Abstract
Higher plants acquire iron (Fe) from the rhizosphere through two strategies. Strategy II, employed by graminaceous plants, involves secretion of phytosiderophores (e.g. deoxymugineic acid in rice [Oryza sativa]) by roots to solubilize Fe(III) in soil. In addition to taking up Fe in the form of Fe(III)-phytosiderophore, rice also possesses the strategy I-like system that may absorb Fe(II) directly. Through mutant screening, we isolated a rice mutant that could not grow with Fe(III)-citrate as the sole Fe source, but was able to grow when Fe(II)-EDTA was supplied. Surprisingly, the mutant accumulated more Fe and other divalent metals in roots and shoots than the wild type when both were supplied with EDTA-Fe(II) or grown under water-logged field conditions. Furthermore, the mutant had a significantly higher concentration of Fe in both unpolished and polished grains than the wild type. Using the map-based cloning method, we identified a point mutation in a gene encoding nicotianamine aminotransferase (NAAT1), which was responsible for the mutant phenotype. Because of the loss of function of NAAT1, the mutant failed to produce deoxymugineic acid and could not absorb Fe(III) efficiently. In contrast, nicotianamine, the substrate for NAAT1, accumulated markedly in roots and shoots of the mutant. Microarray analysis showed that the expression of a number of the genes involved in Fe(II) acquisition was greatly stimulated in the naat1 mutant. Our results demonstrate that disruption of deoxymugineic acid biosynthesis can stimulate Fe(II) acquisition and increase iron accumulation in rice.
Iron (Fe) is an important mineral for both plant production and human nutrition. Fe deficiency is one of the most prevalent nutrient deficiencies in the world, affecting an estimated 2 billion people, especially in the area where vegetable-based diets are the primary food source (World Health Organization, 2002). Rice (Oryza sativa), the dominant cereal crop in many developing countries, particularly contains low Fe in its polished grains (Barry, 2006). To alleviate the problem of severe Fe deficiency worldwide, an international Fe biofortification program has been organized by the HarvestPlus global initiative (http://www.harvestplus.org) to develop Fe-rich rice varieties using both traditional breeding and biotechnological approaches. Most efforts have been directed toward screening for high Fe content materials from various sources of germplasm. To use biotechnological methods for Fe biofortification requires that the molecular mechanisms controlling Fe acquisition in rice be understood.
In aerobic soils, Fe is present mainly as Fe(III) oxides, which have very low solubility and are not readily available to plants (Guerinot and Yi, 1994). To solubilize and absorb Fe efficiently, higher plants have evolved two distinct strategies to acquire Fe from the rhizosphere (Marschner et al., 1986). Strategy I is employed by dicotyledonous and nongraminaceous monocotyledonous species, whereas strategy II is used only by graminaceous monocotyledonous species. Strategy I involves induction of membrane-bound Fe(III)-chelate reductases, which reduce Fe(III) to the more soluble form Fe(II), followed by uptake of Fe(II) via Fe(II) transporters. Genes encoding the Fe(III)-chelate reductases, FRO1, FRO2, and FRO3, and the Fe(II) transporters, IRT1 and IRT2, have been isolated from Arabidopsis (Arabidopsis thaliana; Eide et al., 1996; Robinson et al., 1997, 1999; Vert et al., 2001). Expression of these genes is greatly induced in the roots of Fe-deficient plants.
Fe acquisition by strategy II plants is characterized by secretion of Fe(III)-chelating substances (phytosiderophores) and uptake of intact Fe(III)-phytosiderophore complexes (Takagi et al., 1984; Römheld and Marschner, 1986). Graminaceous plants can synthesize and secrete mugineic acid (MA) family phytosiderophores from their roots to chelate Fe(III) in the soil. The biosynthetic pathway of MA has been studied intensively (Mori and Nishizawa, 1987; Shojima et al., 1989, 1990; Ma and Nomoto, 1993). MA is synthesized from l-Met through nicotianamine (NA). NA aminotransferase (NAAT) is a critical enzyme that transfers the amino group from NA to form the 3″-oxo intermediate of deoxymugineic acid (DMA). Overexpression of the barley (Hordeum vulgare) NAAT gene in rice increased the secretion of MA and consequently enhanced tolerance to low Fe availability (Takahashi et al., 2001). The synthesized MA chelated Fe(III) to form Fe(III)-MA complexes, which can be taken up into the root through Fe(III)-MA transporters (Takagi et al., 1984). ZmYS1 (Curie et al., 2001; Schaaf et al., 2004) and HvYS1 (Murata et al., 2006), which encode high-affinity Fe(III)-MA transporters in maize (Zea mays) and barley, have been cloned and characterized. In rice, 18 homologs of ZmYS1 have been identified and are named OsYSL (rice YS1-like genes; Koike et al., 2004). The closest rice homolog of ZmYS1 is OsYSL15, which is likely to be a Fe(III)-MA transporter.
NA is not only a key intermediate for the biosynthesis of MA for strategy II plants, but also an important metal chelator that can facilitate transport of Fe and other transition metals inside plants (von Wiren et al., 1999). Investigations of the NA-free tomato (Solanum lycopersicum) mutant chloronerva showed that NA is involved in sensing the Fe status within the plant and the translocation of Fe and other metals from the roots to the aerial parts (Pich et al., 1994; Higuchi et al., 1996). NA level in plants is controlled by NA synthase (NAS) and NAAT (Mori, 1999). Genes encoding NAS and NAAT enzymes have been isolated in barley and other plant species (Higuchi et al., 1999; Ling et al., 1999; Takahashi et al., 2001). Transgenic tobacco (Nicotiana tabacum) plants overexpressing HvNAAT showed Fe-deficient symptoms in young leaves and abnormal flower shapes (Takahashi et al., 2003). The phenotype was caused by a shortage of NA as a result of overconsumption of endogenous NA in NAAT-overexpressed transgenic plants. Ectopic expression of the Arabidopsis NAS gene in tobacco resulted in a 10-fold increase in NA level and a significant increase of iron, zinc (Zn), and manganese (Mn) concentrations in leaves of adult plants (Douchkov et al., 2005). NA participates in phloem transport and cytoplasmic distribution of metals through transporters encoded by the YSL genes in plants (Koike et al., 2004). OsYSL2 was demonstrated to be a metal-NA transporter expressed in the phloem (Koike et al., 2004).
Recently, it has been shown that rice possesses not only a strategy II but also a strategy I-like system that may take up Fe(II) via Fe(II) transporters OsIRT1 and OsIRT2 (Ishimaru et al., 2006, 2007). Unlike the typical strategy I plants, the expression and enzymatic activity of rice FROs were not detectable in Fe-deficient roots (Ishimaru et al., 2006), indicating that rice takes up Fe(II) without going through the step of Fe(III) reduction. A reconstructed yeast (Saccharomyces cerevisiae) Fe-chelate reductase gene driven by the Fe-deficient induced promoter led to enhanced tolerance to low Fe availability in calcareous soils in transgenic rice (Ishimaru et al., 2007), indicating that the Fe(II) uptake system is functional in rice.
In this study, we isolated and characterized a rice mutant whose strategy II system was interrupted due to the loss of a functional NAAT enzyme. Surprisingly, the mutant exhibited significant elevation of Fe and other metal concentrations in both seedlings and seeds when plants were supplied with Fe(II). Physiological and molecular analysis showed that diminishing MA synthesis in the naat1 mutant stimulated Fe(II) acquisition system and led to Fe accumulation. Our results demonstrate the importance of the Fe(II) uptake strategy in rice Fe acquisition.
RESULTS
Isolation and Gene Cloning of the naat1 Mutant
Seeds from the M2 generation of an ethyl methane sulfonate-mutagenized population of rice ‘Nipponbare’ were germinated and grown in a nutrient solution [containing Fe(III); Yoshida et al., 1976] to screen mutants with abnormal root development. A rice mutant with much shortened adventitious roots and lateral roots at 10 d after germination was obtained (Fig. 1A). Further experiments showed that the mutant exhibited developmental retardation when grown in a growth chamber under aerobic soil conditions, but not under water-logged conditions (Fig. 1, B and C). Besides, under the same nutrient solution, the mutant exhibited severe chlorotic and growth inhibition symptoms (Fig. 1, E and F). The chlorophyll content (SPAD value) in mutant leaves was 8.6 times lower than that of wild type when plants were supplied with Fe(III) (Fig. 1G).
Figure 1.
Phenotypes of wild-type, naat1 mutant, and complemented transgenic plants. A, Ten-day-old seedlings grown with nutrient solution supplemented with citrate-Fe(III) or EDTA-Fe(II). From left to right, seedlings of wild type and naat1 under citrate-Fe(III), and wild type and naat1 under EDTA-Fe(II). B, Growth of naat1 and wild-type mutant seedlings under aerobic conditions and water logged. C, Mature wild-type (right) and naat1 (left) plants grown under water-logged conditions. D, Seven-day-old seedlings grown with nutrient solution supplemented with citrate-Fe(III). From left to right, wild-type, naat1, and naat1 transgenic lines 1 and 2 expressing NAAT1 cDNA. RT-PCR results of NAAT1 cDNA in wild-type, naat1, and the naat1 transgenic lines 1 and 2 seedlings are presented at the bottom of the corresponding seedlings. E and F, Shoot (E) and root (F) growth of wild-type and naat1 seedlings in solution culture supplemented with citrate-Fe(III) for 10 d. G, Chlorophyll content in the leaves of 10-d-old wild-type and naat1 seedlings grown in solution culture supplemented with citrate-Fe(III) or EDTA-Fe(II). Chlorophyll content of the fully expanded youngest leaf was measured using a SPAD-502 chlorophyll meter. A and D: bar = 2 cm.
With the same nutrient solution, the mutant was unable to survive longer than 20 d after germination. To rescue the mutant, we changed various components in the nutrient solution. When citrate-Fe(III) was replaced by EDTA-Fe(II), growth of roots and shoots of mutant plants was normal and chlorophyll content (SPAD value) of the mutant seedlings was at the same level as that of wild-type seedlings (Fig. 1, A and G; Supplemental Table S1). Similarly, normal growth was observed when FeSO4 was used in the nutrient solution, whereas FeCl3 could not restore normal growth (data not shown). These results suggested that the mutant could utilize Fe(II), but not Fe(III).
Genetic analysis showed that a single recessive gene was responsible for the mutant phenotype. Using 2,900 F2 mutant seedlings selected from a F2 population derived from a cross between the mutant and the Indica cultivar ‘Kasalath’, the mutation was mapped to a 123-kb region between RM13046 and RM13051 (Fig. 2A). This region contains 18 open reading frames (ORFs), including the NICOTIANAMINE AMINOTRANSFERASE1 (NAAT1) gene (LOC_Os02g20360.1; Inoue et al., 2004). NAAT1 is an enzyme required in the biosynthesis of MA, which is secreted by graminaceous plants as chelators to form complexes with Fe(III) in soil and to facilitate absorption of Fe(III) (Kanazawa et al., 1994). The mutant phenotype suggested the NAAT1 gene as a tentative candidate gene for the mutation. To test this hypothesis, both PCR and reverse transcription (RT)-PCR analyses of the NAAT1 gene were conducted using wild-type and mutant genomic DNA and cDNA as templates. Sequence comparison of the PCR products revealed the presence of a mutant-specific point mutation (G to T) occurring at nucleotide position 3,136 bp of the coding sequence within the fourth exon of the gene (Fig. 2A). This mutation results in deletion of the fourth exon from the NAAT1 mRNA, shortening the amino acid sequence in the NAAT1 protein (Fig. 2, B and C). Expression of mutant NAAT1 cDNA in Escherichia coli confirmed that the mutant version of naat1 cDNA directs synthesis of a protein 2 kD smaller than that of the wild type (Fig. 2C).
Figure 2.
Molecular identification of naat1. A, naat1 was mapped between the markers RM13046 and RM13051. The region is covered by two plasmid clones P572A4 and P543C11 and contains 18 ORFs. naat1 has a point mutation at position 3136. Black bars show NAAT1 exons. B, Protein sequence of rice NAAT1. The bold and underlined region is missing in the naat1 mutant. C, RT-PCR of NAAT1 using wild-type or naat1 mutant cDNA as templates (left) and SDS-PAGE analysis of expression of NAAT1 proteins of wild type and naat1 in E. coli BL21 (right). Cell pellet was disrupted by sonication and separated into soluble and insoluble fractions. MW, Protein Mr marker. Arrows indicate the position of NAAT1 proteins. D, Phylogenetic tree (neighbor-joining tree) analysis of six rice OsNAAT, barley HvNAAT-A, and HvNAAT-B. TIGR loci of the OsNAAT1, OsNAAT-L1, OsNAAT-L2, OsNAAT-L3, OsNAAT-L4, and OsNAAT-L5 are Os2g20360, Os2g19924, Os6g23684, Os11g35040, Os11g42510, and Os2g19970, respectively.
Confirmation that the point mutation in naat1 was responsible for the mutant phenotype was achieved by genetic complementation. The wild-type full-length ORF of NAAT1 was inserted into the binary vector pTF101.1 under the control of a maize ubiquitin-1 (Ubi-1) promoter and a nopaline synthase terminator (Frame et al., 2002). The resulting construct was used to introduce the complete NAAT1 gene into the genome of the naat1 mutant via Agrobacterium-mediated transformation (Chen et al., 2003). Transgenic mutant lines expressing both the shortened and the full-length NAAT1 mRNA displayed normal growth phenotype when grown in the presence of citrate-Fe(III) (Fig. 1D).
Significant Accumulation of NA in naat1 Plants
To investigate whether disruption of the NAAT1 enzyme affects accumulation of the NAAT1 substrate NA, we measured NA content in shoots and roots of 10-d-old naat1 and wild-type seedlings with a supply of 35 μm citrate-Fe(III), 125 μm EDTA-Fe(II), or no Fe. Results showed that both shoots and roots of wild-type plants contained a low concentration of NA, ranging from 0.038 to 0.088 μg g−1 fresh weight in all treatments (Fig. 3B). In contrast, NA concentrations in naat1 shoots and roots were greatly elevated compared with those of the wild type. NA concentrations of the mutant were 3- to 43-fold higher than those of the wild type (Fig. 3B).
Figure 3.
The amount of DMA secreted by roots and the concentrations of NA in wild-type and naat1 seedlings. A, Growth of seedlings of wild type (left two pots) and naat1 (right two pots) with EDTA-Fe(II) supply (pots 1 and 3 from the left) or without Fe (pots 2 and 4 from the left). naat1 and wild-type seeds were germinated and grown in tap water for 10 d, then transferred to 1.2-L pots containing one-half-strength Kimura B solution and 125 μm EDTA-Fe(II) (pH 5.5) for an additional 15 d. Thereafter, naat1 and wild-type plants were divided into two groups: one with and the other without Fe. Photos were taken on day 13 after the treatment. B, NA content in shoots and roots of 10-d-old wild-type and naat1 seedlings grown in nutrient solution supplied with either 35 μm citrate-Fe(III) or125 μm EDTA(II) or no Fe. C, Amount of DMA secreted from the roots of wild-type and naat1 mutant seedlings shown in A. D and E, Concentrations of Fe and Zn in shoots and roots of wild-type and naat1 seedlings shown in A. F, Cd concentration in the seedlings grown in medium supplied with or without Fe(II); 1 μm CdCl2 was included in the medium for this experiment. [See online article for color version of this figure.]
Biosynthesis of DMA Is Blocked in naat1 Plants
NAAT is a critical enzyme in the biosynthesis of MA. It catalyzes the transfer of the amino group from NA to form the precursor of DMA, the 3″-oxo form of DMA. To determine the secretion of DMA, seedlings of the wild type and naat1 were precultured with one-half-strength Kimura nutrient solution containing 125 μm Fe(II)-EDTA (pH 5.5; Ma et al., 2001). Plants were then divided into two groups, treated either with or without Fe(II)-EDTA for 13 d. An additional treatment of wild-type plants supplied with 125 μm Fe(III)-EDTA (pH 5.5) was also included to investigate DMA secretion in the wild type.
Results showed that, without Fe(II) supply, both wild-type and naat1 mutant plants developed typical Fe-deficient chlorotic symptoms in the treatment, but were normal in the Fe(II) supply condition (Fig. 3A). Root exudates were collected and measured on day 13 after treatment. Wild-type roots secreted considerable amounts of DMA: 151 and 223 nmol g−1 dry weight in Fe(II) or no-Fe supply treatments, respectively (Fig. 3C). In contrast, naat1 roots secreted negligible amounts of DMA in both Fe(II) or no-Fe treatments, indicating that the loss of function of NAAT1 blocked the production and secretion of DMA. For comparison, wild-type roots supplied with 125 μm Fe(III)-EDTA secreted little DMA (data not shown).
Up-Regulation of Genes Involved in Fe(II) Uptake in the naat1 Mutant
To evaluate the effect of disruption of the NAAT1 enzyme on the rice Fe absorption system, we performed Affymetrix GeneChip analysis on the leaves and roots of the 10-d-old wild-type and naat1 seedlings grown with either 35 μm citrate-Fe(III), 125 μm EDTA-Fe(II), or no Fe. Gene expression patterns from the microarray analysis were verified by RT-PCR for 12 genes encoding Fe uptake or transport proteins. The results from RT-PCR analysis were consistent with those from the microarray analysis (Table I; Fig. 4).
Table I.
Up-regulation of genes possibly involved in Fe-Zn homeostasis in the roots and leaves of the naat1 mutant of rice
Root and leaf RNA were extracted from the10-d-old seedlings. *, naat1 compared to the wild type.
| Genes | TIGR Locus | Induction Ratio*
|
Putative Function | Reference | |||||
|---|---|---|---|---|---|---|---|---|---|
| Leaf
|
Root
|
||||||||
| Fe3+ | Fe2+ | No Fe | Fe3+ | Fe2+ | No Fe | ||||
| Fe-regulated transcription factors | |||||||||
| OsIRO2 | Os01g72370.2 | 67.2 | 1.2 | 0.9 | 14.5 | 9.9 | 1.0 | Regulating expression of Fe-response protein | Ogo et al. (2006) |
| OsIRbHLH1 | Os12g32400.1 | 11.3 | 0.4 | 1.9 | 11.3 | 15.0 | 0.7 | Not reported before | |
| OsIRbHLH2 | Os03g26210.2 | 21.5 | 1.0 | 1.7 | 10.6 | 8.6 | 1.2 | Not reported before | |
| OsIRNLPl | Os12g18410.2 | 220.1 | 1.1 | 1.7 | 30.4 | 24.3 | 1.3 | Ogo et al. (2006) | |
| OsNACl | Os02g36880.3 | 9.8 | 3.0 | 2.9 | 0.9 | 0.7 | 1.1 | Guo et al. (2003) | |
| Metal transporters | |||||||||
| OsNRAMPl | Os07g15460.1 | 91.8 | 4.1 | 1.4 | 12.3 | 10.1 | 1.9 | Ferrous transporter | Belouchi et al. (1997) |
| OsIRT1 | Os03g46470.1 | 6.3 | 0.5 | 4.3 | 29.8 | 2.5 | 0.5 | Zn-Fe transport family protein | Bughio et al. (2002) |
| OsIRT2 | Os03g46454.1 | 12.2 | 3.8 | 3.8 | 63.5 | 3.8 | 1.7 | Ishimaru et al. (2006) | |
| OsZIPL1 | Os05g39550.1 | 45.6 | 1.9 | 42.4 | 4.9 | 2.8 | 1.7 | Putative Zn/Fe transporter | Not reported before |
| OsYSLl | Os04g45860.1 | 2.0 | 1.4 | 3.4 | 4.0 | 1.9 | 1.0 | Fe-phytosiderophore transporter | Koike et al. (2004) |
| OsYSL15 | Os02g43410.2 | 29.4 | 1.1 | 4.0 | 6.8 | 7.2 | 0.7 | Koike et al. (2004) | |
| OsYSL2 | Os02g43370.1 | 6.8 | 10.0 | 9.1 | 46.2 | 0.9 | 7.8 | Metal-NA transporter | Koike et al. (2004) |
| NA biosynthesis and metabolism | |||||||||
| OsNASl | Os03g19436.1 | 2645.5 | 0.1 | 1.5 | 1.8 | 3.9 | 0.7 | MA synthesis and Fe long-distance transportation | Higuchi et al. (2001) |
| OsNAS2 | Os03g19420.2 | 222.0 | 0.4 | 2.0 | 2.0 | 3.5 | 0.7 | Higuchi et al. (2001) | |
| OsNAS3 | Os07g48980.1 | 1.0 | 0.7 | 5.1 | 7.4 | 4.1 | 1.5 | Higuchi et al. (2001) | |
| OsNAATl | Os02g20360.1 | 23.9 | 0.5 | 1.9 | 3.9 | 6.8 | 0.8 | Inoue et al. (2004) | |
Figure 4.
Expression analysis using RT-PCR. RNA was isolated from roots and shoots of 15-d-old seedlings of wild-type and naat1 mutant plants grown in solution culture with Fe(III), Fe(II) supply, or no-Fe conditions. OsActin primers were used as a template control. Primers and amplification numbers of the selected genes and OsActin are listed in Supplemental Table S3.
Table I shows that a number of genes involved in various steps of Fe(II) uptake and transport were up-regulated in the naat1 mutant seedlings under no-Fe, Fe(III), or Fe(II) supply conditions. When Fe(III) was supplied as the sole Fe source, all Fe deficiency-inducible genes examined were up-regulated in roots and/or leaves of the naat1 mutant plants compared with the wild-type counterparts (Table I), which were consistent with an Fe deficiency response (Inoue et al., 2003; Kobayashi et al., 2005). These results confirmed that the naat1 mutant was not able to utilize Fe(III).
Because the naat1 mutant can only absorb Fe in the form of Fe(II), the difference in the gene expression pattern between wild-type and naat1 plants under the Fe(II) supply conditions should reflect their difference in the Fe(II) uptake capacity. Microarray data showed that a group of genes involved in Fe(II) uptake and transport were greatly activated in naat1 seedlings under Fe(II) supply conditions. OsNRAMP1 belongs to the same family as AtNRAMP1 and GmNRAMP1, which function as ferrous transporters in Arabidopsis and soybean (Glycine max; Curie et al., 2000; Kaiser et al., 2003). In the naat1 mutant, expression of OsNRAMP1 (Belouchi et al., 1997) was markedly increased in both leaves and roots under Fe(II) supply conditions (Table I). Similarly, a number of other Fe-Zn cotransporters (Ishimaru et al., 2005), OsIRT1, OsIRT2, and a novel ZIP-like gene, named OsZIPL1, were also induced in the naat1 roots (Table I). These results suggest the existence of a mechanism for enhancement of Fe(II) uptake in the mutant. Under Fe(II) supply conditions, three genes involved in Fe(III)-phytosiderophore or metal-NA chelate transport were up-regulated in the naat1 mutant: OsYSL1 in the roots and OsYSL15 and OsYSL2 in both roots and leaves (Table I). The strong up-regulation of transporters related to Fe and other transition metals in roots and leaves suggests enhanced capacity for uptake and transport of Fe and other metals in naat1.
Significant induction of OsIRO2, which binds preferentially to sequences upstream of Fe deficiency-inducible genes, such as OsNAS1, OsNAS3, and OsIRT1 (Ogo et al., 2006), was observed in mutant roots. This induction may account for, at least partly, the observed up-regulation of the Fe and metal-NA transporters in the mutant. Three other putative transcription factors, including two other basic helix-loop-helix proteins and an unknown nuclear-localizing protein, were significantly induced in both leaves and roots under Fe(III) supply conditions and in roots under Fe(II) supply. Because of their strong response to low Fe status, we name them OsIRbHLH1, OsIRbHLH2, and OsIRUNLP1, respectively (Table I). A NAC domain containing protein OsNAC1 (Guo et al., 2003) was also up-regulated in leaves of the naat1 mutant. It is possible that these transcription factors play a role in the regulation of the expression of the Fe-Zn transporter genes.
Under Fe(II) supply conditions, expression of OsNAS1, OsNAS2, and OsNAS3 in the naat1 roots was 3.9, 3.5, and 4.1 times higher than those in wild-type plants. Induction of NAS genes may have contributed to the significant accumulation of NA in naat1 shoots and roots, in addition to the markedly reduced consumption of NA for MA biosynthesis in the mutant (Fig. 3B).
The naat1 Mutant Takes Up More Divalent Metals Than Wild-Type Plants
We measured the uptake and translocation of Fe and other metal elements in wild-type and naat1 plants harvested from the DMA secretion experiment. In the treatment with Fe(II)-EDTA, Fe concentrations in the naat1 shoots and roots were 15.8% and 41.8% higher than those in wild-type seedlings (Fig. 3D), indicating that the naat1 mutant had enhanced ferrous uptake system. These results were further supported by the finding that the naat1 mutant contained significantly higher (58%–92%) concentrations of Zn in shoots and roots than the wild type (Fig. 3E). Furthermore, when 1 μm cadmium (Cd) was added to the nutrient solution, Cd concentrations in both naat1 roots or shoots were about 50% higher than that those in wild-type seedlings (Fig. 3F).
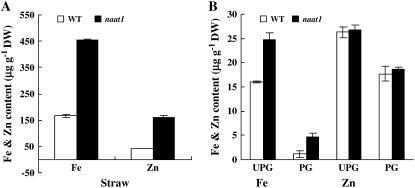
The naat1 mutant grew normally in paddy soil and accumulated more Fe in seeds than wild-type plants in a field experiment.
Despite the fact that the naat1 mutant was unable to absorb Fe(III) through strategy II, plants grew normally in water-logged paddy soils where ferrous iron was abundant (Fig. 1C). Agronomic traits, including the duration of maturation, plant height, tiller number, seed set rate, seed number per panicle, and seed weight of naat1 and wild-type plants, were evaluated in a field experiment. Growing conditions used in the field experiment were the same as those for normal paddy rice, for which continuous flooding water was provided until the late grain-filling stage. No significant (P > 0.05) differences between mutant and wild-type plants were observed in all agronomic traits evaluated (Table II). Elemental analysis showed that the naat1 straw contained 2.7, 3.9, and 1.8 times more Fe, Zn, and Cd, respectively, than wild-type straw (Fig. 5A). Fe concentrations in the naat1 unpolished and polished grains were 24.7 and 4.6 mg kg−1 dry weight, respectively, which were 1.8 and 3.8 times higher than their wild-type counterparts. Although naat1 plants contained more Zn and Cd in the seedlings and straws (Fig. 3, D–F), Zn concentrations in the unpolished and polished grains remained the same as in the wild-type grain, and the Cd concentrations were lower than those of the wild-type grain (Fig. 5B). Accumulation of other divalent metals, including copper (Cu) and Mn, showed a similar pattern as for Zn and Cd between naat1 and wild-type plants (Supplemental Table S2).
Table II.
Agronomic traits (means ± se) of naat1 mutant and wild-type plants
| Genotypes | Duration of Maturation | Plant Height | Tiller Nos. | Seed No. per Panicle | Seed-Setting Rates | Seed Weight |
|---|---|---|---|---|---|---|
| d | cm | % | g/1,000 seeds | |||
| Wild type | 116 | 80.4 ± 1.0 | 19.6 ± 0.7 | 1,091.6 ± 128.6 | 88.5 ± 1.3 | 23.1 ± 0.1 |
| naat1 | 116 | 80.3 ± 0.9 | 20.3 ± 0.7 | 999.1 ± 92.6 | 87.9 ± 1.4 | 23.1 ± 0.1 |
Figure 5.
Concentrations of Fe and Zn in straw, polished grains, and unpolished grains of wild-type and naat1 plants. A, Fe, Zn, and Cd concentrations in straw. B, Fe, Zn, and Cd concentrations in grains. UPG, Unpolished grains; PG, polished grains. Samples were harvested from the field experiment.
DISCUSSION
MA is a natural Fe chelator secreted from roots of graminaceous plants that solubilizes Fe in the soil. Biosynthesis and secretion of MA are therefore crucial to the acquisition of Fe(III) in strategy II plants. NAAT is a critical enzyme in the biosynthesis of MA from l-Met. BLAST searches identified five other putative NAAT genes in rice, named OsNAAT-L1 to OsNAAT-L5. Phylogenetic analysis indicated that NAAT1 is closely clustered with barley NAAT-A and NAAT-B (Fig. 2D; Takahashi et al., 1999). Our microarray and RT-PCR analysis showed that only OsNAAT1, among all NAAT-like genes analyzed, was induced by Fe deficiency (data not shown). We also observed that naat1 mutant plants lost the ability to synthesize and secrete DMA regardless of the status of Fe supply (Fig. 3A). Taken together, we conclude that, among the six rice NAAT-like genes, only OsNAAT1 is involved in the production of DMA. Due to the loss of function of OsNAAT1, the naat1 mutant could not survive in aerobic soils or in hydroponic cultures with Fe(III) as the sole Fe source.
However, Fe(II) is abundant in flooded paddy soils. Rice can utilize Fe(II) directly through the strategy I-like system, even though it has no functional ferric reductase to reduce Fe(III) to Fe(II) (Ishimaru et al., 2006). Ishimaru et al. (2007) expressed a reconstructed yeast ferric reductase gene under the control of the Fe-deficient induced promoter (OsIRT1 promoter) in rice. Transgenic rice conferred enhanced tolerance to Fe deficiency. In this study, we showed that the MA-free mutant, naat1, had higher expressions of Fe(II) transporters IRT1, IRT2, and other putative Fe transporter ZIP-LIKE genes than the wild type. These transporters can mediate Fe(II) uptake when Fe(II) is available. In addition, overaccumulation of NA in the naat1 mutant may also contribute to the increased Fe content in the mutant. A number of studies have demonstrated that NA is an important metal chelator that facilitates the internal transport of metal ions in both strategy I and II plants (Pich et al., 1994; Higuchi et al., 1996). NA participates in phloem transport and cytoplasmic distribution of metals through transporters encoded by YSL genes (Koike et al., 2004). In the naat1 mutant, overaccumulation of NA is likely to be a result of both the loss of function of NAAT and an up-regulation of NAS. Furthermore, the mutant had increased expression of OsYSL2 (Table I), which has been shown to be a metal-NA transporter expressed in the phloem (Koike et al., 2004). The higher expression of OsYSL2 could enhance metal transport through the phloem. The mechanism controlling up-regulation of Fe(II) and NA-metal transporters in the mutant is unclear. We speculate that the accumulation of NA in the mutant may mimic Fe starvation, leading to signals that stimulate expression of these genes. Further studies to manipulate expression of the NAS genes in both the wild type and naat1 through a transgenic approach would provide a more definitive answer to the question.
In the naat1 mutant, although the greatly diminished DMA secretion blocked the absorption of Fe(III) through the strategy II system, activation of the Fe(II) uptake system appears to overcompensate for the loss of strategy II, resulting in enhanced uptake of Fe when Fe(II) was available. Our results concur with the findings of Ishimaru et al. (2006, 2007) that rice possesses the functional machinery of Fe(II) uptake.
In naat1 rice, the Fe(II) uptake system was enhanced, resulting in increased accumulation of Fe, Zn, Cd, and other divalent metals in the mutant roots and shoots in the hydroponic experiments and in the mature straw from the field experiment (Fig. 5, A–C). However, only Fe concentration was significantly enhanced in the grain. It is likely that there is one (or more) possible Fe-specific transporter that transports the accumulated Fe from shoots to grains. Further investigations are needed to identify any Fe-specific transporters involved in the translocation of Fe from shoot tissues to grains.
Data from the HarvestPlus program show that commercial varieties of rice normally contain about 2 mg kg−1 Fe and 12 mg kg−1 Zn in the polished grain (Barry, 2006). Among thousands of rice genotypes investigated, only 30 lines were found to contain more than 5 mg kg−1 Fe in the polished grain (Barry, 2006). Under our field experimental conditions, the Fe concentration in the polished grain of the wild type (‘Nipponbare’) was only 1.2 mg kg−1 (Fig. 5B), which was lower than the reported average value. This low value may be partly due to the stringent grain polishing procedure used in our study. Nevertheless, under the same conditions, the Fe concentration of the polished grain of the naat1 mutant harvested from the field experiment was 4.6 mg kg−1 (Fig. 5B), which is 3.8-fold higher than the wild type and approaches that of the highest naturally existing lines. It would be interesting to test whether targeted mutations in NAAT1 in high-Fe germplasms could result in further enhancement of Fe content in rice grains.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The rice (Oryza sativa) mutant naat1 was identified in an ethyl methane sulfonate-mutagenized population from the cultivar ‘Nipponbare’ in culture solution prepared as described by Yoshida et al. (1976). The solution contained 1.425 mm NH4NO3, 0.323 mm NaH2PO4, 0.513 mm K2SO4, 0.998 mm CaCl2, 1.643 mm MgSO4, 0.009 mm MnCl2, 0.075 μm (NH4)6Mo7O24, 0.019 mm H3BO3, 0.155 μm CuSO4, 0.036 mm FeCl3, 0.070 mm citric acid, and 0.152 μm ZnSO4. A mapping population of 2,900 F2 mutant plants was generated from crosses between heterozygous naat1 mutant plants and Indica var Kasalath.
For NA analysis, Affymetrix GeneChip assay, and SPAD measurement, naat1 mutant and wild-type seeds were germinated in distilled water for 2 d. After germination, 15 seedlings were transferred to a plastic net floating on the Yoshida nutrient solution (Yoshida et al., 1976) containing 35 μm citrate-Fe(III) or 125 μm EDTA-Fe(II). Seedlings were grown in a growth chamber at 30°C/22°C day/night temperatures with a 12-h-light/12-h-dark regime (450 μmol photons m−2 s−1). Wild-type and naat1 seedlings were grown separately in different 3-L plastic pots. Twenty seedlings were grown in each pot.
Seedlings used for measuring metal uptake and DMA secretion were prepared as follows. The naat1 and wild-type seeds were germinated in tap water and placed on a plastic net floating on 0.5 mm CaSO4 solution for 10 d, then transferred to 1.2-L pots (three seedlings/pot) containing one-half-strength Kimura B solution (Ma et al., 2001) and 125 μm EDTA-Fe(II) (pH 5.5) for an additional 15 d. Nutrient solution was renewed once in 2 d. Thereafter, naat1 and wild-type plants were divided into two groups: one with and the other without EDTA-Fe(II), each with three replicate pots, and grown for 13 d before collection of root exudates for DMA measurement. During the last 6 d of plant growth, 1 μm CdSO4 was added to the nutrient solutions of both −Fe and +Fe treatments. In an additional treatment, wild-type plants were supplied with 125 μm Fe(III)-EDTA (pH 5.5) to test its secretion of DMA under Fe-sufficient conditions.
A field experiment was carried out to compare the growth performance and metal uptake of the naat1 mutant and the wild type. Pregerminated seeds of naat1 and wild type were planted in a paddy field on the farm of the Huajia campus, Zhejiang University, with a planting distance of 18 × 18 cm. The soil contained 10.92 g kg−1 total Fe and 87.8 mg kg−1 total Zn, respectively. During the whole growing period, the paddy field was maintained with 5- to 15-cm standing water until the plants reached the late grain-filling stage. Fertilizers (nitrogen, phosphorus, and potassium) were applied at normal rates for paddy rice in the region. At the maturation stage, 10 plants each were harvested for the measurement of the agronomic traits. Dried straws and seeds were used for analysis of metal concentrations.
Mapping and Cloning of NAAT1
The NAAT1 gene was mapped to the short arm of chromosome 2 between simple sequence repeat markers RM301 and RM324 using 81 F2 mutant plants. The locus was further placed within a 123-kb region between RM13046 and RM13051 markers using 2,900 F2 mutant plants and three newly developed simple sequence repeat markers. Based on the phenotype of the mutant, the NAAT1 gene was selected out of the 18 putative proteins coded by the 123-kb DNA region as a candidate gene. Genomic DNAs and cDNAs of the gene were amplified by PCR or RT-PCR from naat1 mutant and wild-type plants. NAAT1 gene-specific forward and reverse primers are 5′-TGTCCCACACCCGTAGAAT-3′ and 5′-CCATTTTGGTGACAACAGG-3′. PCR and RT-PCR products were cloned into pMD18-T vector (TaKaRa) and sequenced.
Expression of NAAT1 Protein in Escherichia coli
NAAT1 cDNA synthesized from the wild-type and naat1 RNA samples were cloned into the expression vector pET-29b (Novagen). The resultant plasmids were expressed using Escherichia coli strain BL21(DE3). Fresh BL21(DE3)-transformed cells were inoculated into 10 mL of Luria-Bertani/ampicillin medium and grown at 37°C until OD600 ranged from 0.7 to 0.8. Expression of NAAT1 proteins was induced by adding isopropyl thio-d-galactoside at a final concentration of 0.5 mm for an additional 4 h at 37°C. Cells were then harvested and lysed by sonication. Soluble and insoluble fractions were fractionated by centrifugation and separated by electrophoresis on 12.5% SDS-PAGE gel.
Complementation Test
The wild-type full-length ORF of NAAT1 was amplified by RT-PCR and inserted into the binary vector pTF101.1 (Frame et al., 2002) under the control of a maize (Zea mays) Ubi-1 promoter and a nopaline synthase terminator. The resulted transformation plasmid pNAAT1-OX was used for Agrobacterium-mediated rice transformation of naat1 mutant as described by Chen et al. (2003).
Measurement of Chlorophyll Content
SPAD values (total chlorophyll content) were determined on the fully expanded youngest leaves of 10-d-old seedlings with a portable chlorophyll meter (SPAD-502; Minolta Sensing).
Measurement of NA
Shoots and roots of 10-d-old naat1 and wild-type seedlings (as described in the section on plant materials and growth conditions) were harvested for NA measurement. Extraction and quantification of NA were performed as described by Weber et al. (2004). Briefly, approximately 150 mg of seedling samples were ground in liquid nitrogen and extracted with 300 μL of water at 80°C for 30 min followed by 10-min centrifugation (18,000g). Twenty microliters of the supernatant solution was injected into an Agilent 1100 series HPLC and fractionated using an Agilent Zorbax SB-aq column (Agilent Technologies) at 35°C. The column effluent was introduced into the ion source of a Finnigan LCQ Deca XP plus electrospray ion trap mass spectrometer (ThermoFinnigan). Quantification was based on the liquid chromatography-mass spectrometry peak area of NA (T. Hasegawa Co.), and a calibration curve constructed by quantifying peak areas of the standard in seven different concentrations (6.5–3,250 ng mL−1).
Measurement of DMA in Root Exudates
Root exudates were collected on day 13 after −Fe(II) or +Fe(II) treatments by immersing roots in deionized water for 5 h from 8 am to 1 pm. Solutions were passed through a cation exchange resin and eluted using 2 m NH4OH. The eluates were concentrated in a rotary evaporator at 40°C. Phytosiderophores in the root exudates were quantified with HPLC using a cation exchange column (Shim-pack; Amino-Li, Shimadzu). The mobile phase was a 0.15 m lithium citrate (pH 2.6), mixed with 0.2 m LiOH at a proportion of 5%. The total flow rate of the mobile phase was 0.4 mL min−1 at 50°C. Detection of fluorescence was conducted after reaction with NaClO and o-phthalaldehyde at emission of 450 nm and excitation of 350 nm. Only DMA was detected in the rice root exudates. The concentration was calculated based on the peak area.
Measurement of Metal Concentrations
To determine the concentrations of Fe, Zn, and Cd of the naat1 and wild-type plants, we performed elemental analysis on 38-d-old seedlings from the DMA secretion experiment and the straw and grain samples from the field experiment. In the DMA secretion experiment, plant roots and shoots were washed three times with deionized water after the collection of root exudates and dried at 70°C for 2 d. Grain samples from the field experiment were dehusked into unpolished grain. Portions of the unpolished grain samples were processed with a rice-milling machine (JNMJ3; Taizhou) for 1 min, three times, to obtain polished grain.
Shoot, root, and straw samples were ground to fine powders and digested with 5 mL of 11 m HNO3 for 5 h at 150°C and metal concentrations measured by atomic absorption spectrometry (Z-2000; Hitachi). Grain samples were ground to powders first, then digested with ultrapure HNO3 and H2O2 in Teflon-coated microwave vessels, and metal concentrations measured using inductively coupled plasma mass spectrometry (Agilent 7500ce).
Affymetrix GeneChip Analysis
Ten-day-old seedlings grown in solution culture with citrate-FeCl3, EDTA-FeSO4, or no Fe were used for RNA sampling (as described in the section on plant materials and growth conditions). RNA samples from three biological replications were prepared according to the procedure recommended by the manufacturer (Affymetrix, 2003). Single-stranded, then double-stranded, cDNA was synthesized using the SuperScript double-stranded cDNA synthesis kit (Invitrogen). A portion of the resulting double-stranded cDNA was used as a template to generate biotin-tagged cRNA using an Affymetrix GeneChip IVT labeling kit (Affymetrix). Fifteen micrograms of the resulting biotin-tagged cRNA were fragmented to a size range of 35 to 200 bases following the manufacturer's instructions (33). Subsequently, 10 μg of this fragmented target cRNA was hybridized at 45°C with rotation for 16 h to probe sets present on an Affymetrix rice genome array. The GeneChip arrays were washed, then stained using streptavidin-phycoerythrin on an Affymetrix Fluidics Station 450 followed by scanning on a GeneChip Scanner 3000. Hybridization data were analyzed using GeneChip Operating Software (GCOS 1.2) and dChipsoftware (Li and Wong, 2001).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Root characters of naat1 and wild-type plants.
Supplemental Table S2. Contents of other metal elements in straw, rough grains, and polished grains.
Supplemental Table S3. Primer sequences of genes in RT-PCR.
Supplementary Material
Acknowledgments
We thank Dr. Stanton Gelvin and Dr. Kan Wang for critical reading of the manuscript. We also thank Mr. Yunrong Wu for field experiment management and Dr. Ming Chen for the annotation of microarray data.
This work was supported by the National Key Basic Research Special Foundation of China (grant no. 2005CB20900), the National Natural Science Foundation (grant nos. 30471118 and 30770191), and the HarvestPlus Program China. Rothamsted Research receives grant-aided support from the Biotechnology and Biological Sciences Research Council (UK).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ping Wu (clspwu@zju.edu.cn).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Affymetrix (2003) GeneChip Expression Analysis Technical Manual. Affymetrix, Santa Clara, CA
- Barry G (2006) High-iron and zinc rice. In Rice Fact Sheet. The International Rice Research Institute, Manila, The Philippines
- Belouchi A, Kwan T, Gros P (1997) Cloning and characterization of the OsNramp family from Oryza sativa, a new family of membrane proteins possibly implicated in the transport of metal ions. Plant Mol Biol 33 1085–1092 [DOI] [PubMed] [Google Scholar]
- Bughio N, Yamaguchi H, Nishizawa NK, Nakanishi H, Mori S (2002) Cloning an iron-regulated metal transporter from rice. J Exp Bot 53 1677–1682 [DOI] [PubMed] [Google Scholar]
- Chen SY, Jin WZ, Wang MY, Zhang F, Zhou J, Jia QJ, Wu YR, Liu FY, Wu P (2003) Distribution and characterization of over 1000 T-DNA tags in rice genome. Plant J 36 105–113 [DOI] [PubMed] [Google Scholar]
- Curie C, Alonso JM, Le Jean M, Ecker JR, Briat JF (2000) Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. Biochem J 347 749–755 [PMC free article] [PubMed] [Google Scholar]
- Curie C, Panaviene Z, Loulergue C, Dellaporta SL, Briat JF, Walker EL (2001) Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409 346–349 [DOI] [PubMed] [Google Scholar]
- Douchkov D, Gryczka C, Stephan UW, Hell R, Baumlein H (2005) Ectopic expression of nicotianamine synthase genes results in improved iron accumulation and increased nickel tolerance in transgenic tobacco. Plant Cell Environ 28 365–374 [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML (1996) A novel iron regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame BR, Shou HX, Chikwamba R, Zhang ZY, Xiang CB, Fonger T, Pegg SE, Li B, Nettleton D, Pei P, et al (2002) Agrobacterium-mediated transformation of maize embryos using a simple binary vector system. Plant Physiol 129 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerinot ML, Yi Y (1994) Iron: nutritious, noxious, and not-readily available. Plant Physiol 104 815–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Rupe MA, Danilevskaya ON, Yang XF, Hu ZH (2003) Genome-wide mRNA profiling reveals heterochronic allelic variation and a new imprinted gene in hybrid maize endosperm. Plant J 36 30–44 [DOI] [PubMed] [Google Scholar]
- Higuchi K, Nishizawa NK, Römheld V, Marschner H, Mori S (1996) Absence of nicotianamine synthase activity in the tomato mutant ‘chloronerva’. J Plant Nutr 19 1235–1239 [Google Scholar]
- Higuchi K, Suzuki K, Nakanishi H, Yamaguchi H, Nishizawa NK, Mori S (1999) Cloning of nicotianamine synthase genes, novel genes involved in the biosynthesis of phytosiderophores. Plant Physiol 119 471–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi K, Watanabe S, Takahashi M, Kawasaki S, Nakanishi H, Nishizawa NK, Mori S (2001) Nicotianamine synthase gene expression differs in barely and rice and Fe-deficient conditions. Plant J 25 159–167 [DOI] [PubMed] [Google Scholar]
- Inoue H, Higuchi K, Takahashi M, Nakanishi H, Mori S, Nishizawa NK (2003) Three rice nicotianamine synthase genes, OsNAS1, OsNAS2, and OsNAS3 are expressed in cells involved in long-distance transport of iron and differentially regulated by iron. Plant J 36 366–381 [DOI] [PubMed] [Google Scholar]
- Inoue H, Suzuki M, Takahashi M, Nakanishi H, Mori S, Nishizawa NK (2004) Rice nicotianamine aminotransferase gene (NAAT1) is expressed in cells involved in long-distance transport of iron. In Abstracts of the XII International Symposium on Iron Nutrition and Interactions in Plants. ISINIP, Tokyo, p 204
- Ishimaru Y, Kim S, Tsukamoto T, Oki H, Kobayashi T, Watanabe S, Matsuhashi S, Takahashi M, Nakanishi H, Mori S, et al (2007) Mutational reconstructed ferric chelate reductase confers enhanced tolerance in rice to iron deficiency in calcareous soil. Proc Natl Acad Sci USA 104 7373–7378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru Y, Suzuki M, Kobayashi T, Takahashi M, Nakanishi H, Mori S, Nishizawa NK (2005) OsZIP4, a novel zinc-regulated zinc transporter in rice. J Exp Bot 56 3207–3214 [DOI] [PubMed] [Google Scholar]
- Ishimaru Y, Suzuki M, Tsukamoto T, Suzuki K, Nakazono M, Kobayashi T, Wada Y, Watanabe S, Matsuhashi S, Takahash M, et al (2006) Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J 45 335–346 [DOI] [PubMed] [Google Scholar]
- Kaiser BN, Moreau S, Castelli J, Thomson R, Lambert A, Bogliolo S, Puppo A, Day DA (2003) The soybean NRAMP homologue, GmDMT1, is a symbiotic divalent metal transporter capable of ferrous iron transport. Plant J 35 295–304 [DOI] [PubMed] [Google Scholar]
- Kanazawa K, Higuchi K, Nishizawa NK, Fushiya S, Chino M, Mori S (1994) Nicotianamine aminotransferase activities are correlated to the phytosiderophore secretions under Fe-deficient conditions in Gramineae. J Exp Bot 5 1903–1906 [Google Scholar]
- Kobayashi T, Suzuki M, Inoue H, Itai RN, Takahashi M, Nakanishi H, Mori S, Nishizawa NK (2005) Expression of iron acquisition-related genes in iron-deficient rice is co-ordinately induced by partially conserved iron-deficiency-responsive elements. J Exp Bot 56 1305–1316 [DOI] [PubMed] [Google Scholar]
- Koike S, Inoue H, Mizuno D, Takahashi M, Nakanishi H, Mori S, Nishizawa NK (2004) OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in the phloem. Plant J 39 415–424 [DOI] [PubMed] [Google Scholar]
- Li C, Wong HW (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 98 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling HQ, Koch G, Bäumlein H, Ganal M (1999) Map-based cloning of chloronerva—a gene involved in iron uptake of higher plants encoding nicotianamine synthase. Proc Natl Acad Sci USA 96 7098–7103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JF, Goto S, Tamai K, Ichii M (2001) Role of root hairs and lateral roots in silicon uptake by rice. Plant Physiol 127 1773–1780 [PMC free article] [PubMed] [Google Scholar]
- Ma JF, Nomoto K (1993) Two related biosynthetic pathways of mugineic acids in Graminaceous plants. Plant Physiol 102 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H, Romheld V, Kissel M (1986) Different strategies in higher plants in mobilization and uptake of iron. J Plant Nutr 9 3–7 [Google Scholar]
- Mori S (1999) Iron acquisition by plants. Curr Opin Plant Biol 2 250–253 [DOI] [PubMed] [Google Scholar]
- Mori S, Nishizawa N (1987) Methionine as a dominant precursor of phytosiderophores in graminaceae plants. Plant Cell Physiol 28 1081–1092 [Google Scholar]
- Murata Y, Ma JF, Yamiji N, Ueno D, Nomoto K, Iwashita T (2006) A specific transporter for iron(III)-phytosiderophore in barley roots. Plant J 46 563–572 [DOI] [PubMed] [Google Scholar]
- Ogo Y, Itai RN, Nakanishi H, Inoue H, Kobayashi T, Suzuki M, Takahashi M, Mori S, Nishizawa NK (2006) Isolation and characterization of IRO2, a novel iron-regulated bHLH transcription factor in graminaceous plants. J Exp Bot 57 2867–2878 [DOI] [PubMed] [Google Scholar]
- Pich A, Scholz G, Stephan UW (1994) Iron-dependent changes of heavy metals, nicotianamine, and citrate in different plant organs in the xylem exudate of two tomato genotypes. Nicotianamine as possible copper translocator. Plant Soil 165 189–196 [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML (1999) A ferric chelate reductase for iron uptake from soils. Nature 397 694–697 [DOI] [PubMed] [Google Scholar]
- Robinson NJ, Sadjuga MR, Groom QJ (1997) The FRO gene family from Arabidopsis thaliana: putative iron-chelate reductases. Plant Soil 196 245–248 [Google Scholar]
- Römheld V, Marschner H (1986) Evidence for a specific uptake system for iron phytosiderophores in roots of grasses. Plant Physiol 80 175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf G, Ludewig U, Erenoglu BE, Mori S, Kitahara T, von Wiren N (2004) ZmYS1 functions as a proton-coupled symporter for phytosiderophore- and nicotianamine-chelated metals. J Biol Chem 279 9091–9096 [DOI] [PubMed] [Google Scholar]
- Shojima S, Nishizawa NK, Fushiya S, Nozoe S, Irifune T, Mori S (1990) Biosynthesis of phytosiderophores. Plant Physiol 93 1497–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shojima S, Nishizawa NK, Mori S (1989) Establishment of a cell-free system for the biosynthesis of nicotianamine. Plant Cell Physiol 30 673–677 [Google Scholar]
- Takagi S, Nomoto K, Takemoto T (1984) Physiological aspect of mugineic acid, a possible phytosiderophore of graminaceous plants. J Plant Nutr 7 1–5 [Google Scholar]
- Takahashi M, Nakanishi H, Kawasaki S, Nishizawa NK, Mori S (2001) Enhanced tolerance of rice to low iron availability in alkaline soils using barley nicotianamine aminotransferase genes. Nat Biotechnol 19 466–469 [DOI] [PubMed] [Google Scholar]
- Takahashi M, Terada Y, Nakai I, Nakanishi H, Yoshimura E, Mori S, Nishizawa NK (2003) Role of nicotianamine in the intracellular delivery of metals and plant reproductive development. Plant Cell 15 1263–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Yamaguchi H, Nakanishi H, Shioiri T, Nishizawa NK, Mori S (1999) Cloning two genes for nicotianamine aminotransferase, a critical enzyme in iron acquisition (strategy II) in graminaceous plants. Plant Physiol 121 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Briat JF, Curie C (2001) Arabidopsis IRT2 gene encodes a root periphery iron transporter. Plant J 26 181–189 [DOI] [PubMed] [Google Scholar]
- von Wiren N, Klair S, Bansal S, Briat JF, Khodr H, Shioiri T, Leigh RA, Hider RC (1999) Nicotianamine chelates both FeIII and FeII: implications for metal transport in plants. Plant Physiol 119 1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Harada E, Vess C, Roepenack-Lahave E, Clemens S (2004) Comparative microarray analysis of Arabidopsis thaliana and Arabidopsis halleri roots identifies nicotianamine synthase, a ZIP transporter and other genes as potential metal hyperaccumulation factors. Plant J 37 269–281 [DOI] [PubMed] [Google Scholar]
- World Health Organization (2002) World Health Report 2002: Reducing Risks, Promoting Healthy Life. World Health Organization, Geneva
- Yoshida S, Forno DA, Cock JH, Gomez KA (1976) Laboratory Manual for Physiological Studies of Rice, Ed 3. The International Rice Research Institute, Manila, The Philippines
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.