Abstract
In natural ecosystems, many plants are able to establish mutually beneficial symbioses with microorganisms. Of critical importance to sustainable agriculture are the symbioses formed between more than 80% of terrestrial plants and arbuscular mycorrhizal (AM) fungi and between legumes and nitrogen-fixing rhizobial bacteria. Interestingly, the two symbioses share overlapping signaling pathways in legumes, suggesting that the evolutionarily recent root nodule symbiosis may have acquired functions from the ancient AM symbiosis. The Medicago truncatula DMI3 (DOESN'T MAKE INFECTIONS3) gene (MtDMI3) and its orthologs in legumes are required for both bacterial and fungal symbioses. MtDMI3 encodes a Ca2+/calmodulin-dependent protein kinase (CCaMK) essential for the transduction of the Ca2+ signal induced by the perception of Nod factors. Putative orthologs of MtDMI3 are also present in non-legumes, but their function in AM symbiosis has not been demonstrated in any non-legume species. Here, we combine reverse genetic approaches and a cross-species complementation test to characterize the function of the rice (Oryza sativa) ortholog of MtDMI3, namely, OsDMI3, in AM symbiosis. We demonstrate that OsDMI3 is not only required for AM symbiosis in rice but also is able to complement a M. truncatula dmi3 mutant, indicating an equivalent role of MtDMI3 orthologs in non-legumes.
More than 80% of vascular flowering plants establish symbiotic associations with arbuscular mycorrhizal (AM) fungi, during which fungal hyphae expand the functional root-soil interface and enhance access to inorganic phosphate and other mineral nutrients (Smith and Read, 1997; Brundrett, 2002). Originating more than 400 million years ago, AM symbiosis likely played a key role in facilitating the movement of plants to land (Remy et al., 1994; Redecker et al., 2000; Heckman et al., 2001). The development of AM symbiosis follows a defined morphological program triggered by yet unknown diffusible fungal signals, termed Myc factors (Genre et al., 2005; Harrison, 2005; Navazio et al., 2007). To initiate AM symbiosis, fungal hyphae first differentiate on the surface of the root to form an appressorium, which in turn gives rise to a penetration peg that facilitates entry into the plant. Once inside the root, fungal hyphae continue to grow until they reach and penetrate the cell wall of an inner cortical cell, where further differentiation yields highly ramified fungal hyphae, termed arbuscules (Harrison, 1997, 2005). In parallel, AM fungi also develop extensive hyphae outside the plant root. The intraradical and extraradical hyphae constitute a filamentous network that bridges rhizosphere and plant roots and consequently facilitates bidirectional nutrient transfer where soil nutrients move to the plant and plant photosynthates flow to the fungus (Jakobsen, 1995; Harrison, 1997; Smith et al., 2001).
In contrast to the ancient AM symbiosis, the nitrogen-fixing root nodule symbiosis between legumes and rhizobial bacteria evolved more recently, approximately 60 to 70 million years ago (Doyle, 1998). The symbiosis begins with a molecular dialog between the host and bacteria (Long, 1996; Spaink, 2000). Flavonoid compounds secreted from legume roots attract the rhizobia to the root and trigger the synthesis and secretion of chitin-like lipochitooligosaccharides of bacterial origin, known as Nod factors. Perception of Nod factors by the plant induces a suite of host responses, including the activation of host gene expression, calcium spiking, root hair deformation and curling, and cortical cell divisions (Downie and Walker, 1999; Oldroyd and Downie, 2004). These molecular, physiological, and morphological changes ultimately result in the formation of the root nodule, within which the differentiated bacteria find an ideal environment for nitrogen fixation.
Despite the remarkable morphological differences between AM and root nodule symbioses, the two share several common features, such as genetically controlled microbial infection of the host plant, transcriptional activation of a common set of host genes, and formation of an intracellular plant-microbe interface where nutrient exchange occurs (Oldroyd and Downie, 2004; Kistner et al., 2005). To date, at least seven genes have been identified in legumes that are required for the establishment of both fungal and bacterial symbioses, the so-called common symbiosis (SYM) genes (Kistner et al., 2005). Examples of common SYM genes include Medicago truncatula MtDMI1 (DOESN'T MAKE INFECTIONS1), MtDMI2, and MtDMI3; all dmi mutants in M. truncatula are blocked at an early stage of both symbiotic interactions (Catoira et al., 2000). MtDMI1 and MtDMI2 act upstream of calcium spiking, while MtDMI3 lies downstream of calcium spiking (Oldroyd and Downie, 2004). The fact that rhizobial and AM symbioses share common signaling components and that the putative orthologs of the common SYM genes are universally conserved in non-legumes (Zhu et al., 2006) support the hypothesis that the nitrogen-fixing root nodule symbiosis in legumes may have evolved from the more ancient AM symbiosis (LaRue and Weeden, 1994; Gianinazzi-Pearson, 1996).
We are particularly interested in investigating the functions of non-legume orthologs of legume genes that are required for both rhizobial and AM symbioses. We hypothesize that if the nitrogen-fixing root nodule symbiosis has co-opted part of the mechanisms initially for the AM symbiosis, then the non-legume orthologs of these common signaling components likely will maintain equivalent biological functions to their legume counterparts. To test this hypothesis, we have chosen putative rice (Oryza sativa) orthologs for functional analysis because rice is a mycorrhizal plant with a completely sequenced genome and abundant genetic and genomic tools. We are employing a dual strategy to accomplish this goal: (1) to perform cross-species complementation tests and (2) to characterize the AM phenotype of rice mutants for which the target genes were knocked out or knocked down. Here, we report the results from functional analysis of OsDMI3, the rice ortholog of MtDMI3, a Ca2+/calmodulin-dependent protein kinase (CCaMK) gene required for both bacterial and fungal symbioses (Levy et al., 2004; Mitra et al., 2004). We demonstrate that OsDMI3 is not only required for AM symbiosis in rice but also is able to complement a M. truncatula dmi3 mutant (TRV25), indicating an equivalent role of DMI3 orthologs in both legumes and non-legumes.
RESULTS
Features of OsDMI3
MtDMI3 orthologs are universally conserved in non-legumes (except for Arabidopsis [Arabidopsis thaliana] and likely members of the Brassica family) that are unable to establish symbiotic associations with AM fungi (Levy et al., 2004; Zhu et al., 2006). OsDMI3 was identified as Os05g41090, a single-copy gene in the rice genome (‘Nipponbare’) that shares high sequence homology (approximately 70% identity at the amino acid level), identical gene structure of seven exons (Fig. 1A), and syntenic chromosomal location with MtDMI3 (Godfroy et al., 2006; Zhu et al., 2006). The conceptual OsDMI3 protein consists of 516 amino acid residues with a domain structure identical to its legume counterparts, including a Ser/Thr kinase domain, a calmodulin-binding domain that overlaps with an autoinhibition domain, and three visinin-like calcium-binding EF-hand motifs (Fig. 1, B and C). In silico analysis of the rice MPSS (massively parallel signature sequencing) database indicated that the expression of OsDMI3 was significantly induced in root tissues under stress conditions (31 transcripts per million versus none in normal roots, leaves, and panicles; Nobuta et al., 2007). Quantitative real-time PCR performed in this study showed that, in comparison with leaves, shoots, and culms, OsDMI3 was expressed at an approximately 30-fold higher level in roots and an approximately 15-fold higher level in panicles (Fig. 1D). This result was further confirmed by analysis of the Dana-Farber Cancer Institute Oryza sativa Gene Index database (http://compbio.dfci.harvard.edu/tgi), from which two of the six ESTs in TC290169 were from flower tissue, while the remaining four came from root tissue. The higher level expression of OsDMI3 in flower organs was in contrast to the expression pattern reported in M. truncatula (Levy et al., 2004), but consistent with that observed for the CCaMK genes in non-legumes such as lily (Lilium longiflorum) and tobacco (Nicotiana tabacum; Poovaiah et al., 1999). The expression level of OsDMI3 in rice roots was not significantly affected by mycorrhization (Fig. 1D).
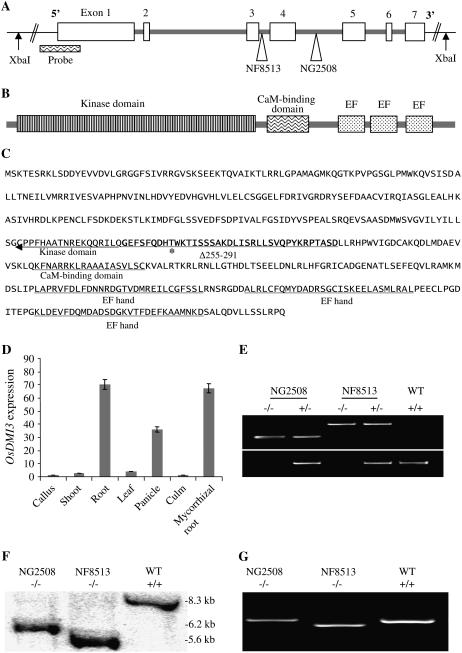
Figure 1.
Isolation and characterization of Tos17 insertion mutants of OsDMI3. A, Gene structure of OsDMI3 and the Tos17 insertion sites in NF8513 and NG2508. The exons and introns are indicated by boxes and lines, respectively. Insertion sites of Tos17 in the two mutant lines are indicated by arrow heads showing the names of the mutant lines. “Probe” indicates the DNA fragment used as a probe for Southern-blot analysis in F. B, Domain structure of the OsDMI3 protein. The kinase domain, calmodulin-binding domain, and three calcium-binding EF hands are indicated. C, Amino acids of OsDMI3. Bold, underlined residues were deleted in NF8513 (Δ255–291). The residue “T” with asterisk indicates the autophosphorylation site within the deleted peptide fragment. Conserved domains indicated in B are also underlined. D, OsDMI3 expression levels in callus, shoots, leaves, culms, roots, panicles, and mycorrhizal roots. Relative transcript abundance was determined by quantitative real-time PCR and normalized against OsUbiquitin1. Error bars represent sds from three independent biological replications. E, Identification of homozygous (−/−) Tos17 insertion mutants by PCR. Top, Identification of positive Tos17 insertion plants (+/− or −/−) by PCR using a pair of Tos17- and OsDMI3-specific primers. Bottom, PCR analysis to distinguish between homozygous (−/−) and heterozygous (+/−) mutant plants using a primer pair flanking the Tos17 insertion site that allowed the amplification of only the wild-type allele under given PCR conditions. WT, Wild-type ‘Nipponbare’. F, Southern blotting of genomic DNA digested with XbaI to confirm the homozygous mutants inferred by PCR. In the wild type, a single 8.3-kb band was detected, as predicted. Based on the restriction map of genomic DNA around OsDMI3 indicated in A and a single XbaI site in Tos17, a 6.2-kb band and a 5.6-kb band were predicted and detected from NG2508 and NF8513 homozygous insertion mutants, respectively. G, RT-PCR analysis of OsDMI3 transcripts in the wild type and homozygous NG2508 and NF8513 mutants. RT-PCR was performed using a primer pair designed from exons flanking the insertion sites. The allele in NG2508 produced a single RT-PCR band of the same size as that of the wild type, while the allele in NF8513 produced a band of a smaller size due to the deletion of the 111-bp exon 3.
Identification and Characterization of the Tos17 Insertion Mutants of OsDMI3
We searched the rice Tos17 insertion mutant database (Miyao et al., 2003) for OsDMI3 insertion lines to be used for testing the function of OsDMI3 in AM symbiosis. We identified two retrotransposon Tos17 insertion alleles for OsDMI3 in the tissue culture-derived lines NF8513 and NG2508. For both lines, Tos17 was inserted into the intron sequence of OsDMI3. In particular, Tos17 was inserted into the third intron and the fourth intron in NF8513 and NG2508, respectively (Fig. 1A). From progeny of the primary mutant lines, positive Tos17 insertion plants were identified by PCR analysis using a pair of Tos17- and OsDMI3-specific primers (Fig. 1E, top). A second-round PCR analysis was performed to distinguish between homozygous mutant (−/−) and heterozygous (+/−) plants using a primer pair flanking the Tos17 insertion site that allowed amplification of only the wild-type allele under given PCR conditions (Fig. 1E, bottom). The homozygous mutant plants were further confirmed by Southern blotting (Fig. 1F). DNA was isolated from putative homozygous mutant plants and digested with XbaI, then hybridized with a DNA probe amplified from the 5′ end of OsDMI3 (Fig. 1A, Probe). Based on the XbaI restriction pattern of genomic DNA around OsDMI3 (indicated in Fig. 1A) and the presence of a single XbaI site in Tos17, cutting with XbaI was predicted to produce one hybridization band of approximately 8.3 kb for the wild type and one smaller band of approximately 5.6 kb and approximately 6.2 kb for the homozygous mutant alleles in NF8513 and NG2508, respectively. As shown in Figure 1F, we observed single predicted bands for the putative homozygous mutant plants from the NF8513 and NG2508 lines.
Reverse transcription (RT)-PCR analyses using a primer pair designed from exons flanking the insertion sites indicated that both mutant alleles were normally expressed in NF8513 and NG2508. The allele in NG2508 produced a single RT-PCR band of the same size as the wild type, while the allele in NF8513 produced a band of a smaller size (Fig. 1G). Sequence analysis of the RT-PCR products revealed that the insertion allele in NG2508 produced a transcript identical to that of the wild type, indicating that the Tos17 insertion in NG2508 did not affect normal intron-exon splicing. In NF8513, however, the insertion of Tos17 closely adjacent (10 bp) to exon 3 (Fig. 1A) resulted in the deletion of the entire 111-bp exon 3, but nevertheless did not disrupt the reading frame. Thus, the resulting protein encoded by the NF8513 mutant allele was predicted to consist of 479 amino acids lacking a 37-amino-acid motif (OsDMI3Δ255–291) near the end of the predicted kinase domain (Fig. 1C). The deleted peptide fragment contains a Thr at position 263 (Thr-263), the putative Ca2+-dependent autophosphorylation site that corresponds to Thr-267 of lily CCaMK (Sathyanarayanan et al., 2001), Thr-265 of Lotus japonicus LjCCaMK (Tirichine et al., 2006), and Thr-271 of MtDMI3 (Gleason et al., 2006). The deletion of the entire exon 3 was expected to abolish OsDMI3 function. Thus, the progeny of NF8513 were subjected to further analyses (described below).
OsDMI3 Is Required for AM Symbiosis in Rice
Despite the conservation of MtDMI3 orthologs in non-legumes, it is unclear whether these orthologous genes are truly required for AM symbiosis in non-legumes. To test the possible role of OsDMI3 in AM symbiosis, the colonization of rice roots by the AM fungus Glomus intraradices was analyzed in progeny of NF8513 and wild-type plants. Seven weeks postinoculation, wild-type plants were densely colonized by G. intraradices, with more than 80% of the entire root system being colonized. As shown in Figure 2 (A–D), all typical symbiotic structures, including intraradical and extraradical hyphae, vesicles, and arbuscules, were readily observed on the wild-type roots. Similar level of fungal colonization was also observed on roots of heterozygous (+/−) and homozygous wild-type (+/+) plants segregated from NF8513 (data not shown). In contrast, vesicles and arbuscules were never observed on roots of a total of 60 homozygous (−/−) mutant plants derived from three different homozygous T1 plants. For homozygous mutant plants, hyphal growth and appressoria formation were infrequently observed on the root surface (Fig. 2, E and F), but further entry between epidermal cells was blocked at the epidermal surface. Occasionally, the fungus was able to penetrate the cortical cells but unable to develop arbuscules (Fig. 2G). The observed defective phenotypes were reminiscent of those observed for the dmi3 mutant (TRV25) in M. truncatula and the corresponding mutants for LjCCaMK in L. japonicus (i.e. sym15-1, sym15-2, sym72-1, and sym72-2; Catoira et al., 2000; Senoo et al., 2000; Demchenko et al., 2004; Kistner et al., 2005; Tirichine et al., 2006). Except for the weak allele of L. japonicus sym15-1, all these legume mutants were characterized by poorly developed external hyphae, the blocking of hyphal penetration at the root epidermis, and the lack of arbuscules and vesicles in roots.
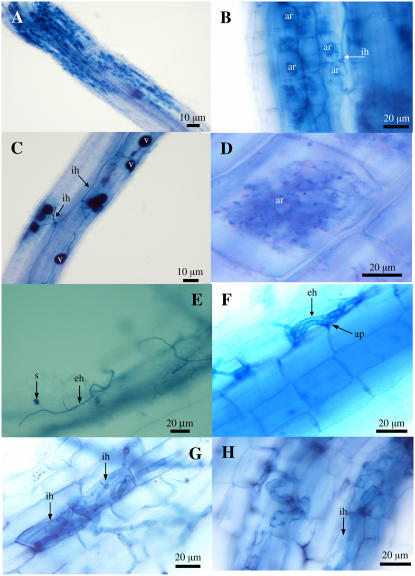
Figure 2.
Cytological characterization of the interaction of the wild type (A–D), mutant (E–G), and the OsDMI3i-2 down-regulated line (H) with G. intraradices. Photographs were taken from roots at 7 weeks postinoculation with G. intraradices. Mycorrhizal colonization was assessed by Trypan Blue staining according to the procedures described by Koske and Gemma (1989) with modification. Stained roots were examined using a light microscope (Olympus BX51) and images were captured by a microscope digital camera system (Olympus DP12-2). A, An overview of arbuscules densely formed on the root of a wild-type plant. B, Intraradical hyphae forming arbuscules on the root of a wild-type plant. C, Vesicles and intraradical hyphae observed on the root of a wild-type plant. D, A mature arbuscule with numerous fine branch hyphae. E, An extracellular hypha with spores (without invasion) on the root surface of a mutant NF8513 plant. F, The fungal hyphae have formed appressoria on the groove between two epidermal cells but failed to enter the root. G, The fungal hyphae have penetrated into cortical cells but failed to form an arbuscule on the root of a mutant plant. H, Similar phenotype as G but from an OsDMI3 down-regulated plant (OsDM3i-2). eh, Extraradical hypha; ih, intraradical hypha; ar, arbuscule; ap, appressorium; v, vesicle.
Numerous rice genes have been identified that were expressed exclusively in G. intraradices-colonized roots and showed no transcriptional activity after treatment with phosphate or after challenge with the fungal pathogens (Guimil et al., 2005). Thus, the expression of these genes can serve as molecular markers for the successful AM symbiosis in rice (Guimil et al., 2005). To further confirm the observed mycorrhizal phenotype at the molecular level, we characterized the expression of OsPT11, a rice mycorrhiza-specific phosphate transporter (Paszkowski et al., 2002), in roots of the wild type, homozygous mutant, and wild-type genotype segregated from a heterozygous (+/−) plant under inoculated and noninoculated conditions. As shown in Figure 3A, OsPT11 was expressed only in the wild-type roots (including the +/+ genotype segregated from NF8513) inoculated with G. intraradices but not in any other treatments.
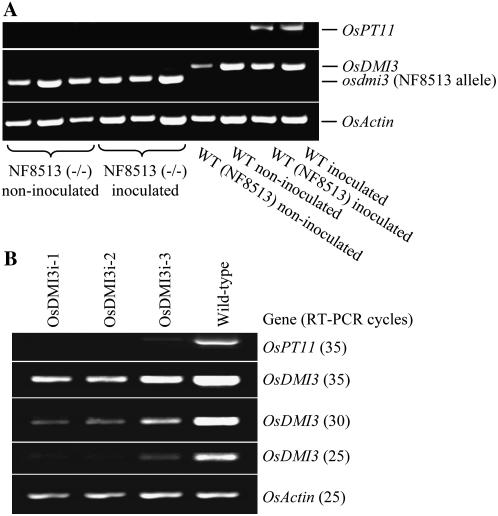
Figure 3.
Molecular characterization of the mutant and RNAi lines of OsDMI3 under inoculated and noninoculated conditions. A, Expression of OsPT11 in the wild-type and mutant roots under inoculated and noninoculated conditions. WT (NF8513) indicates wild-type plants at the OsDMI3 locus segregated from a heterozygous plant. B, Down-regulation of OsDMI3 in the three RNAi lines, OsDMI3i-1, OsDMI3i-2, and OsDMI3i-3. The number in parentheses indicates the cycle number of the RT-PCR. The rice Actin gene was used as a control.
Tissue culture-induced activation of Tos17 can result in an average of 10 insertions in the same genome (Miyao et al., 2007). In NF8513, there are a total of eight listed Tos17 insertions, represented by the Tos17 flanking sequences in the NCBI GenBank (i.e. AB155310, AB156672–AB156675, and AG212632–AG212634), from which OsDMI3 (AB156672) is the only insertion site on rice chromosome 5. To rule out the possibility that the observed mutant phenotype was due to Tos17 insertions into genes other than OsDMI3, we performed a cosegregation analysis consisting of 30 progeny plants from a single heterozygous plant. From the 30 plants, only the six homozygous mutant segregants showed the defective AM phenotype as described above (data not shown).
To gain further evidence that OsDMI3 is essential for AM symbiosis in rice, we generated transgenic rice plants (‘Nipponbare’) expressing an RNA interference (RNAi) construct consisting of an inverted-repeat sequence of the first exon of OsDMI3. A BLAST search using the target sequence as a query did not result in any hits other than OsDMI3 in the rice genome, thus excluding the possibility of off-target gene silencing. We selected three independent transgenic RNAi knockdown lines, designated OsDMI3i-1, OsDMI3i-2, and OsDMI3i-3, for further analysis. OsDMI3 was dramatically down-regulated in the root of OsDMI3i-1 and OsDMI3i-2, but only moderately down-regulated in OsDMI3i-3 (Fig. 3B). It is noteworthy that all the T1 progeny of OsDMI3i-1 (n = 42) were transgenic, which was not uncommon and likely due to multiple independent T-DNA insertions. Nevertheless, the segregation of transgenic versus wild-type plants in T1 progeny of OsDMI3i-2 (n = 50) and OsDMI3i-3 (n = 46) fits the 3:1 ratio expected from a single T-DNA insertion. Semiquantitative RT-PCR analysis indicated that the transgenic T1 plants maintained an equivalent level of gene silencing efficiency to the primary T0 plants. Strikingly, cytological and molecular analysis based on the expression of OsPT11 revealed that all the transgenic plants from OsDMI3i-1 (n = 42) and OsDMI3i-2 (n = 37) showed a defective AM phenotype similar to that of the knockout insertion mutant (Fig. 2H), while all wild-type plants segregated from the T1 progeny were normally colonized by the AM fungus. Interestingly, the expression level of OsDMI3 appeared to correspond well with the colonization level of the AM fungus. In the progeny of OsDMI3i-3 where OsDMI3 was only moderately down-regulated, arbusculars were detected in approximately 5% of the root system. Taken together, cytological, genetic, and molecular evidence indicated that OsDMI3 is required for the establishment of AM symbiosis in rice.
OsDMI3 Can Complement the Defective AM Phenotype of a M. truncatula dmi3 Mutant
An alternative strategy to investigate ortholog functionality is to perform a cross-species complementation test. Rescue of the null phenotypes by expressing putative orthologs from other species provides the strongest possible evidence of conserved molecular function. The successful complementation of a nodulation-defective M. truncatula dmi3 mutant by OsDMI3 and a lily ortholog has been described (Gleason et al., 2006; Godfroy et al., 2006). In both reports, however, the AM phenotype of the transgenic roots was not characterized.
A full-length cDNA of OsDMI3 (AK070533) under the control of the 35S promoter was introduced into the M. truncatula dmi3-1 mutant (TRV25; Catoira et al., 2000) by Agrobacterium rhizogenes-mediated hairy root transformation (Boisson-Dernier et al., 2001). As expected, 35S-OsDMI3 restores nodule formation (Fig. 4A). Consistent with the result reported by Godfroy et al. (2006), the nodules were white and spherical and without bacterial infection. Thus, these nodules were likely nonfunctional. To determine whether OsDMI3 was capable of complementing the defective phenotype of AM symbiosis, we inoculated the transgenic roots with G. intraradices. As shown in Figure 4C, abundant arbuscules were observed from the mutant roots expressing the OsDMI3 gene (n = 18). We also demonstrated that MtPT4, a mycorrhiza-specific phosphate transporter gene in M. truncatula (Harrison et al., 2002; Javot et al., 2007), was expressed in wild-type and transgenic roots inoculated with G. intraradices but not in mutant roots (Fig. 4D). The successful colonization of G. intraradices and expression of MtPT4 in the transgenic roots strongly indicate that OsDMI3 can functionally complement the AM symbiosis of the M. truncatula dmi3-1 mutant.
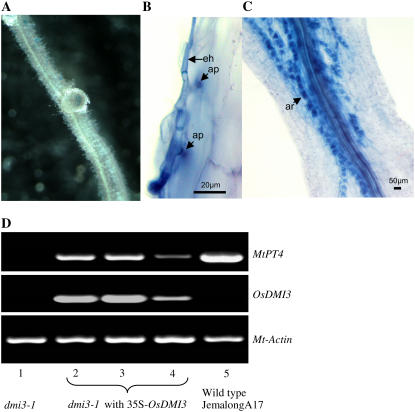
Figure 4.
Complementation of the M. truncatula dmi3-1 mutant (TRV25) by OsDMI3. The dmi3-1 mutant of M. truncatula was transformed with OsDMI3 using A. rhizogenes-mediated hairy root transformation (Boisson-Dernier et al., 2001). A full-length cDNA clone of OsDMI3 (AK070533) was cloned into a binary vector modified from pHellsgate8 driven by the CaMV-35S promoter (Helliwell et al., 2002). The binary vector was introduced into the A. rhizogenes strain, ARqua1, and transformed into the roots of the dmi3-1 mutant. A, Transformation of dmi3-1 roots with 35S-OsDMI3 leads to complementation of the nodulation phenotype. B, Defective phenotype of the M. truncatula dmi3-1 mutant inoculated with G. intraradices. The fungal hyphae formed appressoria on the root surface but failed to penetrate the root. eh, Extraradical hypha; ar, arbuscule; ap, appressorium. C, Transformation of dmi3-1 roots with 35S-OsDMI3 leads to complementation of the AM phenotype. D, Expression of MtPT4 in the transgenic dmi3-1 roots inoculated with G. intraradices. The M. truncatula Actin gene was used as a control.
DISCUSSION
Tremendous progress has been made recently in cloning the genes that are essential for rhizobial and AM symbioses from the two model legumes, M. truncatula and L. japonicus (Oldroyd and Downie, 2004; Riely et al., 2004; Oldroyd et al., 2005). One of the most interesting findings was that nearly all the genes cloned thus far, either the common SYM genes or genes required only for rhizobial symbiosis, have their putative orthologs in non-legumes (Zhu et al., 2006). This finding provides a unique opportunity to study ortholog functionality across legumes and non-legumes and to address important biological questions pertaining to the evolution of the root symbioses in plants. What makes legumes so special for root nodule symbiosis, given that most, if not all, genes required for the process are also present in non-legumes? Do non-legume orthologs play equivalent roles to their legume counterparts? Addressing these questions will not only gain insights into the evolution of root symbioses but also shed light on the evolution of species- or family-specific phenotypes in plants.
MtDMI3 represents one of the common SYM genes that are required for both fungal and bacterial symbioses in legumes (Catoira et al., 2000; Kistner et al., 2005). MtDMI3 is essential for the transduction of the Ca2+ signal induced by the perception of Nod factors (Levy et al., 2004; Mitra et al., 2004). Ca2+ presumably also serves as an intracellular messenger in the AM symbiosis, based on the requirement of MtDMI1, a putative ion channel protein, and MtDMI3 for the symbiosis (Ane et al., 2004; Harrison, 2005; Oldroyd et al., 2005). A recent report has demonstrated the implication of Ca2+ in the early signaling events between AM fungi and soybean (Glycine max) cell cultures (Navazio et al., 2007). To date, MtDMI3 orthologs have been identified in many land plants except for Arabidopsis, suggestive of their potential role in AM symbiosis in non-legumes (Zhu et al., 2006). The missing of a MtDMI3 ortholog in Arabidopsis could be one of the reasons why Arabidopsis is unable to establish symbiosis with AM fungi (Levy et al., 2004; Zhu et al., 2006). However, the role of the orthologs of legume common SYM genes in AM symbiosis has not been demonstrated in any non-legume species.
In this study, we combine reverse genetic approaches and a cross-species complementation test to characterize the function of OsDMI3 in AM symbiosis. The fact that the OsDMI3 loss-of-function mutant abolished the ability of rice to establish AM symbiosis indicates that OsDMI3 could perform an equivalent function to its legume orthologs in AM symbiosis. Furthermore, OsDMI3 was able to transduce mycorrhizal signals when transformed into a M. truncatula dmi3 mutant and functionally complement for mycorrhizal symbiosis. Despite this, the complete complementation of bacterial symbiosis of the same M. truncatula mutant using OsDMI3 was not achieved (Godfroy et al., 2006); in this case, nodule formation was restored but the bacteria could not enter the nodules to form a successful symbiosis. Therefore, it is likely that OsDMI3 may exhibit taxonomic-specific functionality (e.g. rhizobial infection) due to sequence divergence, leading to more accurate deciphering of signals triggered by Myc factors than by Nod factors when transformed into the M. truncatula mutant.
Despite the inability of nearly all non-legumes to form root nodules that accommodate the rhizobia to fix atmospheric nitrogen, it seems likely that rhizobial Nod factors could be sensed by non-legume plants and trigger downstream signaling pathways that allow, for example, endophytic colonization of bacteria that benefits the plant (Chi et al., 2005; Singh et al., 2006). Support of this possibility is that the legume genes encoding the putative Nod factor receptors (e.g. LjNFR1/MtLYK3 and LjNFR5/MtNFP) and Nod factor response factors (e.g. the GRAS family of transcription factors MtNSP1 and MtNSP2) are all conserved in non-legumes, despite that these genes are only required for nodulation symbiosis in legumes (Madsen et al., 2003; Radutoiu et al., 2003; Limpens et al., 2003; Kalo et al., 2005; Smit et al., 2005; Arrighi et al., 2006; Zhu et al., 2006; Zhang et al., 2007). Experimental data also suggested that Nod factors may be perceived by the non-legume plant. In rice, it was reported that Nod factors can induce the expression of a reporter gene under the control of the M. truncatula ENOD12 promoter (Reddy et al., 1998). Prithiviraj et al. (2003) also showed that application of Nod factors enhanced germination and early growth of rice and other non-legumes. Taken together, the available evidence suggests that the Nod factor signaling pathway is at least partially present in non-legume species.
Normally, infection by rhizobia is prerequisite for the development of nitrogen-fixing root nodules in legumes. Interestingly, the specific removal of the autoinhibition domain or mutation of the autophosphorylation site of the legume MtDMI3 orthologs can lead to autoactivation of the nodulation signaling pathway, resulting in spontaneous nodulation in the absence of bacterial infection (Gleason et al., 2006; Tirichine et al., 2006). Although the exact mechanisms underlying spontaneous nodulation are still not clear, this finding has led to the speculation that the root nodule formation may be transferred to non-leguminous crops (Gleason et al., 2006; Tirichine et al., 2006). To test this possibility in rice, we transferred a construct that contains the OsDMI3 kinase domain under the control of the 35S promoter, analogous to what was done by Gleason et al. (2006). The roots of transgenic rice plants did not show any obvious nodule organogenesis (data not shown). These observations suggest that the downstream pathway required for nodule organogenesis in legume may not be perfectly conserved in non-legumes, which may represent a major challenge toward the transfer of nodule development to non-legumes.
In conclusion, our data indicate that OsDMI3 is essential for AM symbiosis in rice and was able to complement an AM-defective phenotype of a M. truncatula dmi3 mutant. Thus far, it is unknown if other non-legume orthologs of legume common SYM genes are also required for AM symbiosis. Further characterization of functions of those genes is of critical importance to elucidate common mechanisms underlying symbiotic plant-microbe interactions and the evolution of legume-rhizobia symbiosis.
MATERIALS AND METHODS
Rice and Medicago truncatula Mutants
The rice (Oryza sativa) mutant lines (NF8513 and NG2508) containing the Tos17 insertion in the OsDMI3 gene in the ‘Nipponbare’ background were provided by the Rice Genome Resource Center of the National Institute of Agrobiological Sciences (RGRC-NIAS), Japan. The M. truncatula dmi3-1 mutant (TRV25) was obtained from Dr. Doug Cook's lab at the University of California, Davis.
Screening for Homozygous Mutant Lines of OsDMI3
Seeds of the Tos17 insertion lines from RGRC-NIAS were T1 progeny of a primary (T0) heterozygous plant. To screen for homozygous mutants, two rounds of PCR were performed. The first-round PCR was to identify plants with Tos17 insertion using the Tos17-specific primer (5′-ATTGTTAGGTTGCAAGTTAGTTAAGA-3′) and the OsDMI3-specific primer (5′-CATCACGGTTGTTGTCGAAC-3′). The second-round PCR was performed to identify homozygous mutant plants using the primer pair flanking the Tos17 insertion (5′-CACAAAAGACACATGGATTGG-3′ and 5′-CATCACGGTTGTTGTCGAAC-3′). Southern-blotting analysis was carried out to confirm the putative homozygous plants by cutting the genomic DNA with XbaI and probing with a DNA fragment amplified from 5′ of the OsDMI3 gene using the primer pair 5′-GAAGGAGCTTGCTTTGTACTC-3′ and 5′-GAGATCGATACCTGTTTCCAC-3′.
Transgenic Rice Lines
A 551-bp DNA fragment corresponding to the first exon of OsDMI3 was cloned into the RNAi vector pMCG161 (AY572837). pMCG161 consists of a chloramphenicol resistance gene for bacterial selection, a basta resistance gene (bar) for plant selection, a CaMV (Cauliflower mosaic virus)-35S promoter to drive the expression of the inverted-repeat target sequence, and a rice waxy intron to stabilize the inverted repeat of the target gene fragment. The construct was introduced into Agrobacterium tumefaciens strain GV3101 and transformed to ‘Nipponbare’ rice as described by Hiei et al. (1997). Primary transgenic plants (T0) were selfed to obtain T1 generation for phenotypic analysis of AM symbiosis.
Hairy Root Transformation of M. truncatula
The dmi3-1 mutant of M. truncatula was transformed with OsDMI3 by using Agrobacterium rhizogenes-mediated hairy root transformation (Boisson-Dernier et al., 2001). A full-length cDNA clone of OsDMI3 (AK070533) was cloned into a binary vector modified from pHellsgate8 driven by the CaMV-35S promoter (Helliwell et al., 2002). The binary vector was introduced into the A. rhizogenes strain ARqua1 and transformed into the roots of the dmi3-1 mutant. Transformed roots were selected on Färhaeus medium (Färhaeus, 1957) containing 20 mg/L kanamycin for 2 weeks at 20°C.
Inoculation of Rice and M. truncatula Roots with Glomus intraradices
The fungus G. intraradices used in this research was ordered from Premier Tech Biotechnologies (Canada) and grown in aseptic conditions according to the procedure described by Bécard and Fortin (1988). The rice plants were grown in an 11-cm pot with sterilized Turface covered with 3-cm depth of sand in a growth chamber with a 13-h-light, 28°C/11-h-dark, 24°C regime. The plants were fertilized twice weekly with half-strength Hoagland solution (Arnon and Hoagland, 1940) supplemented with 100 μm KH2PO4 (a phosphorus-limiting condition). Roots of 2-week-old rice plants were inoculated by adding 1,000 spores to the sand at 1.5-cm depth. Roots were harvested at intervals of 5 and 7 weeks postinoculation. A random sample of the root tissues was assessed for mycorrhizal colonization. The remaining tissue were frozen in liquid N2 and stored at −80°C for subsequent RNA isolation. Inoculation of M. truncatula roots was performed according to the procedures described by Liu et al. (2003).
Mycorrhizal colonization was assessed by Trypan Blue staining according to the procedures described by Koske and Gemma (1989) with modification. Roots were fixed and stored in 50% (v/v) ethanol. The fixed roots were incubated at 90°C in 10% KOH for 20 min. After rinsing with water, the roots were soaked in 1% HCl at room temperature for overnight. The roots were then stained at 90°C for 30 min in an acidic glycerol solution containing 0.1% Trypan Blue. The Trypan Blue solution was poured off and the roots were destained in acidic glycerol. Stained roots were examined using light microscope (Olympus BX51) and images were captured by a microscope digital camera system (Olympus DP12-2).
Inoculation of A. rhizogenes-Transformed M. truncatula Roots with Sinorhizobium meliloti
Nodulation assay was performed as described by Limpens et al. (2003). Three weeks after transformation, composite plants were starved for nitrate for 3 d (21°C; 16/8 h light/dark) on Färhaeus medium [without Ca(NO3)2]. The plants were then transferred to sterile Turface saturated with Färhaeus medium [without Ca(NO3)2]. Each plant was inoculated with 1 mL of culture (OD600 0.1) of S. meliloti strain RCR 2011 carrying the lacZ reporter gene in plasmid pXLGD4 (Catoira et al., 2000). The nodulation was scored 2 weeks after inoculation.
Analysis of Gene Expression
Total RNA was isolated by the Qiagen Plant RNeasy. Two micrograms of RNA was used to perform RT reactions using M-MLV reverse transcriptase (Invitrogen) in a 20-μL reaction mixture. Two microliters of the RT reaction was used as a template in a 20-μL PCR reaction solution. The PCR primers were as follows: OsActin, 5′-GCGATAATGGAACTGGTATG-3′ and 5′-CTCCATTTCCTGGTCATAGTC-3′; OsDMI3, 5′-GCTTTTTGATCGGATTGTGG-3′ and 5′-CGCAGATTATCCAGCTCCTC-3′; OsPT11, 5′-ATGGCTCGACGGACAGTAAG-3′ and 5′-GATCAGCTGGATCATGTACCT-3′; MtActin, 5′-GGAGAAGCTTGCATATGTTG-3′ and 5′-TTAGAAGCACTTCCTGTGGA-3′; MtPT4, 5′-GCTCTGGTCTTTCTTTTGGT-3′ and 5′-ACCAACAACTCATTGTACCG-3′; and transgenic OsDMI3, 5′-GAGCCTCCGGTGAAACATAA-3′ and 5′-GAGGGGAGTGAGCAAGTCTG-3′. Quantitative real-time PCR was performed on a DNA Engine Opticon 2 (Bio-Rad) using iQ SYBR Green Supermix (Bio-Rad) with 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s. Rice ubiquitin1 (AK121590) was used as an internal standard. The PCR primers used for the real-time PCR experiments were: for rice ubiquitin1, 5′-CCAGGACAAGATGATCTGCC-3′ and 5′-AAGAAGCTGAAGCATCCAGC-3′; and for OsDMI3, 5′-CGCAGATTATCCAGCTCCTC-3′ and 5′-AGGCCAACAGCAAGTGATCT-3′.
Acknowledgments
We thank Dr. Douglas Cook for providing seeds of the M. truncatula mutants and the RGRC (Japan) for providing the rice mutant seeds and full-length cDNA clones. We also thank Dr. Jean-Michel Ané and Muthusubramanian Venkateshwaran for help with the nodulation assay.
This work was supported by the Kentucky Science and Engineering Foundation (grant to H.Z.) and by the U.S. National Science Foundation (grant no. IOS 0640197 to H.Z. and J.L.). This article (07–06–111) is published with the approval of the Director of the Kentucky Agricultural Experiment Station.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Hongyan Zhu (hzhu4@uky.edu).
Open Access articles can be viewed online without a subscription.
References
- Ane JM, Kiss GB, Riely BK, Penmetsa RV, Oldroyd GE, Ayax C, Levy J, Debelle F, Baek JM, Kalo P, et al (2004) Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303 1364–1367 [DOI] [PubMed] [Google Scholar]
- Arnon DI, Hoagland DR (1940) Crop production in artificial culture solutions and in soils with special reference to factors influencing yields and absorption of inorganic nutrients. Soil Sci 50 463–483 [Google Scholar]
- Arrighi JF, Barre A, Ben Amor B, Bersoult A, Soriano LC, Mirabella R, de Carvalho-Niebel F, Journet EP, Gherardi M, Huguet T, et al (2006) The Medicago truncatula lysine motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol 142 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bécard G, Fortin JA (1988) Early events of vesicular-arbuscular mycorrhiza formation on Ri T-DNA transformed roots. New Phytol 108 211–218 [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A, Chabaud M, Garcia F, Becard G, Rosenberg C, Barker DG (2001) Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact 14 695–700 [DOI] [PubMed] [Google Scholar]
- Brundrett MC (2002) Coevolution of roots and mycorrhizas of land plants. New Phytol 154 275–304 [DOI] [PubMed] [Google Scholar]
- Catoira R, Galera C, de Billy F, Penmetsa RV, Journet EP, Maillet F, Rosenberg C, Cook D, Gough C, Denarie J (2000) Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12 1647–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi F, Shen SH, Cheng HP, Jing YX, Yanni YG, Dazzo FB (2005) Ascending migration of endophytic rhizobia, from roots to leaves, inside rice plants and assessment of benefits to rice growth physiology. Appl Environ Microbiol 71 7271–7278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demchenko K, Winzer T, Stougaard J, Parniske M, Pawlowski K (2004) Distinct roles of Lotus japonicus SYMRK and SYM15 in root colonization and arbuscule formation. New Phytol 163 381–392 [DOI] [PubMed] [Google Scholar]
- Downie JA, Walker SA (1999) Plant responses to nodulation factors. Curr Opin Plant Biol 2 483–489 [DOI] [PubMed] [Google Scholar]
- Doyle JJ (1998) Phylogenetic perspectives on nodulation: an evolving view of plants and symbiotic bacteria. Trends Plant Sci 3 473–478 [Google Scholar]
- Färhaeus G (1957) The infection of white clover root hairs by nodule bacteria studied by a simple slide technique. J Gen Microbiol 16 374–381 [DOI] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Timmers T, Bonfante P, Barker DG (2005) Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell 17 3489–3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianinazzi-Pearson V (1996) Plant cell responses to arbuscular mycorrhizal fungi: getting to the roots of the symbiosis. Plant Cell 8 1871–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason C, Chaudhuri S, Yang T, Munoz A, Poovaiah BW, Oldroyd GE (2006) Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441 1149–1152 [DOI] [PubMed] [Google Scholar]
- Godfroy O, Debelle F, Timmers T, Rosenberg C (2006) A rice calcium- and calmodulin-dependent protein kinase restores nodulation to a legume mutant. Mol Plant Microbe Interact 19 495–501 [DOI] [PubMed] [Google Scholar]
- Guimil S, Chang HS, Zhu T, Sesma A, Osbourn A, Roux C, Ioannidis V, Oakeley EJ, Docquier M, Descombes P, et al (2005) Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proc Natl Acad Sci USA 102 8066–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MJ (1997) The arbuscular mycorrhizal symbiosis: an underground association. Trends Plant Sci 2 54–56 [Google Scholar]
- Harrison MJ (2005) Signaling in the arbuscular mycorrhizal symbiosis. Annu Rev Microbiol 59 19–42 [DOI] [PubMed] [Google Scholar]
- Harrison MJ, Dewbre GR, Liu J (2002) A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14 2413–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman DS, Geiser DM, Eidell BR, Stauffer RL, Kardos NL, Hedges SB (2001) Molecular evidence for the early colonization of land by fungi and plants. Science 293 1129–1133 [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Wesley SV, Wielopolska AJ, Waterhouse PM (2002) High-throughput vectors for efficient gene silencing in plants. Funct Plant Biol 29 1217–1225 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Komari T, Kubo T (1997) Transformation of rice mediated by Agrobacterium tumefaciens. Plant Mol Biol 35 205–218 [PubMed] [Google Scholar]
- Jakobsen I (1995) Transport of phosphorus and carbon in VA mycorrhizas. In A Varma, B Hock, eds, Mycorrhiza Structure, Function, Molecular Biology and Biotechnology. Springer-Verlag, Berlin, pp 297–325
- Javot H, Penmetsa RV, Terzaghi N, Cook DR, Harrison MJ (2007) A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 104 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalo P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 17 1786–1789 [DOI] [PubMed] [Google Scholar]
- Kistner C, Winzer T, Pitzschke A, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Webb KJ, et al (2005) Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell 17 2217–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koske RE, Gemma JN (1989) A modified procedure for staining roots to detect VA mycorrhizas. Mycol Res 92 486–505 [Google Scholar]
- LaRue TA, Weeden NF (1994) The symbiosis genes of the host. In GB Kiss, G Endre, eds, Proceedings of the 1st European Nitrogen Fixation Conference. Officina Press, Szeged, Hungary, pp 147–151
- Levy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet EP, Ane JM, Lauber E, Bisseling T, et al (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303 1361–1364 [DOI] [PubMed] [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R (2003) LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302 630–633 [DOI] [PubMed] [Google Scholar]
- Limpens E, Ramos J, Franken C, Raz V, Compaan B, Franssen H, Bisseling T, Geurts R (2003) RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J Exp Bot 55 983–992 [DOI] [PubMed] [Google Scholar]
- Liu J, Blaylock LA, Endre G, Cho J, Town CD, VandenBosch KA, Harrison MJ (2003) Transcript profiling coupled with spatial expression analyses reveals genes involved in distinct developmental stages of an arbuscular mycorrhizal symbiosis. Plant Cell 15 2106–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SR (1996) Rhizobium symbiosis: nod factors in perspective. Plant Cell 8 1885–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425 637–640 [DOI] [PubMed] [Google Scholar]
- Mitra RM, Gleason CA, Edwards A, Hadfield J, Downie JA, Oldroyd GE, Long SR (2004) A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: gene identification by transcript-based cloning. Proc Natl Acad Sci USA 101 4701–4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao A, Iwasaki Y, Kitano H, Itoh J, Maekawa M, Murata K, Yatou O, Nagato Y, Hirochika H (2007) A large-scale collection of phenotypic data describing an insertional mutant population to facilitate functional analysis of rice genes. Plant Mol Biol 63 625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao A, Tanaka K, Murata K, Sawaki H, Takeda S, Abe K, Shinozuka Y, Onosato K, Hirochika H (2003) Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 15 1771–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navazio L, Moscatiello R, Genre A, Novero M, Baldan B, Bonfante P, Mariani P (2007) A diffusible signal from arbuscular mycorrhizal fungi elicits a transient cytosolic calcium elevation in host plant cells. Plant Physiol 144 673–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobuta K, Venu RC, Lu C, Belo A, Vemaraju K, Kulkarni K, Wang W, Pillay M, Green PJ, Wang GL, et al (2007) An expression atlas of rice mRNAs and small RNAs. Nat Biotechnol 25 473–477 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA (2004) Calcium, kinases and nodulation signalling in legumes. Nat Rev Mol Cell Biol 5 566–576 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Harrison MJ, Udvardi M (2005) Peace talks and trade deals: keys to long-term harmony in legume-microbe symbioses. Plant Physiol 137 1205–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkowski U, Kroken S, Roux C, Briggs SP (2002) Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 99 13324–13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poovaiah BW, Xia M, Liu Z, Wang W, Yang T, Sathyanarayanan PV, Franceschi VR (1999) Developmental regulation of the gene for chimeric calcium/calmodulin-dependent protein kinase in anthers. Planta 209 161–171 [DOI] [PubMed] [Google Scholar]
- Prithiviraj B, Zhou X, Souleimanov A, Khan WM, Smith DL (2003) A host-specific bacteria-to-plant signal molecule (Nod factor) enhances germination and early growth of diverse crop plants. Planta 216 437–445 [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Gronlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425 585–592 [DOI] [PubMed] [Google Scholar]
- Reddy PM, Ladha JK, Ramos MC, Maillet F, Hernandez RJ, Torrizo LB, Oliva NP, Datta SK, Datta K (1998) Rhizobial lipochitooligosaccharide nodulation factors activate expression of the early nodulin gene ENOD12 in rice. Plant J 14 693–702 [Google Scholar]
- Redecker D, Kodner R, Graham LE (2000) Glomalean fungi from the Ordovician. Science 289 1920–1921 [DOI] [PubMed] [Google Scholar]
- Remy W, Taylor TN, Hass H, Kerp H (1994) Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc Natl Acad Sci USA 91 11841–11843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riely BK, Ane JM, Penmetsa RV, Cook DR (2004) Genetic and genomic analysis in model legumes bring Nod-factor signaling to center stage. Curr Opin Plant Biol 7 408–413 [DOI] [PubMed] [Google Scholar]
- Sathyanarayanan PV, Siems WF, Jones JP, Poovaiah BW (2001) Calcium-stimulated autophosphorylation site of plant chimeric calcium/calmodulin-dependent protein kinase. J Biol Chem 276 32940–32947 [DOI] [PubMed] [Google Scholar]
- Senoo K, Solaiman MZ, Kawaguchi M, Imaizumi-Anraku H, Akao S, Tanaka A, Obata H (2000) Isolation of two different phenotypes of mycorrhizal mutants in the model legume plant Lotus japonicus after EMS-treatment. Plant Cell Physiol 41 726–732 [DOI] [PubMed] [Google Scholar]
- Singh RK, Mishra RP, Jaiswal HK, Kumar V, Pandey SP, Rao SB, Annapurna K (2006) Isolation and identification of natural endophytic rhizobia from rice (Oryza sativa L) through rDNA PCR-RFLP and sequence analysis. Curr Microbiol 52 345–349 [DOI] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debelle F, Gough C, Bisseling T, Geurts R (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308 1789–1791 [DOI] [PubMed] [Google Scholar]
- Smith SE, Dickson S, Smith FA (2001) Nutrient transfer in arbuscular mycorrhizas: How are fungal and plant processes integrated? Aust J Plant Physiol 28 683–694 [Google Scholar]
- Smith SE, Read DJ (1997) Mycorrhizal Symbiosis. Academic Press, San Diego
- Spaink HP (2000) Root nodulation and infection factors produced by rhizobial bacteria. Annu Rev Microbiol 54 257–288 [DOI] [PubMed] [Google Scholar]
- Tirichine L, Imaizumi-Anraku H, Yoshida S, Murakami Y, Madsen LH, Miwa H, Nakagawa T, Sandal N, Albrektsen AS, Kawaguchi M, et al (2006) Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441 1153–1156 [DOI] [PubMed] [Google Scholar]
- Zhang XC, Wu X, Findley S, Wan J, Libault M, Nguyen HT, Cannon SB, Stacey G (2007) Molecular evolution of lysin motif-type receptor-like kinases in plants. Plant Physiol 144 623–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Riely BK, Burns NJ, Ane JM (2006) Tracing nonlegume orthologs of legume genes required for nodulation and arbuscular mycorrhizal symbioses. Genetics 172 2491–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]