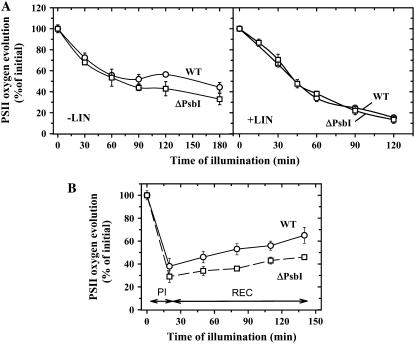
Although the PsbI protein is relatively stable, the turnover of D1 is significantly accelerated in its absence in ΔPsbI. Pulse-chase labeling revealed a half-life of D1 in wild type of about 2 h, while in the mutant it was reduced to approximately 30 min (Fig. 6, top). To find out if this high rate of D1 turnover in the mutant was related to a higher sensitivity of PSII photochemistry to photodamage, we also evaluated the time course of light-induced inhibition of oxygen evolution in wild-type and mutant cultures subjected to white light of 500 μmol photons m−2 s−1 either in the absence or presence of lincomycin. Cultures of wild type and ΔPsbI treated with lincomycin did not exhibit any differences in the decline of oxygen evolution (Fig. 7A, right), while in the absence of lincomycin, the decrease in activity in ΔPsbI was somewhat faster than in wild type (Fig. 7A, left). Recovery from photoinhibition under low-light conditions was also slightly slower in ΔPsbI as evidenced by assessment of oxygen evolution in photoinhibited cells of wild type and ΔPsbI during subsequent incubation at 50 μmol photons m−2 s−1 (Fig. 7B). The results showed that the oxygen-evolving PSII complexes of the mutant are equally sensitive to light-induced inactivation as complexes of wild type, while the regeneration of active PSII is less efficient despite the accelerated turnover of D1.
Figure 7.
PSII repair under high and low irradiance in cells of wild type and ΔPsbI. A, Cells of wild type (circles) and ΔPsbI (squares) were illuminated with 500 μmol photons m−2 s−1 of white light for 180 min in the absence (left) or for 120 min in the presence (right) of 100 μg mL−1 lincomycin and during illumination PSII oxygen-evolving activity was assayed in whole cells. B, Cells of wild type (circles) and ΔPsbI (squares) were illuminated at 2,000 μmol photons m−2 s−1 for 20 min (PI), then the cells were transferred to low irradiance of 50 μmol photons m−2 s−1 and incubated for an additional 120 min (REC). During these light regimes, PSII oxygen-evolving activity was assayed in whole cells. Values in the plots represent means of three measurements ± sd. Initial values for wild type and ΔPsbI were in the range of 372 ± 27 and 258 ± 13 μmol O2 mg Chl−1 h−1, respectively.
DISCUSSION
Role of PsbI in the Formation of the PSII Core and RC Complexes
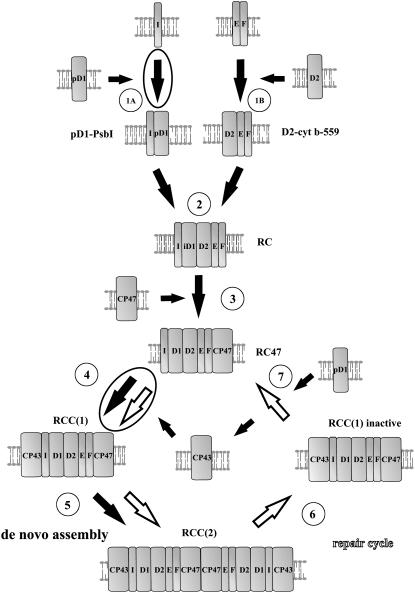
Although the PsbI protein is known to be a component of the isolated PSII RC complex (Ikeuchi et al., 1989; Webber et al., 1989), it has been unclear at what stage PsbI actually binds to PSII during assembly. Here we show that PsbI binds to PSII at an early stage. In particular, we have shown that PsbI is a component of the three RC complexes found in the ΔCP47 strain. All three complexes lack CP47 and CP43 and contain D1, D2, Cyt b-559, and PsbI and so resemble the isolated PSII RC complex in composition. Given that the complexes show different electrophoretic mobilities, it is possible that they differ with respect to the presence of additional assembly factors not found in the final PSII complex. These proteins remain unknown but the modified stoichiometry of these RC complexes induced by the absence of PsbI suggests that PsbI might affect binding of these proteins. Despite binding to PSII early in assembly it is clear, however, that PsbI is not crucial for accumulation of RC complexes.
In contrast, there is a significant stabilization effect of PsbI on the binding of CP43 within the PSII core complex (Fig. 8, step 4), in excellent agreement with the close proximity of PsbI and CP43 in the recent structural studies models of PSII (Ferreira et al., 2004; Loll et al., 2005). The increased levels of RC47 complexes in the PsbI mutant also help to explain the reduced oxygen-evolving activity displayed by the mutant.
Figure 8.
Illustration of function of PsbI during PSII assembly and repair. De novo assembly of PSII (large black arrows) starts with association of PsbI (I) with pD1 (step 1A) forming pD1-PsbI precomplex on one side, and association of Cyt b-559 protein subunits α and β (E and F) with the D2 protein forming Cyt b-559-D2 precomplex on the other side (step 1B). Assembly continues by formation of the RC complex from both precomplexes (step 2). Then CP47 (step 3) and CP43 (step 4) are attached and resulting PSII core monomers RCC(1) form the PSII core dimer [RCC(2), step 5]. PSII repair cycle (white arrows) starts with inactivation of the D1 protein and monomerization (step 6). Then it continues by detachment of CP43 and selective replacement of the D1 protein (step 7). The final two steps, the CP43 attachment and PSII dimerization, seem to be common with the de novo assembly pathway. PsbI is important for processes of formation of pD1-PsbI precomplex (step 1A) and attachment of CP43 to RC47 (step 4) as indicated by encircled arrows. For simplicity, other small and extrinsic PSII subunits were omitted. pD1, Unprocessed forms of D1; iD1, partially processed forms of D1.
Relationship between D1 and PsbI during de Novo Assembly of PSII
Our analysis of the ΔCYT and ΔD1 strains unable to form the PSII RC complex allowed us to look at assembly steps that precede the formation of the RC complex. One of these steps is the formation of a D2-Cyt b-559 precomplex (Fig. 8, step 1B) originally observed in plant etioplasts (Müller and Eichacker, 1999). In this study, we observed the formation of another RC precomplex consisting of D1 and PsbI (Fig. 8, step 1A). Support for the existence of this specific precomplex was based on three lines of evidence: (1) identification of D1-PsbI complexes in thylakoids of wild type, ΔCP47, and ΔCYT by 2D analysis; (2) coimmunoprecipitation of PsbI and D1 from the ΔCYT strain; and (3) copurification of D1 with PsbI-His. The first approach also showed that removal of PsbI led to a decrease in the level of unassembled D1 protein in wild type (Fig. 2, C [arrow] and D), in the CP47-less strain (Fig. 3C), and in the ΔCYT strain (Fig. 4A), suggesting an important role of PsbI in D1 stabilization before its incorporation into PSII. In ΔCYT, which lacks D2 due to effects of deleting the psbEFLJ operon, PsbI could be coimmunoprecipitated not only with mature D1 but also the unprocessed forms, pD1 and iD1, indicating an early interaction of PsbI with D1 even before its maturation. In this respect, it is interesting to note that the antibodies against PsbI and D1 also precipitated the 23-kD N-terminal part of D1 (Fig. 4B, designated by *). This D1-related species seems to be a D1 synthesis intermediate as it was not precipitated by antibody against the C terminus of D1 and disappears during the chase. This finding raises the interesting possibility that binding of PsbI to D1 occurs during D1 synthesis on membrane-bound ribosomes. In agreement with this idea, a significant level of PsbI was found in membrane-bound polysomes (data not shown). We therefore assume that PsbI may stabilize the D1 nascent chain before it is incorporated into the de novo assembled PSII.
2D analysis of wild-type thylakoids showed a fraction of PsbI that is not assembled in any PSII complex. This fraction cannot originate from PSII disassembly or from release of PsbI from complexes during native electrophoresis, as no unassembled D1 and D2 are found in wild type at growth temperature and unassembled PsbI is also found in the strain lacking the D1 protein, the regular binding partner of PsbI (Fig. 1C). In this respect, PsbI is reminiscent of the α-subunit of Cyt b-559. A significant fraction of this protein also remains unassembled in wild-type thylakoids (Komenda et al., 2004). It is possible that small stabilizing partners of D1 and D2 like PsbI, PsbE, or PsbF exist in a surplus nonstoichiometric number of copies in comparison with D1 and D2. In this way, the membrane might be prepared for the prompt initiation of de novo assembly of PSII under variable environmental conditions that require a fast response by the organism.
Relationship between D1 and PsbI during the Repair of PSII
Despite the close structural relationship between D1 and PsbI, their half-lives are very different. The D1 protein is known as a fast turning-over protein, while PsbI is much more stable. Because D1 turnover is controlled by the same FtsH (slr0228) protease as the free pool of PsbI (Supplemental Fig. S1), D1 turnover must be tightly regulated to prevent parallel degradation of PsbI. On the other hand, when PsbI is absent in the psbI deletion mutant, D1 degradation is accelerated in comparison with wild type even though the effectiveness of PSII repair in the mutant is lower. The accelerated D1 turnover seen in the PsbI mutant is most probably related to the destabilization of CP43 binding to PSII in the absence of PsbI. The 2D analysis of the pulse-chase labeled ΔPsbI strain showed that fast D1 replacement occurs in the RC47 complex, as observed for the wild type (Komenda et al., 2006), but that RC47 complexes containing the newly replaced D1 were impaired in the rebinding of CP43 to form active core complexes and instead underwent further cycles of D1 degradation and reinsertion. The resulting frequent but futile replacement of D1 would represent a significant energy demand for the organism that would be another reason for the slower photoautotrophic growth of the mutant in comparison with wild type. On the other hand, the efficient insertion of the D1 protein into RC47 of the ΔPsbI and ΔCP43/ΔPsbI strains indicate that the PsbI protein is not involved in insertion of the newly synthesized D1 protein during selective D1 replacement.
MATERIALS AND METHODS
Construction and Cultivation of Cyanobacterial Strains
The strains used in the study were derived from the Glc-tolerant strain of Synechocystis sp. PCC 6803 (Williams, 1988) referred to as wild type. The following, previously described strains were used in the study: (1) the CP43-less strain ΔCP43 with the psbC gene inactivated by kanamycin resistance cassette (Vermaas et al., 1988); (2) the CP47-less strain ΔCP47 with the psbB gene inactivated by spectinomycin resistance cassette (Eaton-Rye and Vermaas, 1991); (3) the ΔCYT strain with the psbEFLJ operon replaced by kanamycin resistance cassette (Pakrasi et al., 1988); and (4) the D1-less strain ΔD1 with psbA1, psbA2, and psbA3 genes inactivated by CAP, kanamycin, and spectinomycin resistance cassettes (Tichý et al., 2003).
The ΔPsbI strain was prepared by replacement of most of the psbI gene (nucleotides 6–105) by a zeocine resistance cassette using megaprimer PCR method (Burke and Barik, 2003). We have adapted this method to generate linear deletion constructs in vitro containing upstream and downstream regions of the psbI gene with the zeocine resistance cassette in the middle. In the first step, upstream and downstream regions of psbI were separately amplified using long fusion primers recognizing in one direction the psbI gene and in the other direction the zeocine cassette: 5′-ACATTAATTGCGTTGCG CTCACTGCTTAACATAAATTCTCCTTAG-3′ and 5′-CAACTTAATCGCCTTGCAGCACATCGACTTTGAATAAGCTTTAGC-3′(the psbI part is underlined). These fusion primers were used in pairs with psbI forward and reverse primers: 5′-GGTAATTCTCGATTCAGTTG-3′ and 5′-GGTGTGATCAAATACTCCTG-3′. In the second step, the zeocine resistance cassette (Streptoalloteichus hindustanus, Invitrogen) was amplified using PCR products from the first step as primers. Finally, the complete deletion construct was amplified using psbI forward and reverse primers and used for transformation of Synechocystis 6803 cells. Transformants were selected and segregated on zeocine-containing agar plates; their full segregation was confirmed by PCR.
Multiple psbI deletion strains were obtained by transformation of single mutants lacking psbC, psbB, psbEFLJ, and psbA genes using chromosomal DNA from ΔPsbI and their selection for the additional resistance to zeocine.
The PsbI-His strain expressing PsbI tagged with the 6× His tag at its N terminus under control of the psbA2 promoter (PsbI-His/ΔPsbI) was constructed using the pSBA plasmid (Lagarde et al., 2000) in a procedure analogous to that described in Tichý et al. (2003). The resulting strain synthesizing both wild-type and His-tagged forms of PsbI was transformed with chromosomal DNA from the psbI deletion mutant and selected for the resistance to zeocine. The complete deletion of the psbI wild-type copies in the PsbI-His strain was confirmed by PCR. The strain PsbI-His/ΔPsbI/ΔCYT was obtained by transformation of PsbI-His/ΔPsbI using chromosomal DNA from the psbEFLJ deletion strain and selection for the additional resistance to kanamycin.
Liquid cultures were grown in 100 to 200 mL BG11 using 500-mL conical flasks, aerated using an orbital shaker, irradiated with 30 μmol photons m−2 s−1 of white light at 29°C, and were used when they reached a Chl concentration of about 5 μg mL−1. Solid medium contained in addition 10 mm Tes/NaOH, pH 8.2, 1.5% agar, and 0.3% sodium thiosulphate (Pakrasi et al., 1988). Media for cultivation of nonautotrophic strains contained in addition 5 mm Glc.
Photoinhibition experiments were performed with cells cultivated in double-wall, thermoregulated cultivation cylinders (internal diameter 35 mm). Here, the culture was maintained at Chl concentration of 6 to 8 μg mL−1 by regularly diluting with approximately 10 mL of BG11 medium every 150 min. The culture was bubbled with air containing 2% (v/v) CO2 and illuminated with white light at 40 μmol photons m−2 s−1 at 29°C. For the large-scale cultivation used for isolation of PSII complexes, cultures were grown in 10-L flasks (culture volume 6–8 L), stirred by a magnetic stirrer, and bubbled with air.
Measurements of autotrophic growth rates were performed in microtitration plates (culture volume 0.25 mL) under intensive shaking and illumination at 25 μmol photons m−2 s−1. Optical densities at 750 nm were measured every 6 h using microplate reader (Tecan Sunrise). Values plotted against time were used for calculation of the doubling time.
Fluorometric and Polarographic Methods
The maximum photochemical efficiency of PSII in the dark-adapted state parameter and kinetics of Chl variable fluorescence decay were measured in dark-adapted cultures (2.5 μg Chl mL−1) using a modulation PAM101 fluorometer (Walz) with an ED-101US cuvette and the Dual-Modulation Kinetic fluorometer (Photon Systems Instruments; Tichý et al., 2003). The light-saturated steady-state rate of oxygen evolution in cell suspensions was measured polarographically in BG11 medium containing 10 mm HEPES/NaOH, pH 7.0, using 0.5 mm p-benzoquinone and 1 mm potassium ferricyanide as artificial electron acceptors.
Preparation of Membranes and Their Protein Analysis
Cyanobacterial membranes were prepared by breaking the cells using glass beads (Komenda and Barber, 1995) with the following modifications: cells were washed, broken, and finally resuspended in 25 mm MES/NaOH, pH 6.5, containing 10 mm CaCl2, 10 mm MgCl2, and 25% glycerol. The large-scale isolation for chromatographic purification of PSII complexes was performed by a similar procedure, only the cells were resuspended in 20 mL of thylakoid buffer (25 mm MES/NaOH, pH 6.5, 100 mm NaCl) containing the protease inhibitor cocktail (Roche), the same volume of glass beads was added, and the cells were broken eight times for 15 s in the smallest container of beadbeater (Biospec Products) with a 5-min interruption for cooling on ice. Glass beads were subsequently removed by filtering and thylakoids were obtained by differential centrifugation.
For analysis of protein complexes, isolated membranes were solubilized with DM (DM/Chl = 40; w/w) and analyzed by BN electrophoresis at 4°C in 5% to 14% polyacrylamide gel according to Schägger and von Jagow (1991). Samples with the same Chl content (6 μg for gel staining and 1 μg for western blot) were loaded onto the gel.
Protein composition of complexes was assessed by electrophoresis in a denaturing 12% to 20% linear gradient polyacrylamide gel containing 7 m urea (Komenda et al., 2002). The whole lanes from the native gel were excised, incubated for 30 min in 25 mm Tris/HCl, pH 7.5, containing 1% SDS (w/v), and placed on the top of the denaturing gel; two lanes were analyzed in a single denaturing gel. Proteins separated in the gel were either stained by Coomassie Blue or transferred onto polyvinylidene difluoride (PVDF) membrane. Membranes were incubated with specific primary antibodies and then with secondary antibody-horseradish peroxidase conjugate (Sigma). The primary antibodies used in the study were raised in rabbits against: (1) residues 58 to 86 of the spinach (Spinacia oleracea) D1 polypeptide; (2) the last 12 residues of the D2 polypeptide from Synechocystis 6803; (3) residues 380 to 394 of barley (Hordeum vulgare) CP47; (4) the whole isolated CP43 from Synechocystis 6803; and (5) the last 14 residues of the PsbI protein from Synechocystis 6803. For autoradiography, the gel or the membrane with labeled proteins was exposed to x-ray film at laboratory temperature for 2 to 3 d or to a PhosphorImager plate (GE Healthcare) overnight.
Samples used for direct comparison and quantification of stained or labeled proteins were loaded with the same Chl content and were run on a single gel. Bands of α- and β-subunits of ATP synthase were used as an internal standard to which intensity of bands was related. Quantification of bands was done using ImageQuant 5.2 software (GE Healthcare).
Radioactive Labeling of the Cells
For the radioactive labeling, cells containing 75 μg of Chl were resuspended in 250 μL of BG11 in a microcentrifuge tube, shaken at 60 μmol photons m−2 s−1 for 30 min, and then a mixture of [35S]Met and [35S]Cys (Trans-label, MP Biochemicals) was added (final specific activity 400 μCi mL−1). The suspension was exposed to light at irradiance and temperature indicated and afterward the cells were frozen in liquid nitrogen and used for isolation of thylakoids.
Coimmunoprecipitation of PsbI and D1
Coimmunoprecipitation was performed using antibodies against the PsbI and the D1 protein as described previously (Komenda et al., 2005). Briefly, thylakoids isolated from radioactively labeled cells of the ΔCYT and ΔCYT/ΔPsbI strain were solubilized with DM and after overnight incubation with the specific antibodies, the immunoglobulins bound to Protein A-Sepharose 4B (Sigma) were released by the Tris buffer containing 2% SDS and 2% dithiothreitol at 50°C. The eluate was analyzed by one-dimensional SDS-PAGE, transferred to PVDF membrane, and used for autoradiography and immunodetection.
Isolation of PSII Complexes
The D1-PsbI-His complex was isolated from thylakoids of PsbI-His/ΔPsbI/ΔCYT using affinity chromatography on immobilized Ni2+ ions. Thylakoid membranes were solubilized with DM at a final concentration of 2% (w/v) in thylakoid buffer (25 mm MES/NaOH, pH 6.5, 100 mm NaCl) for 30 min in the dark and on ice. The Chl concentration was 1 mg mL−1. The unsolubilized material was removed by centrifugation for 20 min at 60,000g at 4°C and the supernatant was loaded to a chromatography column with Fractogel EMD Chelate (Merck) charged with Ni2+ and equilibrated with thylakoid buffer containing 0.04% DM (w/v; Bumba et al., 2005). The column was subsequently washed with the same buffer until the eluate remained colorless and the PSII was eluted using the isolation buffer with an added 20 mm imidazole and 0.04% DM.
Chl Content
For the measurement of Chl concentration, sedimented cells or membranes were extracted with 100% methanol and Chl content in the extract was calculated from the absorbance at 666 and 720 nm (Wellburn and Lichtenthaler, 1984).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Level of the unassembled PsbI protein is controlled by the protease FtsH (slr0228).
Supplemental Figure S2. PsbI does not affect assembly of the PSII RC47 complex in the CP43-less strain.
Supplemental Figure S3. Effect of PsbI absence on levels of the D1 protein in the wild type, psbB (ΔCP47), and psbEFLJ (ΔCYT) deletion mutants.
Supplemental Table S1. Characteristics of the wild-type strain and the mutant ΔPsbI.
Supplemental Table S2. Distribution of the stained and labeled D1 protein among PSII complexes after pulse and pulse-chase labeling of wild type and the ΔPsbI mutant.
Supplementary Material
Acknowledgments
The authors are grateful to Professors E.-M. Aro, L.A. Eichacker, and R. Barbato for donation of specific antisera; E. Prachová, P. Zelík, and J. Knoppová for help with strain cultivations, measurements of growth curves, and 2D gels; and Professor P.J. Nixon for providing the psbA/slr0228 strain and critical reading of the manuscript.
This work was supported by the Grant Agency of the Czech Republic (project no. 206/06/0322), by the Ministry of Education, Youth and Sports of the Czech Republic (project no. MSM6007665808), and by the Czech Academy of Sciences (Institutional Research Concept no. AV0Z50200510).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Josef Komenda (komenda@alga.cz).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Anbudurai PR, Mor TS, Ohad I, Shestakov SV, Pakrasi HB (1994) The CtpA gene encodes the C-terminal processing protease for the D1 protein of the photosystem-II reaction-center complex. Proc Natl Acad Sci USA 91 8082–8086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber J (2006) Photosystem II: an enzyme of global significance. Biochem Soc Trans 34 619–631 [DOI] [PubMed] [Google Scholar]
- Bumba L, Tichý M, Dobáková M, Komenda J, Vácha F (2005) Localization of the PsbH subunit in photosystem II from the Synechocystis 6803 using the His-tagged Ni-NTA nanogold labeling. J Struct Biol 152 28–35 [DOI] [PubMed] [Google Scholar]
- Burke E, Barik S (2003) Megaprimer PCR: application in mutagenesis and gene fusion. Methods Mol Biol 226 525–532 [DOI] [PubMed] [Google Scholar]
- Eaton-Rye JJ, Vermaas WFJ (1991) Oligonucleotide-directed mutagenesis of psbB, the gene encoding CP47, employing a deletion mutant strain of the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol Biol 17 1165–1177 [DOI] [PubMed] [Google Scholar]
- Ferreira KN, Iverson TN, Maglaoui K, Barber J, Iwata S (2004) Architecture of the photosynthetic oxygen-evolving center. Science 303 1831–1838 [DOI] [PubMed] [Google Scholar]
- Ikeuchi M, Koike H, Inoue Y (1989) Identification of psbI and psbL gene products in cyanobacteria photosystem II reaction center preparation. FEBS Lett 251 155–160 [DOI] [PubMed] [Google Scholar]
- Ikeuchi M, Shukla VK, Pakrasi HB, Inoue Y (1995) Directed inactivation of the psbI gene does not affect photosystem II in the cyanobacterium Synechocystis sp. PCC 6803. Mol Gen Genet 249 622–628 [DOI] [PubMed] [Google Scholar]
- Inagaki N, Yamamoto Y, Satoh K (2001) A sequential two-step proteolytic process in the carboxyl-terminal truncation of precursor D1 protein in Synechocystis sp. PCC 6803. FEBS Lett 509 197–201 [DOI] [PubMed] [Google Scholar]
- Komenda J, Barber J (1995) Comparison of psbO and psbH deletion mutants of Synechocystis PCC 6803 indicates that degradation of D1 protein is regulated by the QB site and is dependent on protein synthesis. Biochemistry 34 9625–9631 [DOI] [PubMed] [Google Scholar]
- Komenda J, Barker M, Kuviková S, DeVries R, Mullineaux CW, Tichý M, Nixon PJ (2006) The FtsH protease slr0228 is important for quality control of photosystem II in the thylakoid membrane of Synechocystis PCC 6803. J Biol Chem 281 1145–1151 [DOI] [PubMed] [Google Scholar]
- Komenda J, Kuviková S, Granvogl B, Eichacker LA, Diner BA, Nixon PJ (2007) Cleavage after residue Ala352 in the C-terminal extension is an early step in the maturation of the D1 subunit of photosystem II in Synechocystis PCC 6803. Biochim Biophys Acta 1767 829–837 [DOI] [PubMed] [Google Scholar]
- Komenda J, Lupínková L, Kopecký J (2002) Absence of the psbH gene product destabilizes the photosystem II complex and bicarbonate binding on its acceptor side in Synechocystis PCC 6803. Eur J Biochem 269 610–619 [DOI] [PubMed] [Google Scholar]
- Komenda J, Reisinger V, Müller BCh, Dobáková M, Granvogl B, Eichacker LA (2004) Accumulation of the D2 protein is a key regulatory step for assembly of the photosystem II reaction center complex in Synechocystis PCC 6803. J Biol Chem 279 48620–48629 [DOI] [PubMed] [Google Scholar]
- Komenda J, Tichý M, Eichacker L (2005) The PsbH protein is associated with the inner antenna CP47 and facilitates D1 processing and incorporation into photosystem II in the cyanobacterium Synechocystis PCC 6803. Plant Cell Physiol 46 1477–1483 [DOI] [PubMed] [Google Scholar]
- Künstner P, Guardiola A, Takahashi Y, Rochaix JD (1995) A mutant strain of Chlamydomonas reinhardtii lacking chloroplast photosystem II psbI gene grows photoautotrophically. J Biol Chem 270 9651–9654 [DOI] [PubMed] [Google Scholar]
- Lagarde D, Beuf L, Vermaas W (2000) Increased production of zeaxanthin and other pigments by application of genetic engineering techniques to Synechocystis sp. strain PCC 6803. Appl Environ Microbiol 66 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loll B, Kern J, Saenger W, Zouni A, Biesiadka J (2005) Towards complete cofactor arrangement in the 3.0 Å resolution structure of photosystem II. Nature 438 1040–1044 [DOI] [PubMed] [Google Scholar]
- Müller B, Eichacker LA (1999) Assembly of the D1 precursor in monomeric photosystem II reaction center precomplexes precedes chlorophyll a-triggered accumulation of reaction center II in barley etioplasts. Plant Cell 11 2365–2377 [PMC free article] [PubMed] [Google Scholar]
- Nanba O, Satoh K (1987) Isolation of a photosystem II reaction center consisting of D-1 and D-2 polypeptides and cytochrome b-559. Proc Natl Acad Sci USA 84 109–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon PJ, Trost JT, Diner BA (1992) Role of the carboxy terminus of polypeptide-D1 in the assembly of a functional water-oxidizing manganese cluster in photosystem-II of the cyanobacterium Synechocystis sp. PCC 6803: assembly requires a free carboxyl group at C-terminal position 344. Biochemistry 31 10859–10871 [DOI] [PubMed] [Google Scholar]
- Pakrasi HB, Williams JGK, Arntzen CJ (1988) Targeted mutagenesis of the psbE and psbF blocks photosynthetic electron transport: evidence for a functional role of cytochrome b-559 in photosystem II. EMBO J 7 325–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, von Jagow G (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem 199 223–231 [DOI] [PubMed] [Google Scholar]
- Schwenkert S, Umate P, Bosco CD, Volz S, Mlčochová L, Zoryan M, Eichacker LA, Ohad I, Herrman RG, Meurer J (2006) PsbI affects the stability, function and phosphorylation patterns of photosystem II assemblies in tobacco. J Biol Chem 281 34227–34238 [DOI] [PubMed] [Google Scholar]
- Takahashi M, Shiraishi T, Asada K (1988) COOH-terminal residues of D1 and the 44 kDa CPa-2 at spinach photosystem II core complex. FEBS Lett 240 6–8 [DOI] [PubMed] [Google Scholar]
- Tichý M, Lupínková L, Sicora C, Vass I, Kuviková S, Prášil O, Komenda J (2003) Synechocystis 6803 mutants expressing distinct forms of the photosystem II D1 protein from Synechococcus 7942: relationship between the psbA coding region and sensitivity to visible and UV-B radiation. Biochim Biophys Acta 1605 55–66 [DOI] [PubMed] [Google Scholar]
- Vermaas WFJ, Ikeuchi M, Inoue Y (1988) Protein composition of the photosystem II core complex in genetically engineered mutants of the cyanobacterium Synechocystis sp. PCC 6803. Photosynth Res 17 97–113 [DOI] [PubMed] [Google Scholar]
- Webber AN, Packham L, Chapman DJ, Barber J, Gray JC (1989) A fifth chloroplast-encoded polypeptide is present in the photosystem II reaction centre complex. FEBS Lett 242 259–262 [Google Scholar]
- Wellburn AR, Lichtenthaler K (1984) Formulae and programme to determine total carotenoids and chlorophyll a and b of leaf extracts in different solvents. In C Sybesma, ed, Advances in Photosynthesis Research. Martinus Nijjhoff, Dordrecht, The Netherlands, pp 10–12
- Williams JGK (1988) Construction of specific mutations in PSII photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol 167 766–778 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.