Abstract
We analyzed the role of the sucrose (Suc) synthase MtSucS1 during nodulation of the model legume Medicago truncatula, integrating data for the developmental, transcriptional, and metabolic processes affected downstream of an impaired Suc cleavage in root nodules. To reduce carbohydrate supply to nodule tissues, transgenic plants expressing a p35S-driven MtSucS1-antisense fusion were constructed. These plants displayed an up to 90% reduction of MtSucS1 proteins in roots and nodules. Phenotypic studies of two independent MtSucS1-reduced lines demonstrated that only under conditions depending on nodulation, these plants appeared to be impaired in above-ground growth. Specifically plant height, shoot weight, leaf development, flowering, as well as seed maturation were reduced, and the efficiency of photosynthesis was affected. Concomitantly, a significantly enhanced root to shoot ratio with a marked increase in root tip numbers was observed. Root nodule formation was found retarded and the impaired nodulation was accompanied by a less efficient nitrogen (N) acquisition. The decreased total N content of MtSucS1-antisense lines and an enhanced carbon to N ratio in roots, nodules, and shoots correlated with the extent of MtSucS1 knockdown. On the level of transcription, effects of an MtSucS1 reduction were evident for genes representing important nodes of the nodule carbon and N metabolism, while metabolite profiling revealed significantly lower levels of amino acids and their derivatives particularly in strongly MtSucS1-reduced nodules. Our results support the model that nodule-enhanced Suc synthase 1 of the model legume M. truncatula is required for the establishment and maintenance of an efficient N-fixing symbiosis.
In plants, a substantial portion of photosynthetically fixed carbon (C) is channeled into the synthesis of the disaccharide Suc. As a major assimilate, Suc is used for long-distance transport through the phloem in the majority of higher plants, and a variety of heterotrophic sink organs, such as seeds, developing leaves, internodes, tubers, or roots are net importers. The utilization and channeling of Suc into sink metabolism requires cleavage of the O-glycosidic bond by either Suc synthase (SucS; EC 2.4.1.13) and/or different invertases (Inv; EC 3.2.1.26). SucS catalyzes the UDP-dependent cleavage of Suc into UDP-Glc and Fru (Akazawa and Okamoto, 1980), and although SucS is able to synthesize Suc under appropriate conditions, in vivo the enzyme is thought to function predominantly in the cleavage direction (Avigad, 1982). Most commonly, SucS was found in heterotrophic tissues, such as storage or reproductive organs (Zrenner et al., 1995; Ruan et al., 2003), and these enzymes may also be directly allied with phloem function by facilitating loading and unloading of Suc (Nolte and Koch, 1993).
In most plant species, different SucS isoforms are encoded by a small gene family, consisting of at least two differentially expressed genes (e.g. Zea mays, Carlson et al., 2002; Oryza sativa, Yu et al., 1992; Solanum, Fu and Park, 1995; Arabidopsis [Arabidopsis thaliana], Bieniawska et al., 2007; Leguminosae, Hohnjec et al., 1999; Barratt et al., 2001; Horst et al., 2007). It is generally assumed that the regulation of SucS activity correlates with the C-consuming metabolism that is either associated with the supply of hexoses (Carlson and Chourey, 1996) or provides UDP-Glc and ADP-Glc as precursors of cellulose, hemicellulose, callose, and starch (Nolte and Koch, 1993; Salnikow et al., 2001). Since SucS takes over pivotal metabolic, structural, and storage functions in diverse sink tissues, the regulation of SucS genes is markedly specific on the transcriptional and posttranscriptional level (Fu and Park, 1995). Several cases of exquisite spatial and temporal transcriptional regulation have been reported (Winter and Huber, 2000), including distinct developmental-, cell-, or organ-specific expression patterns for particular genes (Chen and Chourey, 1989; Ruan et al., 1997; Barratt et al., 2001). Common SucS expression regulation also depends on the carbohydrate status of the tissues (Koch et al., 1992) or specific environmental conditions, such as drought stress (Gordon et al., 1999; Larrainzar et al., 2007). Interestingly, SucS from various species are posttranslationally modified by seryl phosphorylation, determining catalytic activity and subcellular enzyme localization either as soluble cytoplasmic enzymes (Winter et al., 1997) or enzymes associated with the plasmalemma, the tonoplast, or the cytoskeleton (Amor et al., 1995; Winter et al., 1998).
With respect to plant-microbe interactions, the heterotrophic microbes represent additional sinks. In N2-fixing root nodules, the microsymbionts reduce N2 to ammonia (NH4+), using dicarboxylic acids, primarily malate, as C sources (Rosendahl et al., 1990; Udvardi and Day, 1997). Dicarboxylic acids are derived from glycolysis and ultimately from Suc breakdown. In nodules, Suc cleavage is catalyzed by either SucS or alkaline Inv, but presumably not by apoplastic Inv (Peiter and Schubert, 2003). In soybean (Glycine max), Lotus japonicus, and M. truncatula root nodules, an alkaline/neutral Inv gene with a nodule-enhanced transcription was described, indicating a Suc-degrading function for those Inv in developing root nodules (Flemetakis et al., 2006; Tesfaye et al., 2006). By contrast, SucS is considered to be principally responsible for Suc degradation in mature nodules (Anthon and Emmerich, 1990; Gordon et al., 1999). In particular under hypoxia, which is essential for nitrogenase activity, there is a specific need for SucS-mediated Suc cleavage (Rolletschek et al., 2002; Albrecht and Mustroph, 2003), rather than for Inv-mediated, ATP-dependent Suc hydrolysis (Zeng et al., 1999). Finally, whereas SucS-mediated Suc breakdown on the one hand delivers the energy required for symbiotic N2 fixation (SNF) via hexose supply to nodule metabolism, SucS activity on the other hand generates C skeletons for an assimilation of the produced NH4+ into amino acids (Udvardi and Day, 1997; Gordon et al., 1999). To date, nodule-enhanced SucS isoforms associated with nodule development and metabolism were reported in a range of legume species (pea [Pisum sativum], Barratt et al., 2001; M. truncatula, Hohnjec et al., 1999; broad bean [Vicia faba], Küster et al., 1993; soybean, Gordon et al., 1992; L. japonicus, Colebatch et al., 2002). In root nodules, SucS expression was detected in the vasculature, in adjacent endodermal and cortical cells, in the meristem, and finally in cells of the central tissue (Gordon et al., 1992; Hohnjec et al., 2003).
The pea rug4 mutant (Wang and Hedley, 1993) bears a mutation in the SucS gene Sus1, resulting in a decreased enzyme activity in developing embryos and nodules down to 5% of wild-type levels (Craig et al., 1999). The Sus1 reduction in rug4 nodules led to a defective SNF and a premature senescence of these symbiotic sinks, accompanied by metabolic and phenotypic changes in seeds (Wang et al., 1990; Gordon et al., 1999). Interestingly, to obtain comparably severe effects on SNF in the model legume L. japonicus, forming determinate nodules, the two nodule-enhanced SucS genes LjSUS1 and LjSUS3 had to be knocked out concomitantly (Horst et al., 2007).
Despite accumulating reverse genetics data on the role of different nodulation-related genes in the model legume M. truncatula, a functional characterization of the nodule-enhanced SucS MtSucS1 is still missing. We have isolated and characterized the MtSucS1 gene in a previous study (Hohnjec et al., 1999), and have subsequently analyzed MtSucS1 and four additional M. truncatula SucS genes on the level of gene expression (Hohnjec et al., 2003). Reporter gene studies revealed that apart from an MtSucS1 induction in the whole plant vasculature and in diverse sink tissues, the MtSucS1 promoter directed intense expression to root tissues under endosymbiotic conditions (Hohnjec et al., 2003). In mature root nodules, strong promoter activity occurred in the infected cells of the N2-fixing zone and was additionally observed in the meristem, the prefixing zone, and in the inner cortex including the nodule vasculature. Concerning the expression analysis of the M. truncatula SucS gene family, only MtSucS1 was significantly activated and expressed at a high level in root nodules. In addition, MtSucS1 was induced in roots colonized by arbuscular mycorrhizal fungi, indicating a general function of MtSucS1 in different legume-microbe interactions (Hohnjec et al., 2003).
Although nodule-enhanced SucSs were shown to be relevant for an efficient SNF, functional studies reported so far did not address downstream effects on nodule metabolism. We here report the construction of MtSucS1-antisense plants and reveal the consequences of MtSucS1 reduction on nodule C and nitrogen (N) metabolism under nodulation-dependent conditions. On the basis of two independent MtSucS1-antisense lines, we demonstrate the effect of MtSucS1 reduction on plant growth and development, and specifically on nodule formation and metabolism. In addition to our phenotypic studies, transcriptional and metabolite profiling indicated physiological adaptations associated with an MtSucS1 reduction in root nodules. By studying the C and N metabolism downstream of MtSucS1-mediated Suc cleavage, the integrated evaluation of our phenotypic, transcriptional, and metabolite data points to an important role for MtSucS1 during nodulation and SNF.
RESULTS
Generation and Molecular Characterization of Transgenic MtSucS-Antisense Lines
We generated transgenic M. truncatula R108 plants carrying an antisense sequence that covered 1,205 bp of the 5′ end of the MtSucS1 gene encoding a nodule-enhanced SucS (Hohnjec et al., 1999). This antisense sequence is expressed from the cauliflower mosaic virus 35S promoter (p35S) and was terminated by the 3′ nopaline synthase terminator (nosT). Primary transformants were initially confirmed by selective kanamycin resistance, by PCR analysis for the presence of transgenes, and by GUS staining (data not shown). Twenty initially screened MtSucS1-antisense plants were transferred to the greenhouse and 15 lines, deriving from independently transformed plants, were characterized in the subsequent generations.
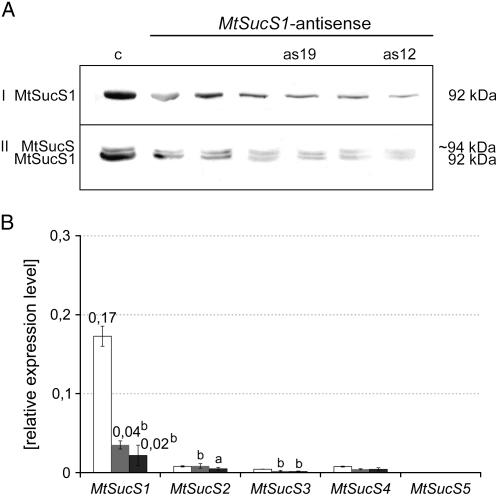
MtSucS1 protein abundance in root nodules, roots, and leaves of at least six independent T2 plants from each line and their progenies was evaluated, using a polyclonal antibody raised against a broad bean SucS (Ross and Davies, 1992). As shown in Figure 1A, several regenerated antisense lines (as lines) exhibited alterations in the MtSucS1 protein content in nodule extracts. In case of the line subsequently named as12, very low MtSucS1 protein levels (approximately 10%) were observed, whereas the MtSucS1 protein level in line as19 was only reduced to approximately 20% (Fig. 1A). Apart from MtSucS1, a second MtSucS protein of slightly higher Mr was consistently detected in root material of all MtSucS1-antisense and control lines. Since this band represented the exclusively detected SucS protein in flowers (Hohnjec et al., 1999), and real-time reverse transcription (RT)-PCR experiments revealed that the MtSucS3 gene (TC94447, Dana-Farber Cancer Institute Mt Gene Index 8) is predominantly expressed in flower tissues (Hohnjec et al., 2003), the second band observed in roots is probably MtSucS3. This isoform shows a sequence identity of 88% to MtSucS1 (TC100410), significantly higher than the identity to other MtSucS genes (Hohnjec et al., 2003). The high sequence homology between MtSucS1 and MtSucS3 might thus explain why both genes are targeted by the MtSucS1-antisense construct (Fig. 1A).
Figure 1.
Relative quantitation of SucS protein and transcript levels. A, Relative quantitation of SucS abundance via western immunoblotting of root nodule (I) and root extracts (II) from MtSucS1-antisense and control lines. Complete transfer of protein extracts onto blotting membrane was checked via Ponceau S protein stain (data not shown). The approximate molecular masses of the polypeptides are indicated. The second band of approximately 94 kD in roots corresponds to a different SucS isoform, probably MtSucS3. B, Relative transcript abundances of SucS genes, as determined via real-time RT-PCR in root nodule total RNA extracts at 37 dpi. The se is indicated. Bar color code: MtSucS1-antisense line as12 (dark gray); MtSucS1-antisense line as19 (gray); controls (white). Significant differences are indicated by a and b (P < 0.05 and P < 0.01, respectively). c, Transformation control; as12 and as19, MtSucS1-antisense lines 12 and 19, respectively.
In root nodule total RNA extracts from both antisense lines, significantly reduced steady-state MtSucS1 transcript levels were confirmed by real-time RT-PCR, being more pronounced in line as12 (12% of the MtSucS1 transcript abundance in the controls) than in line as19 tissues (24%; Fig. 1B). In addition to nodules, MtSucS1 expression was altered in leaves and roots, corresponding to the results on the protein level (data not shown). We verified the stable genomic insertion of the MtSucS1-antisense construct by Southern hybridizations, that way excluding that divergent MtSucS1 protein levels of lines as12 and as19 were caused by different numbers of insertions. These analyses not only revealed a characteristic MtSucS1-antisense fragment additional to the endogenous MtSucS1 fragments (Supplemental Fig. S1C), but also detected a transgene tandem insertion in each antisense line (Supplemental Fig. S1D). In both lines, the fragment patterns detected remained constant in the T1 to T5 progenies of selfed MtSucS1-antisense plants (data not shown), indicating a stable inheritance of the antisense fusion.
The Reduction of MtSucS1 Levels Impairs Above-Ground Biomass Production under Nodulation-Dependent Conditions
To analyze the phenotypic changes in MtSucS1-reduced lines, the transgenic plants were grown in the greenhouse and their growth behavior was monitored. In addition to MtSucS1-antisense lines as12 and as19, two independent transgenic M. truncatula lines, expressing promoter MtSucS1-gusAint fusions (Hohnjec et al., 2003), were used as controls (subsequently named c). Under nitrate-limited, nodulation-dependent conditions, plants expressing the MtSucS1-antisense construct showed overall stunted vegetative growth (Fig. 2). The impaired phenotypes, in terms of reduced plant height, leaf development, as well as flower and pod numbers, were analyzed over a time course from 0 to 58 d postinoculation (dpi). In general, distinct phenotypic alterations of plant size were more pronounced in as12 than in as19 plants, corresponding to the level of MtSucS1 reduction. To quantify phenotypic changes in the timing of the establishment of vegetative organs, a framework for precise characterizations of plant development was adopted from Moreau et al. (2006). This work takes temporal developmental patterns of vegetative and reproductive organs into account, such as leaf initiation, appearance, growth rate, and endurance. An impact of MtSucS1 reduction on vegetative above-ground development was found in the average developmental timing of the main axis and its five primary branches (B0–B4). When the rate of leaf initiation (RLi), i.e. the mean number of leaves originated per dpi, was calculated over the entire experimental period (0–58 dpi), this growth parameter was reduced in as12 (66% for RLias12) and as19 (54% for RLias19) in relation to controls (RLic; Table I; Supplemental Fig. S2A). Accordingly, the mean plastochrons, i.e. the time intervals to initiate two chronological leaves of MtSucS1-reduced lines are significantly 1.5- and 1.9-fold extended in as12 and as19, respectively (Table I). Similarly, the average number of leaves appeared per dpi (RLa) was 1.4- to 1.7-fold reduced in MtSucS1 transformants. This corresponds to significantly elongated phyllochrons (Table I; Supplemental Fig. S2B). In addition, we observed reductions in the timing and rate of primary branching of the axes in both MtSucS1-transformed lines (data not shown).
Figure 2.
Phenotype of transgenic lines expressing the p35S-MtSucS1-antisense construct. Plants, grown under nitrate-limiting growth conditions, were inoculated with S. meliloti 1021 in pots (left section; A–D) and with S. meliloti RCR2011 pXLGD4 in aeroponic caissons (right section; E–G), respectively. A to D, Phenotype of MtSucS1-antisense lines as12 and as19, and corresponding controls at 29 (A), 32 (B), 37 (C), and as an overview at 32 dpi (D). Ordering of plant lines, starting from the left position: control lines (position 1 and 2, respectively), MtSucS1-antisense line as19 (position 3), MtSucS1-antisense line as12 (position 4). E, Phenotype of control (left) and MtSucS1-antisense as12 (right) lines, grown in aeroponic caissons at 26 dpi. F and G, Above-ground biomass production of control (F) versus MtSucS1-antisense as12 (G) lines at 26 dpi. H and I, Phenotype of MtSucS1-antisense lines as12 and as19, and controls under fully supplemented conditions. Ordering of plant lines in E to I is as in A to D. J to M, Dark-field micrographs of longitudinal, cleared sections of root nodules at 22 dpi for control lines c1 and c2 (J and K), MtSucS1-antisense line as19 (L), and MtSucS1-antisense line as12 (M). Root nodule zoning and the frequency of infected cells in the central symbiotic region were comparable for both the MtSucS1-antisense and the control lines. Pronounced senescing was evident in as12 nodules that displayed an increased yellowish accumulation of collapsed cells. N to Q, Fluorescence imaging of S. meliloti-derived mRFP in longitudinal nodule sections at 18 dpi using confocal laser-scanning microscopy, indicating a similar degree of colonization in nodules of both the MtSucS1-antisense and the control lines. N and O, control lines c1 and c2. P, MtSucS1-antisense line as19. Q, MtSucS1-antisense line as12. Scale bars = 200 μm (J–Q).
Table I.
Above-ground biomass development in MtSucS1-reduced and control lines
Values given are means ± se. Significant differences are indicated by a and b (P < 0.05 and P < 0.01, respectively). n, Number of replicates; RFi, rate of flower initiation.
| Line | n | RLia (Leaf Primordia/dpi) | RLaa (Leaves/dpi) | RFibc (Flower Primordia/dpi) | Plastochrona | Phyllochrona | Florochronbc |
|---|---|---|---|---|---|---|---|
| dpi | |||||||
| as12 | 6 | 0.300b (±0.014) | 0.239b (±0.016) | 0.037a (±0.016) | 3.3b (±0.2) | 4.2b (±0.3) | 26.7a (±8.7) |
| as19 | 6 | 0.247b (±0.027) | 0.197b (±0.018) | 0.014b (±0.011) | 4.1b (±0.5) | 5.1b (±0.5) | 30.9b (±8.9) |
| c | 6 | 0.454 (±0.043) | 0.337 (±0.027) | 0.157 (±0.035) | 2.2 (±0.3) | 3.0 (±0.3) | 6.4 (±1.1) |
Time interval between 0 and 58 dpi.
Time interval between 35 and 58 dpi.
Mean only for individuals already started flowering.
Using the decimal code for leaf developmental stages, the entire averaged leaf production (as leaf unit) in the time interval between 0 and 58 dpi was quantified (data not shown). The initial slopes of the leaf rate curves from 0 to approximately 20 dpi were similar for MtSucS1 transformants and controls, indicating equal growth rates at those early stages of the symbiosis. At 27 dpi for as12 and 38 dpi for as19, respectively, leaf units of the MtSucS1-antisense lines started to be significantly reduced. Concerning the assimilating surface produced, total leaf area of line as12 tended to decrease, while leaves of as19 were comparable to those of controls (data not shown). In particular for line as12, we observed a 1 to 2 weeks earlier senescence of older leaves (data not shown).
The impact of MtSucS1 reduction on photosynthetic performance was determined via measuring the photosynthetic yield (ΦPSII), calculated from chlorophyll a fluorescence transients on the youngest and the oldest fully expanded leaf pairs. As pictured in Supplemental Figure S3, ΦPSII during steady-state photosynthesis showed a tendency to decline with the leaf age. Photosynthetic performance of leaves initiated by the stronger MtSucS1-reduced line as12 was lowered (Supplemental Fig. S3) and the percentage decrease in the rate of photosynthesis was particularly pronounced in old leaves (78.6%).
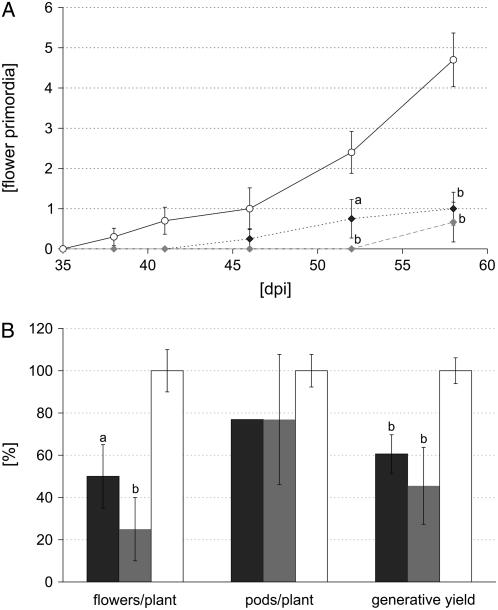
Finally, the reproductive development of MtSucS1-reduced lines lagged behind controls and morphological markers such as flowering and seed maturation were found significantly delayed under nodulation-dependent conditions. In general, the date of appearance of the primary reproductive node, bearing at least one emerged flower primordium, was retarded 9 d for line as12 and 21 d for line as19 (Fig. 3A). The initiation of flowering correlated with the delay in leaf development. After the appearance of 11.5 and 12.5 leaves, respectively, MtSucS1-antisense lines as12 and as19 started to produce flowering buds, whereas controls already started the reproductive phase when 10.8 leaves were completely unfolded. This tendency was even more pronounced for flower development in relation to leaf initiation (data not shown). Concerning the chronological development of reproductive nodes, the time interval to initiate two successive flowers (florochron) rapidly elongated in a range of 4- and 5-fold for as12 and as19, respectively (Table I). Since the delay in the establishment of florescence was linked to reduced and decelerated seed maturity (data not shown), the generative yield (i.e. the total amount of flowers and pods) was significantly reduced to 61% and 46% in MtSucS1-as12 and -as19 at time point 58 dpi, respectively, while the number of aborted floral organs was found to be highest for as12 plants (Fig. 3B).
Figure 3.
Generative development in nodulated MtSucS1-reduced and control lines. A, Time course of flower production (35–58 dpi). B, Generative yield (total flower and pod numbers) at 58 dpi. Data are given as averages ± se for at least six individuals per line. Significant differences are indicated by a and b (P < 0.05 and P < 0.01, respectively). Relative values are in comparison to control levels set to 100%. Color code in A: MtSucS1-antisense line as12 (dark gray squares); MtSucS1-antisense line as19 (gray squares); controls (white circles). Bar color code in B: MtSucS1-antisense line as12 (dark gray); MtSucS1-antisense line as19 (gray); controls (white).
Reduced MtSucS1 Levels Enhance Root to Shoot Ratios under Nodulation-Dependent Conditions
Similar to the above-ground biomass development, MtSucS1 reduction led to a decreased production of root material under nodulation-dependent conditions. Here, absolute root fresh weights (FWs) for MtSucS1-antisense line as12 and as19 averaged 0.27 and 0.28 g, respectively, in comparison to those of controls averaging 0.47 g (Table II). Since root and shoot weights are maintained at a certain balance that is characteristic for each species, root growth has to be related to above-ground biomass production. The root to shoot ratio (RSR), a commonly used parameter for the biomass partitioning to above- and below-ground material, is one measure to assess not only growth, but also the overall health status of plants. In nodulated MtSucS1-antisense lines, relative root growth increased, revealed by significant 1.8- and 1.7-fold increases in the RSR for MtSucS1-antisense lines as12 and as19, respectively (Table II; Supplemental Fig. S5). By contrast, for plants grown under nonstressed, nonsymbiotic conditions with sufficient nutrient and water supply (Fig. 2, H and I), the RSR based on FW for MtSucS1-antisense lines averaged 0.73, not significantly different from the value monitored for control plants (0.66; data not shown) and similar to the known average of approximately 0.8 for a wide range of fully supplemented plants (Waring et al., 1998). Like FW parameters, we measured significant differences in the percentage of total dry weight (DW) for the below-ground material of as lines, indicated by increases of 1.3- and 1.4-fold in root weight ratios (RWRs; DWroot per unit DWplant), an effect that was most pronounced in line as19 (Table II).
Table II.
Plant weight in MtSucS1-reduced and control lines
Values given are means ± se. Significant differences are indicated by a and b (P < 0.05 and P < 0.01, respectively). n, Number of replicates.
| Line | n | Total Plant Mass
|
Root Mass
|
RSR | RWR | ||
|---|---|---|---|---|---|---|---|
| FW | DW | FW | DW | ||||
| g | |||||||
| as12 | 6 | 0.44b (±0.04) | 0.045b (±0.004) | 0.27a (±0.03) | 0.012b (±0.001) | 1.50b (±0.10) | 0.26b (±0.10) |
| as19 | 6 | 0.49a (±0.07) | 0.045b (±0.004) | 0.28a (±0.04) | 0.016b (±0.002) | 1.40b (±0.09) | 0.29a (±0.09) |
| c | 12 | 1.05 (±0.18) | 0.130 (±0.021) | 0.47 (±0.08) | 0.026 (±0.004) | 0.83 (±0.04) | 0.21 (±0.04) |
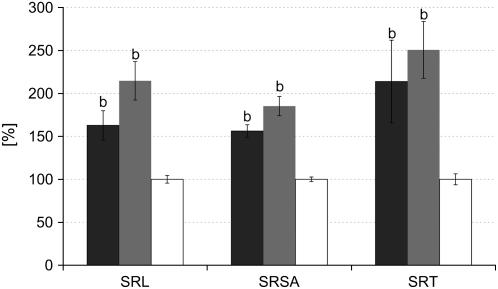
Morphological markers of root architecture, i.e. root length, root surface area, and root tip numbers are important parameters, indicating critical functions such as nutrient and water acquisition. Although total root masses were reduced at 47 dpi, most parameters tended to increase in MtSucS1-reduced lines (Table III). In particular, as19 plants exhibited significant increases in total root length, surface area, and root tip numbers. This corresponds to a more pronounced relative allocation of dry matter to the as19 root system (Table II). On the basis of root parameters normalized to the DWshoot, a highly significant increase in root system formation in both MtSucS1-reduced lines was found under nodulation-dependent conditions (Fig. 4; Supplemental Fig. S4). Here, the shoot-controlled regulation of root formation led to a markedly increased specific root elongation accompanied by enhanced lateral root growth. This was indicated by a 3.1- to 3.5-fold enrichment of root tips per unit DWshoot (specific root tip numbers) and a significant 1.6- to 2.1-fold increase in the specific root length in lines as12 and as19, respectively. In contrast, in fully N-supplemented plants of all lines studied, no significant increases in these parameters were observed (Supplemental Fig. S6), indicating that the MtSucS1-antisense lines are not significantly altered in C partitioning to the roots under conditions of sufficient external N supply.
Table III.
Root morphology markers root length, surface area, volume, and root tip numbers in MtSucS1-reduced and control lines
Values given are means ± se. Significant differences are indicated by a and b (P < 0.05 and P < 0.01, respectively). RL, Root length; RSA, root surface area; RV, root volume; RT, root tip number; n, number of replicates.
| Line | n | Root Morphology Markers
|
Mass Parameters
|
||||
|---|---|---|---|---|---|---|---|
| RL | RSA | RV | RT | RSR | DWshoot | ||
| cm | cm2 | cm3 | mg | ||||
| as12 | 11 | 229.5 (±15.5) | 38.2 (±3.8) | 0.518 (±0.072) | 240 (±37) | 1.46b (±0.05) | 66.06b (±8.22) |
| as19 | 11 | 293.8a (±22.6) | 44.9 (±5.3) | 0.567 (±0.092) | 282a (±27) | 1.47b (±0.07) | 66.77b (±9.71) |
| c | 22 | 253.6 (±16.7) | 41.4 (±2.7) | 0.545 (±0.039) | 212 (±5) | 0.88 (±0.03) | 113.10 (±7.58) |
Figure 4.
Root development in nodulated MtSucS1-reduced and control lines at time point 44 dpi. Data are given as averages ± se for 12 individuals of MtSucS1-antisense line as12 and as19, respectively, and 22 control plants. Relative values are in comparison to control levels set to 100%. Significant differences are indicated by a and b (P < 0.05 and P < 0.01, respectively). Bar color code, MtSucS1-antisense line as12 (dark gray); MtSucS1-antisense line as19 (gray); controls (white). SRSA, Specific root surface area; SRT, specific root tip number.
Reduced MtSucS1 Levels Affect Root Nodule Development and Symbiotic Nitrogen Fixation
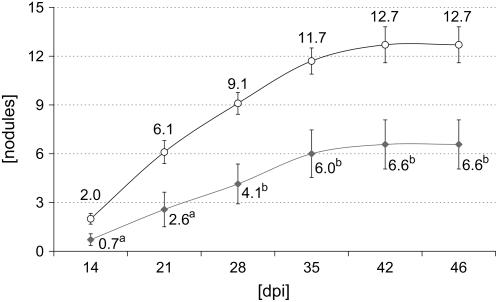
We examined root nodule formation in both MtSucS1-antisense lines over a time period of 47 dpi, using N-limited conditions. At time point 47 dpi, the number of nodules formed was 1.4- and 1.9-fold reduced in antisense line as12 and as19, respectively (P < 0.05; Table IV). In addition, specific nodule numbers SNshoot and SNroot (nodule number normalized on DWshoot and DWroot, respectively) showed an increase with the level of MtSucS1 reduction and were therefore significantly altered in antisense line as12 (Table IV). Based on the chronology of nodulation of MtSucS1-antisense line as19 and the control lines in a time interval between 0 and 46 dpi, our data suggest an impaired nodule formation in response to MtSucS1 reduction. A significant reduction in nodule numbers was apparent from time point 14 dpi on and remained constant, leading to a 1.9-fold reduction of nodule numbers in line as19 at 46 dpi (Fig. 5). This nodulation phenotype on plates corresponded to results derived from open pot experiments (Table IV).
Table IV.
Nodulation parameters in MtSucS1-reduced and control lines
Nodulation data are for plants at 47 dpi. Values given are means ± se. Significant differences are indicated by a and b (P < 0.05 and P < 0.01, respectively). n, Number of replicates; SNshoot, specific nodule number normalized on DW shoot; SNroot, specific nodule number normalized on DW root.
| Line | n | Nodule Parameters
|
Specific Nodulation Parameters
|
|||
|---|---|---|---|---|---|---|
| Nodules/Plant | DWnodules | Specific DWnodule | SNshoot | SNroot | ||
| mg | mg | mg−1 | mg−1 | |||
| as12 | 11 | 23.7b (±1.9) | 1.411a (±0.085) | 0.063 (±0.005) | 0.450a (±0.052) | 1.570 (±0.128) |
| as19 | 11 | 18.1b (±1.5) | 1.444 (±0.133) | 0.082a (±0.008) | 0.324 (±0.041) | 1.118 (±0.108) |
| c | 22 | 33.4 (±2.5) | 1.670 (±0.097) | 0.057 (±0.006) | 0.319 (±0.021) | 1.171 (±0.103) |
Figure 5.
Nodulation time course of MtSucS1-antisense line as19 in comparison to control lines, using plants grown on plates. Color code: MtSucS1-antisense line as19 (gray squares); controls (white circles). Significant differences are indicated by a and b (P < 0.05 and P < 0.01, respectively, for n = 10).
Concerning their morphology as well as the number of infected cells, root nodules of MtSucS1-antisense and control lines were comparable (Fig. 2, J–M), and the usage of a Sinorhizobium meliloti strain expressing a constitutive red fluorescent protein (Smit et al., 2005) demonstrated that also the degree of infection was similar (Fig. 2, N–Q). In contrast, MtSucS1-antisense nodules displayed symptoms of an earlier senescence in the central part of the distal symbiotic zone, in particular in the strongly MtSucS1-reduced line as12 (Fig. 2, J–M), suggesting consequences for the efficiency of SNF.
An indirect method of determining specific nitrogenase activity via acetylene reduction (AR) assays was applied to date the onset of N2 fixation, using complete plants grown on nodulation plates. MtSucS1-as19 and -as12 root nodules achieved initial AR (>5% of the AR from control plants) at time points 13 and 17 dpi, respectively, whereas control nodules started to support N2 fixation significantly earlier at 11 dpi (Supplemental Fig. S7). To overcome known shortcomings of the AR assay (Witty and Minchin, 1988) and since this method can only measure N2 fixation capacity over a short time period at the day of sampling, the total C and N content of plant tissues was determined. The calculation of C to N (C/N) ratios is able to derive more long term views of the N2 fixation activity of transgenic plant lines (Carlsson and Huss-Danell, 2003), and C/N ratios include structural as well as storage components.
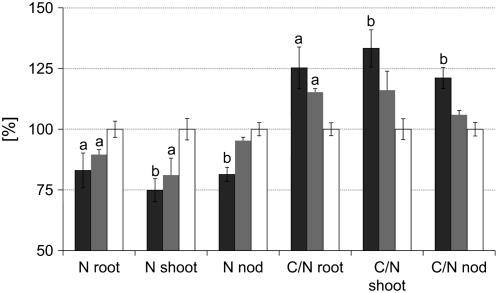
Both in control and wild-type plants, C and N accounted for about 39% and 2.8% in whole-plant DW, respectively, whereas MtSucS1-antisense lines were significantly affected in their contents of C (36% in as12; 38% in as19) and N (2.2% in as12; 2.5% in as19), in line with earlier senescence observed in the distal symbiotic zone of MtSucS1-antisense nodules. When a more detailed analysis was performed, using the plant organs shoot, root, and root nodules, the N content decreased with the level of MtSucS1 reduction in both antisense lines. As shown in Table III, after 47 dpi of N deprivation, the N content significantly decreased approximately 1.2-fold in root and nodule tissues of as12 plants. Here, the less efficiently MtSucS1-reduced line as19 reached the control nodule N level, but in root dry matter a significant 1.1-fold decrease was detected. The N differences were most pronounced and significantly reduced in shoot material of both antisense lines (1.3- and 1.2-fold, respectively). Taking DWshoot into account, the total amount of N accumulated in the above-ground material is 4.3- and 3.2-fold reduced in MtSucS1-antisense lines as12 and as19, respectively (Table V). Considering our results for the root morphology markers and the N yield, especially for the root length normalized to the above-ground N content, a highly significant increase of 181% and 234% was determined for as12 and as19, respectively (data not shown). While the C content of antisense and control lines largely was in the same order for the tissues studied (data not shown), declined N levels in MtSucS1-antisense material led to a significantly increased C/N ratio in all tissues tested from as12 and in root and shoot material of as19 (Table V; Fig. 6).
Table V.
C and N content in nodule, root, and shoot tissues of MtSucS1-reduced and control lines
Percentage N and C content in dry matter of root, shoot, and nodule tissue samples grown under nodulation-dependent conditions at 47 dpi. Values given are means ± se. Significant differences are indicated by a and b (P < 0.05 and P < 0.01, respectively). RL, Root length.
| Line | n | Nodules
|
Roots
|
Shoots
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| N | C/N | N | C/N | N | C/N | Nshoot | RL/Nshoot | ||
| % | % | % | mg | m/mg | |||||
| as12 | 5 | 5.05b (±0.18) | 8.13b (±0.29) | 1.65a (±0.14) | 22.33b (±1.53) | 2.50b (±0.16) | 16.89b (±0.97) | 0.82b (±0.06) | 2.14a (±0.26) |
| as19 | 5 | 5.91 (±0.09) | 7.11 (±0.12) | 1.78a (±0.04) | 20.54b (±0.27) | 2.71a (±0.24) | 14.71 (±0.99) | 1.08b (±0.25) | 2.60a (±0.49) |
| c | 10 | 6.20 (±0.17) | 6.71 (±0.19) | 1.99 (±0.07) | 17.82 (±0.47) | 3.34 (±0.15) | 12.67 (±0.55) | 3.50 (±0.72) | 0.83 (±0.07) |
Figure 6.
C-N balance of root, shoot, and nodule tissue samples of MtSucS1-antisense lines as12 and as19 in comparison to control lines. C and N were measured in dried tissues and results are expressed as relative values of the DW. The remaining components of the DW include elemental hydrogen, oxygen, and salts. Data represent means ± se from six plants per MtSucS1-antisense line and 12 controls. Significant differences are indicated by a and b (P < 0.05 and P < 0.01, respectively). Relative values are in comparison to control levels set to 100%. Bar color code: MtSucS1-antisense line as12 (dark gray); MtSucS1-antisense line as19 (gray); controls (white). nod, Nodule.
Under the constant experimental growth conditions used, these results point to specific differences in N uptake among MtSucS1-antisense and control lines. In particular with respect to the whole-plant N levels, our analyses demonstrate that root nodule formation is reduced and SNF is impaired in MtSucS1-antisense lines (Table IV; Fig. 5).
MtSucS1 Reduction Affects the Expression of Genes Connected to Nodule C and N Metabolism
To link the phenotypical data with transcriptional alterations elicited during MtSucS1 reduction in N2-fixing root nodules, we examined the expression profiles of 78 M. truncatula and 13 S. meliloti (Sm) genes, coding for proteins associated with nodule metabolism and maintenance of SNF (for genes and primer sequences see Supplemental Table S1). Apart from all known members of the MtSucS gene family, we focused on several Inv as well as other genes of the C and N metabolism downstream of Suc breakdown. These primarily included genes involved in the catabolism of photosynthates, the assimilation of fixed N, molecular transport, and cell wall synthesis, all of them reported to be predominantly or exclusively expressed in mature M. truncatula nodules. Additionally, transcript levels of 13 S. meliloti genes with assigned functions for an efficient N2 fixation have been determined. Gene expression patterns in fully developed nodules were compared between MtSucS1-reduced and control lines. Nodules without any adherent root tissues were harvested at 26 dpi from plants grown in parallel in aeroponic caissons, at a stage of plant development shown in Figure 2, E to G.
The analysis of gene expression ratios revealed that 48 plant and eight Sm genes appeared to be >1.5-fold differentially regulated at least in one MtSucS1-antisense line, with 27 plant and four bacterial genes being altered more than 2-fold (Supplemental Table S2). The expression of about 74% and 62% of the plant and Sm genes tested, respectively, was therefore affected by MtSucS1 reduction in root nodule tissues. For easier visualization in a hierarchical clustering, only the >1.5-fold differentially expressed genes were taken into consideration, almost all of them significantly regulated at P < 0.05 in at least one of the two antisense lines (Fig. 7A). Using this criterion, four clusters of expression profiles were distinguished. Genes were either identified as consistently down-regulated (cluster I) or up-regulated (cluster III) in nodules of both MtSucS1-reduced lines. In cluster II, transcript levels were different between the two antisense lines studied. Cluster IV represents Sm genes, identified as being differentially expressed in bacteroids of MtSucS1-reduced root nodules.
Figure 7.
Hierarchical clustering of the expression of selected M. truncatula and S. meliloti genes as well as the steady-state levels of metabolites in root nodules of MtSucS1-antisense lines as12 and as19. A, Cluster analysis of 48 M. truncatula and eight S. meliloti genes that change expression at least 1.5-fold in MtSucS1-antisense line as12 and as19 root nodules at 26 dpi in comparison to controls, as determined by real-time RT-PCR (expression ratios in Supplemental Table S3). M. truncatula genes are clustered according to the expression patterns: I, consistent down-regulation; II, MtSucS1-antisense line-dependent expression patterns; III, consistent up-regulation. Cluster IV includes genes differentially expressed in S. meliloti bacteroids of MtSucS1-reduced root nodules. Significant differences are indicated by a and b (P < 0.05 and P < 0.01, respectively) for as12 and as19 (as12/as19). Fold-changes represent ratios from mean nodule expression data derived from at least three biological replicates per MtSucS1-antisense line and six per controls. Each biological replicate was based on nodule pools derived from three individual plants. Hierarchical clustering was carried out using average linkage values. B, Cluster analysis of 80 metabolites from 21 dpi root nodules of MtSucS1-antisense lines as12 and as19 that change response values at least 1.2-fold, as determined by GC-MS (metabolite ratios in Supplemental Table S3). For this analysis, a sensitive visualization scale was used where a metabolite ratio of antisense versus control root nodules of ±3 was assigned the strongest color intensity, allowing to better depict general trends of changes in metabolite levels. The color code relates to the metabolite ratios (green, decreased levels; red, increased levels). Metabolites are clustered according to steady-state levels and classified into groups based on altered levels in root nodules of both MtSucS1-antisense lines: I, lower abundance; II, MtSucS1-antisense line-dependent abundances; III, higher abundance with MtSucS1 reduction. Significant differences are indicated by a and b (P < 0.05 and P < 0.01, respectively). Fold-changes represent ratios of mean normalized data from at least six biological replicates per MtSucS1-antisense and 10 per control line. Each biological replicate was based on pools of nodules derived from three individual plants. Hierarchical clustering was carried out using average linkage values.
The expression of 18 out of 65 genes included in this study was significantly down-regulated in both MtSucS1-antisense lines (Fig. 7A; cluster I). Among those were the MtSucS1 (TC100410) transcripts that were found sharply reduced via MtSucS1-antisense expression (reduction levelas12 −7.9; reduction levelas19 −3.0). These transcriptional reductions observed coincide with the level of MtSucS1 protein reduction, estimated from western-blot analyses on root nodules of lines as12 and as19, respectively (Fig. 1). Other MtSucS genes (TC95820, TC9447, TC98648, TC99016) appeared to follow MtSucS1 reduction. Nevertheless, threshold cycle values for these SucS genes were generally high in control nodules, in line with a low abundance of these transcripts in nodule tissues (Hohnjec et al., 2003) and confirming that MtSucS1 is the major SucS gene expressed in the root nodule symbiosis.
Among the genes that were repressed in MtSucS1-reduced nodules, several are involved in Suc breakdown, such as cell wall Inv (TC107108), vacuolar acid Inv PsI-1 (NF062D01), neutral Inv (TC106499), and acid Inv (TC99506) genes. In coincidence with transcriptional reductions of genes functioning in Suc cleavage, the expression of Suc transporter genes (TC107845, EST332714), several genes encoding monosaccharide transporters (TC99699, TC101691), and an H+/monosaccharide cotransporter (TC107440) were turned down. In the MtSucS1-antisense lines, down-regulation was also observed for genes specifying the myb-related, tuber-specific, and Suc-responsive element binding protein (TC103015) and a β-amylase (TC94273), previously shown to be induced in mature, starch-degrading root nodules (El Yahyaoui et al., 2004). Finally, a gene encoding a nodule-enhanced phosphoenolpyruvate carboxylase (PEPC; TC107075) has been found to be repressed in MtSucS1-antisense lines, indicating a reduced glycolytic activity and an impaired formation of oxalacetate.
Cluster II comprised 11 genes that were activated in line as19 and repressed in as12 (Fig. 7A). Among those were genes related to Suc breakdown in several legume tissues, e.g. encoding an acid β-fructofuranosidase precursor (TC96236) and the cell wall Inv2 (TC103706; Weber et al., 1995). In addition, genes specifying a H+/hexose cotransporter (TC95549), a Suc-P synthase (TC107513), a trehalose-6-P phosphatase (TC94692), a UDP-Glc-6-dehydrogenase (TC100484), and a UDP-sugar pyrophosphorylase (TC94709) showed marked repression in the strongly MtSucS1-reduced line as12, whereas these genes related to C metabolism appear to be transcribed more similar in as19 and control nodules. Interestingly, a 2.1-fold induction of an enolase gene (TC100309) was observed in MtSucS1-antisense nodules of as19, whereas the expression level of this member of the glycolytic pathway was slightly repressed in line as12.
Based on the criterion of >1.5-fold relative induction, 14 tentative consensus sequences (TCs) showed increased transcript levels in MtSucS1-antisense root nodules (Fig. 7A, cluster III). Among those were primarily genes involved in amino acid biosynthesis such as cytosolic Asp aminotransferase (AAT) 1 (TC106918), Gln synthetase (TC106913), and Gln synthetase 1 (TC96622), all of them induced in efficient Medicago root nodules (Mathis et al., 2000; Tesfaye et al., 2006). Strikingly, comparisons of expression profiles revealed that both MtSucS1 transformants exhibited a substantial induction of a nitrate reductase (NR) gene (TC107357), indicating that the nonsymbiotic N acquisition pathway might be transcriptionally activated in MtSucS1-reduced nodules. Since there was no activation of further SucS isogenes, other genes specifying enzymes for Suc breakdown might have been recruited in MtSucS1-antisense nodules. In this respect, it is interesting to note that a gene encoding an alkaline/neutral Inv (TC106886) was 2.6- and 1.6-fold induced in root nodules of lines as12 and as19, respectively. A similar increase in transcript levels was seen for genes involved in the formation of dicarboxylic acids such as malate dehydrogenase (MDH; TC107189) and carbonic anhydrase (CA; TC100150).
S. meliloti genes important for N2 fixation processes were tested for transcriptional activity in both antisense lines. Included in this set were genes that encode components implicated in the nutritional requirements of the bacteria, proteins potentially involved in transcriptional regulation, and those with significance for N2 fixation and transport processes. Five out of 13 Sm genes tested were significantly repressed in nodules of both MtSucS1-altered lines (Fig. 7A, cluster IV), none of the genes tested was found induced in as12 and only two (SMa0831 and SMa0817) were slightly enhanced in bacteroids of line as19 root nodules.
The dctA gene, encoding an ion-coupled membrane-spanning C4-dicarboxylate transporter (SMb20611; Jording and Pühler, 1993), responsible for uptake of dicarboxylates across the inner membrane of the microsymbiont (Bolton et al., 1986), was 1.7-fold repressed in nodules of line as12. In contrast, its expression was equal to controls in the less MtSucS1-reduced line as19. Notably, dctA expression in response to C4 dicarboxylates and the activation of the N2 fixation (nif) and fixation (fix) operons are thought to be controlled via regulatory transcriptional activator NifA (SMa0815; Gong et al., 2006). In transgenic line as12, nifA expression was similar to controls, whereas a minor gene induction was found in as19 root nodules (Supplemental Table S2). Accordingly, genes organized in the nif/fix operons such as genes encoding the N2 fixation protein NifX (SMa0831), the oxidoreductase protein FixC (SMa0817), the nitrogenase NifD (SMa0827), and NifH (SM a0825) showed similar expression patterns (data not shown), while only the transcription of a gene coding for the electron transfer flavoprotein FixB (SMa0819) consistently decreased in nodules of both MtSucS1-reduced lines (Supplemental Table S2). Interestingly, S. meliloti pyruvate dehydrogenase (SMb20020) and diphosphopyridine nucleotide-dependent malic enzyme (SMc00169) gene expression was strongly reduced, indicating impaired acetyl-CoA production in bacteroids of MtSucS1-antisense lines. Concerning transport between macro- and microsymbionts, significant transcriptional repression in bacteroids of MtSucS1-reduced nodules were measured for genes encoding the putative Glu/Asp transport protein (SMa0677) and the putative ATP-binding cassette transporter periplasmatic amino acid binding protein (SMb20263), both purposed to be involved in the connection of the metabolisms of the two symbiotic partners (Lodwig et al., 2003).
MtSucS1 Reduction Is Accompanied by Changes in the Metabolite Pools of Root Nodules
Since plant development is obviously altered and genes related to the C and N metabolism are affected by MtSucS1 reduction in transgenic M. truncatula root nodules, we tried to obtain a more detailed insight into nodule metabolism by measuring metabolic alterations via gas chromatography-mass spectrometry (GC-MS). A comparison of the metabolite composition in symbiotic root nodules of MtSucS1-altered and control plants harvested at 21 dpi covered changes in 80 metabolites that mainly represent the primary metabolic pool, ranging from 1.1- to 4.3-fold differences (Supplemental Table S3). Based on the quantitative data, these metabolite changes were hierarchically clustered for visualization (Fig. 7B).
Several metabolic compounds, representing important nodes in glycolysis, the tricarboxylic acid (TCA) cycle, or amino acid biosynthesis, appeared to be preferentially and coordinately down-regulated in MtSucS1-reduced nodules (Fig. 7B; cluster I). The most apparent differences were found for certain proteinogenic and nonproteinogenic amino acid pools. Significant metabolic changes, in a range of −1.2 to −4.0 were monitored for Met, Trp, Leu, Phe, Gly, Pro, Arg, Ile, Lys, Tyr, Thr, His, HSer, and HCys. Several precursors and intermediates of the amino acid biosynthesis pathways (e.g. d/l-diaminopimelate, 2-aminoadipate, l,l-cystathionine) and amino acid derivatives such as Orn or N-acetyl-l-Glu also exhibited significantly reduced levels in nodules of both MtSucS1-antisense lines. Val and Cys decreased preferentially in nodules of the strongly MtSucS1-reduced line as12, while these compounds remained largely unchanged in as19 nodules (Fig. 7B, cluster II). Intriguingly, Asn as the main transport form of fixed N, showed a 1.6- and Gln a 1.4-fold reduction in as12 nodules, while these compounds were measured at 1.2- and 1.7-fold enhanced levels, respectively, in line as19. Glu was marginally altered in as12 root nodules and showed increased abundance in as19 extracts.
Comparing the summarized content of all amino acids measured, the nodule tissue of line as12 only comprised 69% of the levels detected in controls, whereas line as19 exhibits a total amino acid content equally to controls (data not shown). These results are in accordance with the total N levels measured in root nodules presented earlier (Table V; Fig. 7B). Thus, the extent of reduced amino acid biosynthesis differed between the MtSucS1-transformed lines, correlating with the degree of SucS1 reduction. Among the lower abundant pools in both MtSucS1-reduced lines, metabolites representing the urea cycle (urea, Orn, Arg, citrulline) and diverse N-containing compounds (e.g. norvaline, agmatine, purines, and pyrimidines) were found. Also sugar acids derived from Glc such as glucarate, gluconate, and glycerate as well as dicarboxylic acids (e.g. malonate, maleate) consistently decreased in line as12 and as19 root nodules. In addition to the amino acids mentioned above, we found a considerable reduction of β-Ala, which can serve as an intermediate in CoA synthesis via the likewise repressed pantothenate. This seems to be in line with the fact that intermediates involved in the first part of the TCA cycle, such as isocitrate and 2-ketoglutarate, as well as (to a less extent) the bacteroid-fueling dicarboxylic acid malate were also reduced. Finally, the pools of the glycolytic intermediates dihydroxyacetone-P, phosphoenolpyruvate, and pyruvate were 1.5- and 1.4-fold decreased in both transgenic lines.
In MtSucS1-reduced nodules, substantial changes in levels of Suc, raffinose, Fru, and Glc were found, and the accumulation of Suc can be directly correlated with the level of MtSucS1 protein reduction. In contrast, the amounts of Fru-6-P and Glc-6-P, either metabolized in the glycolytic pathway and/or used for starch or cell wall synthesis, were slightly enhanced in MtSucS1-antisense nodules (Fig. 7B). Interestingly, the major hexose sugars Glc and Fru were found at higher abundances in MtSucS1-reduced lines, but these monosaccharides were not detected in approximately equal amounts, as would be expected from Suc breakdown. Notably, the MtSucS1-antisense lines as12 and as19 showed a 2.2- to 2.3-fold excess of Glc over Fru in nodule extracts, whereas in controls, the Glc to Fru ratio was build up to 3.4 (data not shown).
The metabolic changes measured for 21 dpi root nodules can also be observed at later stages of nodulation (Supplemental Table S3), but amino acid pools were less affected in MtSucS1-reduced root nodules at 26 dpi. This observation may coincide with reduced nodule numbers in the MtSucS1-antisense lines (Fig. 5) or could reflect a long-term adaptation of transgenic nodule tissues to MtSucS1-reduced conditions.
DISCUSSION
The goal of this work was to elucidate the relevance of the SucS MtSucS1 for N nutrition during symbiotic N fixation in the model legume M. truncatula. To this end, we constructed M. truncatula antisense plants that displayed a reduced expression of MtSucS1 at both the transcript and the protein level. Among those plants, the two independent, homozygous lines as12 and as19 were analyzed in detail, exhibiting up to 90% MtSucS1 reduction in roots as well as root nodules. Since it is well known that the nodule-enhanced (ne)-SucS amounts directly reflect SucS activity (Gordon and James, 1997; Horst et al., 2007), we analyzed whether the pronounced reduction of MtSucS1 levels in our transgenic lines had an effect on physiological parameters such as plant growth, organ formation, as well as C and N metabolism during nodulation of M. truncatula.
MtSucS1-Reduced Lines Suffer from N Starvation under Nodulation-Dependent Conditions
The repression of MtSucS1 led to a significantly impaired vegetative above-ground growth, when M. truncatula plants relied on N supply via nodulation. Differences between nodulated MtSucS1-reduced and control lines occurred in developmental timing, plant size, and leaf development. Additionally, a lowered photosynthetic performance and an enhanced leaf senescence accompanied with early abscission of older leaves were observed. These significant alterations in primary productivity and tissue maintenance are likely caused by N limitation, since it was reported that constrained N supply affects leaf development, reducing the rate of leaf expansion, the specific leaf area, and photosynthetic activity (McDonald et al., 1992).
Apart from reduced total plant weights, both nodulated antisense lines showed a delayed flowering and elongated florochron, reduced numbers of flowers, early abscissions of floral parts or immature pods, and a reduced number of seeds per pod. Consequently, the reproductive yields were found significantly reduced, while average seed masses increased in MtSucS1-reduced plants, particularly in line as19. The transition from vegetative growth to flowering, as well as seed size, pod, and seed numbers are determined by several biotic and abiotic factors (Bernier et al., 1993). In fact, there is strong evidence for a negative correlation between seed size, seed quality, and nutrient supply, accompanied by a selection for larger seeds under N- and P-limited growth (Mustart and Cowling, 1992). Particularly in legume species, low abundances of total N from soil, fertilizer, or SNF caused low seed yields, while sufficient SNF or N supply increased not only total plant weight, but also seed nutritional quality and yield (Hardarson et al., 1993; Burstin et al., 2007); effects were also observed for fully supplemented MtSucS1-antisense lines. The delayed and negatively affected above-ground development of nodulated antisense lines may thus be a direct effect of nonoptimal N acquisition under MtSucS1 reduction and at later developmental stages a consequence of exhausting N demands during flowering and seed maturation. In our study, we measured significantly decreased N levels not only in shoots and roots, but also in nodules of MtSucS1-reduced lines. These lowered N contents corresponded to the extent of MtSucS1 protein reduction and seemed to be directly correlated with reduced nodule formation as well as a delayed SNF. This leads to the conclusion that an impaired nodule initiation and maintenance accounts for the N starvation symptoms observed in MtSucS1-antisense nodules, possibly supported by the earlier senescence observed in comparison to control nodules. In line with this observation, a defective SNF and a premature senescence of root nodules was also described for the rug4 mutant of pea (Gordon et al., 1999).
Concerning root development, both nodulated antisense lines displayed significantly increased root-to-shoot and root-to-weight ratios. Plant root-shoot balance is considered a functional equilibrium, determining the nutrient uptake capacity of roots and being indicative of plant health. An extended root growth, e.g. resulting from a reduced soil fertility and nutrient shortage, thus indicates a common physiological stress condition (Marschner, 1995; Jørgensen and Ledgard, 1997). From this, we conclude that relative root growth of MtSucS1-reduced lines geared toward maximized soil contact to further improve nutrient acquisition under N-limiting conditions. Hence, root formation in MtSucS1-antisense lines might mirror a morphological feedback to N limitation and the relative increase in root growth can probably be valued as an attempt to overcome lower N acquisition caused by an inefficient SNF in MtSucS1-reduced nodules, at the immediate expense of shoot growth. Alternatively, since Suc catabolism in root nodules is impaired as a consequence of antisensing MtSucS1, Suc might in addition become increasingly available for root growth. Consequently, this higher level of C partitioning to the roots could result in less Suc being allocated to shoot growth in MtSucS1-reduced plants.
Since no growth differences were detectable between MtSucS1-antisense and control lines under N supplemented growth conditions, and since signs of N limitation are also well known from ineffective legume-Rhizobium interactions (Zahran, 1999) as well as from SucS-reduced root nodules during stress responses (Gálvez et al., 2005; Larrainzar et al., 2007), the observed symptoms of N starvation in nodulated MtSucS1-reduced lines primarily point to a handicapped SNF. Consequently, the observed phenotypes under conditions depending on an efficient symbiosis can be correlated with a lack of MtSucS1 in root nodules, a phenotype also observed for SucS knockout mutants in pea and L. japonicus (Craig et al., 1999; Gordon et al., 1999; Horst et al., 2007).
Transcriptional and Metabolic Profiling of MtSucS1-Reduced Nodules Support an Important Function of MtSucS1 during Nodulation
Having demonstrated the relevance of MtSucS1 for SNF, we focused on the elucidation of transcriptional and metabolic alterations downstream of Suc breakdown, to derive better insights into the M. truncatula root nodule physiology under conditions of MtSucS1 reduction. In root nodules, both antisense lines showed an up to 90% decline in SucS1 transcript and protein levels. Nevertheless, residual amounts may still lead to a basic Suc-cleaving activity in transgenic nodules, probably more pronounced in line as19.
The stress-inducing condition of an MtSucS1 reduction led us to monitor putative alternatives for Suc breakdown that might be induced to compensate for a reduced glycolysis. In this respect, we studied the expression of genes encoding different Inv. Interestingly, a moderate induction of an alkaline/neutral Inv gene (TC106886) occurred in MtSucS1-reduced root nodules, showing high sequence similarities to the Inv1 gene that is expressed during L. japonicus nodule development (Flemetakis et al., 2006), an observation similar to the activation of other alkaline/neutral Inv genes in soybean and M. truncatula nodules (Manthey et al., 2004; Tesfaye et al., 2006). By contrast, the M. truncatula neutral Inv gene (TC106499), formerly shown to be induced in root nodules (Tesfaye et al., 2006), was found to be moderately down-regulated under conditions of MtSucS1 reduction and the acid and cell wall Inv expression was not affected at all. Although not all M. truncatula Inv genes might be known to date and therefore could not be taken into account here, both the transcript profiles and the impaired plant development indicate a decreased total Suc cleavage capacity of MtSucS1-reduced nodules, being more pronounced in line as12. This is supported by the fact that an accumulation of Suc and the storage sugar raffinose occurs in nodule tissues. Thus, whether or not currently unknown Inv genes might be induced, they are clearly not able to fully substitute MtSucS1 enzymes to circumvent severe N starvation effects. Since Inv enzyme activity is not believed to be favored under hypoxia (Rolletschek et al., 2002; Albrecht and Mustroph, 2003), a condition prerequisite for efficient SNF in infected cells, Inv gene induction might be limited to the uninfected cells. We here cannot distinguish between the activation of genes in infected and uninfected nodule cells, but the induced Inv gene expression observed might only occur in the latter one.
It is well known that SNF activity relies on photosynthate supply to the nodule (Gordon et al., 1986) and in addition, the root nodule N metabolism is linked to C metabolism by the requirements for ATP, reductant, as well as C skeletons to assimilate ammonium (NH4+), all being derived from glycolysis and the TCA cycle (Vance et al., 1994). Thus, one could expect that due to the limited ability for Suc degradation in transgenic nodules, successive metabolic steps of SNF are impaired. Consequently, the reduced Suc breakdown might explain the significant decreases we observed for the metabolite pools of glycolysis and the TCA cycle. A similar Suc accumulation accompanied with malate depletion was also reported for the LjSUS3 mutant (Horst et al., 2007) and for wild-type nodules as a result of a stress-induced SucS down-regulation (Gálvez et al., 2005; Larrainzar et al., 2007), showing a direct correlation to stress intensity.
MtSucS1 reduction was also found to influence the transcriptome of the microsymbiont, since S. meliloti pyruvate dehydrogenase and diphosphopyridine nucleotide-dependent malic enzyme gene expression was strongly reduced in both antisense lines. The encoded enzymes are both known key players in the C catabolism of actively N2-fixing bacteroids, producing acetyl-CoA for an efficient TCA cycle in a concerted action. In addition, bacteroids in MtSucS1-reduced nodules exhibited decreased transcriptional activity of those genes encoding proteins for the transport of ketoacids and amino acids between microsymbiont and plant (Lodwig et al., 2003). From this, we conclude that an inefficient functioning of the microsymbiont respiratory chain and a subsequent down-regulation of the highly energy demanding N2 fixation process occurred in MtSucS1-antisense root nodules, resulting in a handicapped SNF.
Our transcriptional and metabolite results indicate a decreased respiratory C flow through glycolysis and TCA cycle, necessary for the production of C skeletons for NH4+ assimilation, or dicarboxylic acids to fuel bacteroid respiration (Fig. 8). The simultaneous responses of macro- and microsymbionts to MtSucS1 reduction in antisense nodules again strengthens the argument that SucS1 is of physiological importance for C supply during efficient root nodule endosymbioses.
Figure 8.
Scheme of the main metabolic pathways occurring in the cytosol, symbiosomes, and plastids of an infected nodule cell with a focus on amino acid biosynthesis. Alterations in MtSucS1-antisense line as12 metabolite profiles at 21 dpi are shown. Changes in metabolite levels were calculated as the ratios of MtSucS1-antisense line as12 versus controls and are listed in Supplemental Table S4. The n-fold changes in metabolite levels are indicated as followed: unaltered, 1- to 1.1-fold increase or decrease (gray letters); 1.1- to 1.3-fold (decrease, light green; increase, light red; respectively); >1.3-fold (decrease, green; increase, red). Nonmonitored substrates are represented in black. Note that not all reactions and reaction products are shown. Significant differences are indicated by a and b (P < 0.05 and P < 0.01, respectively; n: 10× as12; 23× c). Main amino acid families sharing a common precursor are grouped. DHAP, Dihydroxyacetone phosphate; N2ase, nitrogenase.
Metabolic Plasticity Could Alleviate the Effects of MtSucS1 Reduction in Transgenic Nodules
In root nodules, genes encoding a ne CA, a ne-MDH, and an AAT were up-regulated under MtSucS1 reduction. Nodule cytosolic PEPC catalyzes the conversion of PEP and bicarbonate into oxalacetate, involving the reversible hydration of CO2 via ne-CA for bicarbonate production (de la Pena et al., 1997). Subsequently, a ne-MDH produces malate, the dicarboxylic acid primarily supplied to the bacteroids (Miller et al., 1998). The C recycling from PEPC and CA activity consequently replenishes TCA cycle intermediates and therefore delivers a significant saving of the CO2 liberated during respiration (Warembourg and Roumet, 1989). Thus, the enhanced levels of MDH and AAT expression that coincide with MtSucS1 reduction could ensure a rapid use of PEPC-delivered oxalacetate, which may avoid feedback inhibition of the CO2-recycling pathway. Together with the CA induction, this could point to an enhanced C use efficiency via CO2 recycling under limited Suc cleavage capacity in MtSucS1-antisense root nodules. In addition, since the pentose phosphate pathway is involved to a minor extent in nodule C utilization and the provision of substrates for purine synthesis (Hong and Copeland, 1990), a moderate increase of pentose phosphate pathway metabolites could be considered as a further alternative to provide C substrates. Finally, it is noteworthy that Gln synthetase (GS) genes were found to be activated, while the levels of corresponding Gln and all other amino acids decreased. Together with the induction of MDH and AAT genes, this could reflect an attempt to drive the nodule metabolism toward an optimal nutrient supply for bacteroids and a fast incorporation of fixed N under conditions of limited C supply (Fig. 8). A limited C provision could either result from reduced C partitioning to the nodules specifically under conditions of SNF or from an impaired Suc cleavage in nodule tissues. The latter is more likely to contribute to C limitations in root nodules, since only under conditions where the plants depend on SNF, MtSucS1-antisense lines displayed significant phenotypes.
Assuming that an increased flux via a particular metabolite is indicated by its accumulation, our metabolite data generally support the conclusion from transcript profiling that amino acid biosynthesis is reduced in MtSucS1-antisense nodules. Decreases in certain amino acid are (at least in the stronger MtSucS1-reduced line as12) reflected in the expression of genes encoding key biosynthetic enzymes. According to previous reports (Vance et al., 1994), the major amino acid produced in amide-transporting N2-fixing root nodules is Asn. In nodules of MtSucS1-antisense line as12, significant reductions of Asn and Asn synthetase 1 (AS1) transcript levels presumably are signs of an impaired SNF, additionally stressed by the consistent depletion of all amino acids tested in this study. Similarly, the major amino acid residues of storage proteins in sinks, such as Arg, Gln, Glu, and citrulline (Allona et al., 1994) are most strongly reduced in the efficiently MtSucS1-reduced line as12.
Despite the obviously lowered amino acid biosynthesis, we observed enhanced transcription of ne-GSs and AAT1. The observation of an early senescence of MtSucS1-reduced (Fig. 2) as well as rug4 mutant nodules (Craig et al., 1999) indicates that the gene inductions measured could account for enhanced reassimilation of NH4+ into Gln during processes of protein turnover, e.g. using NH4+ derived from the urea cycle. Furthermore, NH4+ can be produced by the concerted action of nitrite and NR—the latter being markedly transcriptionally activated in MtSucS1-antisense nodules. The specific NR gene induction implies an enhanced activity of the direct, nonsymbiotic N acquisition pathway, possibly to overcome limitations in SNF.
Taken together, the metabolic plasticity observed in MtSucS1-antisense nodules (Fig. 8) apparently targets at the production of sufficient levels of key players in the linked C and N metabolism, to partially compensate for an MtSucS1 reduction during nodulation. This might explain the more alleviated MtSucS1 knockdown phenotype compared to the deletion mutant rug4 in pea (Gordon et al., 1992; Craig et al., 1999) as well as the LjSUS1/LjSUS3 double mutant in L. japonicus (Horst et al., 2007).
CONCLUSION
Although a loss of or a strong reduction in sink-specific SucS can force severe phenotypes in plants (Chourey et al., 1998; Ruan et al., 2003), several studies failed to define exclusive roles for distinct SucS isoforms in different plant species (Weckwerth et al., 2004; Bieniawska et al., 2007). In this context, it is interesting to note that an 80% to 90% reduction of MtSucS1 proteins in roots and root nodules specifically limited plant growth and organ development under nodulation-dependent conditions, leading to marked changes in expression of genes involved in the nodule C and N metabolism and consequently to a reduced performance of amino acid biosynthesis. Thus, in line with the observations in pea and L. japonicus knockout mutants, the integration of our developmental, transcriptional, and metabolic data not only supports an association between ne-SucS and an efficient SNF, but provides deeper insights into the nodule metabolism under conditions of MtSucS1 reduction. It will be interesting to evaluate the relative contributions of a reduced C allocation to the central nodule tissues and an impaired Suc cleavage therein. Finally, the availability of MtSucS1-reduced lines now allows to investigate, whether MtSucS1 is also critical for other endosymbiotic conditions of the model legume M. truncatula, e.g. the interaction with beneficial arbuscular mycorrhizal fungi.
MATERIALS AND METHODS
Isolation of Nucleic Acids, Recombinant DNA Techniques, Southern-, and Western-Blot Analysis
Recombinant DNA experiments were carried out as described in Hohnjec et al. (2003). Total RNA was isolated using the RNeasy Plant mini kit and DNase I on-column digestion according to the manufacturer's instructions (Qiagen). RNA preparations were tested for their integrity on agarose gels, were spectrophotometrically checked for yield and quality using a NanoDrop ND-1000 (NanoDrop Technologies), and were stored at −80°C until use.
For Southern-blot analyses of transgenic MtSucS1-antisense and control lines, approximately 25 μg of genomic plant DNA was digested with appropriate restriction enzymes, separated by electrophoresis through 0.8% (w/v) agarose gels, transferred to hybond N membranes, and hybridized to digoxygenin (DIG)-labeled MtSucS1 PCR fragments and a DIG-gus probe, respectively, at 42°C. The filters were washed three times in 2× SSC (1× SSC is 0.15 mm NaCl, 0.015 mm Na citrate), 0.1% SDS, twice in 1× SSC, 0.1% SDS, and once in 0.5× SSC, 0.1% SDS for 15 min each at 65°C. Chemiluminescence's documentation (via CDP-Star substrate, Roche Diagnostics GmbH) of the probed fragments was performed via alkaline phosphatase-linked anti-DIG antibody (Roche Diagnostics GmbH).
According to Hohnjec et al. (2003), proteins from 27 dpi old plant material were extracted, separated via acrylamide SDS-PAGE electrophoresis, and electroblotted onto nitrocellulose membranes. Subsequently, tissues SucS amounts were determined in immunohistologically via primary multiclonal antibody raised against broad bean (Vicia faba) SucS (Ross and Davies, 1992). Subsequently, an alkaline phosphatase conjugated secondary antibody was used and detected with the AP-substrate X-phosphate/nitroblue tetrazolium (Amersham Biosciences) as described by the manufacturer.
Construction of Transgenic MtSucS1-Antisense Lines
A 1,205 bp fragment covering the 5′ end of the previously isolated, 2,728 bp long, complete coding sequence of MtSucS1 (Hohnjec et al., 1999) was cloned into the binary vector MF2 (provided by F. Frugier, Centre National de la Recherche Scientifique). The insertion downstream of the constitutively active cauliflower mosaic virus 35S promoter (p35S) was done using native XbaI/NotI sites, resulting in an antisense orientation of the partial MtSucS1 sequence (Supplemental Fig. S1). The construct cloned was verified by DNA sequencing. Using standard protocols, the binary vector MF2∷p35S-MtSucS1 antisense was electroporated into the Agrobacterium tumefaciens strain EHA105 (Hood et al., 1993) to start leaf-disc transformation into Medicago truncatula ecotype R108-1 (Hoffmann et al., 1997). In accordance to the transformation protocols described by Hohnjec et al. (2003), M. truncatula R108-1 leaf segments were transformed with the MF2∷p35S-MtSucS1 antisense constructs. Transgenic F0 plants were grown in pots in sterile soil/vermiculite/sand (1:1:1) substrate in the growth chamber with 16 h light (22°C) and 8 h dark (18°C) cycles. For subsequent in planta transformation control, a p35S-driven gusAint (Vancanneyt et al., 1990) marker gene upstream of the p35S-MtSucS1 antisense construct in the MF2 vector was used. In the phenotypic analyses of MtSucS1-modulated plants under endosymbiotic conditions, two independently transformed, homozygous pMtSucS1-gusAint lines (Hohnjec et al., 2003) were used as control lines.
Conditions of Plant Growth and Microsymbiont Inoculation
Seeds from transgenic M. truncatula MtSucS1-antisense and control lines were scarified and surface sterilized (Trieu et al., 2000), spread on 0.8% water agar (agar-agar; Sigma) in inverted petri dishes, and stratified 4 d at 4°C, 2 d at RT in the dark, before being finally exposed to the light for 1 d to synchronize germination. For phenotypic characterizations, transgenic T1 to T4 descendants of the MtSucS1-antisense and control lines were grown in 27 cm3 pots filled with sterile clay granules (Seramis, Masterfoods), until the plantlets developed at least one trifoliate, before they were transferred into 600 cm3 pots. The growth chamber conditions were set to an 16 h light (22°C) and 8 h dark (18°C) cycle, providing a photosynthetically active radiation of 500 μE m−2 s−1 and a relative humidity of 60%. Twice a week, the plants were supplemented with N-free nodulation solutions. The nutrition solution contained 1 mm KNO3 only in the first week after sowing. Otherwise, the composition was: 1) 1 mm CaCl2 × 2 water; 2) 0.5 mm MgSO4 × 7 water; 3) 0.5 mm K2SO4; 4) 0.5 μm Na-Fe EDTA; 5) 2 mm phosphate out of 0.76 m K2HPO4 and 0.34 m KH2PO4 stock solutions; 6) 1 μm Na2MoO4 × 2 water, 15 μm H3BO3, 1 μm ZnSO4 × 7 water, 10 μm MnSO4 × water, 3.2 μm CuSO4 × 5 water, 0.2 μm CoSO4 × 7 water. For nutrition of potted plants, solutions were added to sterilized water or to 18 g/liter plant agar (Duchefa) for use in petri dish studies.
For phenotyping, defined transgenic MtSucS1-antisense and control seeds were germinated as described above and subsequently transferred onto petri dishes containing nodulation buffer, into seramis filled pots, or aeroponic containers, respectively. For nodulation studies, plants were fertilized with N-free nodulation solutions (composition as above and aeroponic solutions [Journet et al., 2001], respectively). The microsymbionts Sinorhizobium meliloti strains Sm2011 (Casse et al., 1979), Sm1021 (Meade and Singer, 1977), and Sm2011-mRFP (Smit et al., 2005) were used in open pot and petri dish approaches, and SmRCR2011 pXLGD4 (Journet et al., 2002) inoculated aeroponically grown plants. Rhizobia were cultured aerobically at 30°C in tryptone yeast (Saccharomyces cerevisiae) medium with appropriate antibiotics to OD 580 0.8 to 1.0.
Plants were grown in aeroponic caissons essentially as described by Journet et al. (2001). In short, growth conditions were set to 22°C, 60% relative humidity, and an 18 h light period. For nodule induction, after 2 weeks the N-rich pregrowth medium (Journet et al., 2001) was replaced by fresh N-free medium, 3 d before inoculation with S. meliloti culture diluted to a final concentration of approximately 5× 105 cells/mL. Subsequently, plant development was monitored over a time period of up to 9 weeks.
Plant Phenotyping and Histological Analyses
The phenotyping scheme of Moreau et al. (2006) for alterations in above-ground vegetative and reproductive development was used. The sequential leaf appearance was monitored by visual inspection and photo documentation (Olympus GmbH). Leaves were rated as initiated when primordia were firstly visible under 10× magnifications, and a leaf was considered present once it was completely unfolded with a planar central leaflet. Intermediate leaf development was categorized into four stages of appearance and coupled to a decimal code for detailed quantifications: unfolded, leaflets separated, and acute or right angle on central leaflet (leaf code 0.25, 0.5, respectively); code 0.75 for leaves, forming an obtuse angle with central leaflet; and appeared, completely unfolded mature leaf with planar central leaflet (code 1.00). For this study, notations of chronological reproductive development were modified: 1) flower primordium without visible petals (flower code 0.3); 2) opening petals on developing flower (0.50); 3) completely opened, mature flower (1.00). The nondestructive plant screenings were performed at routine intervals of 3 to 4 d during the course of development from inoculated seedling (day 7 after sowing) up to 58 dpi. RLi and RLa were calculated as the slope of the linear regression of average leaf number and time on the basis of at least six individuals per line. Time intervals between the initiation (plastochron) and appearance (phyllochron) of two chronological leaves are reciprocal to RLi and RLa, respectively. Accordingly, the rate of flower initiation and florochrons were calculated. Total leaf area was measured with an area meter (LI-COR Inc.).
Parameters of the root architecture were obtained from pot-grown, cleaned root samples, thoroughly spread on a glass-recording device (Richner et al., 2000), and determined with a transparency-scanning module (Snap Scan 1236, AGFA). Root parameters such as root length, root tip numbers, and the root surface area were quantified with the WinRhizo Pro software (Instruments Régent Inc.) on the basis of 300 dpi grayscale images. RSR was based on FW material, monitored via analytic balance (Sartorius AG). RWR was calculated as ratio of root weight per unit total plant weight on bases of DWs. To obtain dry matter, plant components were lyophilized or dried at 70°C until constant weight was achieved via precision balance (Sartorius AG).
The results on plant development presented in this work are based on three independent greenhouse growth experiments carried out in time intervals of 6 to 8 months.
Semithin sections of root nodules were obtained as described in Hohnjec et al. (2003), using a Leica VT 1000S vibratome (Leica). Light microscopy was performed on semithin, cleared longitudinal root nodule sections at 22 dpi (described in Hohnjec et al., 2003) using an Olympus BH-2 light microscope and micrographs were taken with an Olympus C-2000Z digital camera (Olympus Optical GmbH). In vivo visualization of the S. meliloti-expressed mRFP (Smit et al., 2005) was performed by confocal laser-scanning microscopy with a Leica TCS SP2 AOBS confocal laser-scanning microscopy system equipped with a helium laser (emission at 543 nm; Leica). For mRFP fluorescence, laser scanning was performed using settings for excitation/emission wavelengths (543/600–630 nm) and bright-field micrographs were collected simultaneously. Image processing was carried out with the Leica confocal software lite version 2.61 (Leica) and Adobe Photoshop 7.0 (Adobe Systems Inc.).
Nitrogenase Activity and C/N Ratio
In accordance to Somasegaran and Hoben (1985), nitrogenase activity (EC 1.7.9.92) was determined via AR assay, measuring representative ethylene evolution. Transgenic M. truncatula seedlings (germinated as above) were placed on petri dishes (three individuals/plate), containing nodulation solutions (as described above), cultured in a growth chamber under 18 h light (22°C) and 6 h dark (18°C) cycles, and inoculated with S. meliloti Sm1021 (Meade and Singer, 1977). For subsequent AR normalization, at the day of measurement, each nondestructed plant was checked for FW and nodule number before being transferred individually into 10 mL reaction vials containing 10% acetylene in air (v/v). After 4 h of incubation, 0.1 mL aliquots were analyzed for ethylene, using the 5710A gas chromatograph (Hewlett-Packard GmbH). AR (as ethylene/acetylene peak integration via 3380A HP integrator) of six to 10 replicates per line and point of harvest were averaged. Transformant N2 fixation was rated as active, when ethylene evolution was found >5% of control AR. The absolute amount of AR was estimated by comparison with an acetylene standard (Linde AG).
Total C and N yields and C/N ratio studies on several plant tissues were maintained from oven dried (3 d at 80°C), homogenized (ball mill grinder Retsch MM2, Retsch GmbH), and weighted material using an elemental analyzer spectrometer (Thermo) according to the manufacturer's instructions.
Chlorophyll a Fluorescence
To examine photosynthetic performances, fluorescence parameters were determined as reported in Schreiber et al. (1994) with a portable chlorophyll fluorometer (PAM 2000, Walz GmbH). These measurements were undertaken on the youngest and oldest appeared leaf pair (leaf decimal code 1.00) of greenhouse-grown plants at 47 dpi in the middle of photoperiod. A modulated measuring beam of <0.1 μmol quanta m−2 s−1, an actinic light of 120 μmol m−2 s−1 (FS), and a saturating beam of about 5,000 μmol m−2 s−1 was used to close all PSII reaction centers and to determine the maximum fluorescence level FM. The photosynthetic yield of PSII (ΦPSII) was calculated as (FM′ − FS)/FM′.
Harvest of Plant Material for Transcriptional and Metabolite Profiling
Aeroponically cultivated, N2-fixing nodules were harvested precisely in the middle of the light cycle at 11, 15, 21, and 26 dpi, immediately frozen in liquid N2, and stored at −80°C. Biological replicates were derived from at least three different root nodule pools, consisting of three to five plant individuals each. Nodule tissue samples for metabolite as well as corresponding total RNA extractions were taken from liquid N2 frozen and homogenized nodule pools. For each replicate, in total 60 individuals per transgenic line were studied during four harvesting points, with an emphases on the N2-fixing stages 21 and 26 dpi.
Quantitative Real-Time RT-PCR
Using in silico electronic northern analyses of the DFCI M. truncatula Gene Index 8 (http://compbio.dfci.harvard.edu/), 65 M. truncatula tentative consensus sequences (TCs), representing genes relevant for the Rhizobium-legume interaction or with a possible role in sink metabolism were selected for real-time RT-PCR experiments, carried out as described in Hohnjec et al. (2003). The results for genes differentially expressed between MtSucS1-antisense lines and controls in roots and root nodules, respectively, were averaged over five biological replicates. Every reaction was performed in two technical replicates per biological replicate and results for gene expression were averaged. The relative expression ratio (2−ΔCT; with ΔCT = CTgene − CTTefα) was calculated as the ratio of normalized gene expression of the gene of interest against the constitutively expressed M. truncatula gene for the translational elongation factor 1-α (MtTefα, TC106485, DFCI M. truncatula Gene Index 8). The gene-specific primers used (Supplemental Table S1) had a calculated melting temperature of 53°C ± 0.5°C and a length of 21 bp; amplification products were 315 ± 35 bp long. All primer sequences were unique in the DFCI M. truncatula Gene Index 8 and S. meliloti database (http://bioinfo.genopole-Toulouse.prd.fr), respectively.
Polar Metabolite Extraction and GC-MS Analysis
Polar metabolite extraction from 2 to 4 μg lyophilized samples was performed in accordance to Barsch et al. (2006). By comparison with purified standards, the National Institute of Standards and Technology 98 database and the freely available Golm metabolite database (Kopka et al., 2005), 80 metabolites were identified. Selected metabolite peak areas were quantified automatically, using the processing setup implemented in the Xcalibur software (ver. 1.4; Thermo). Normalizing the respective peak areas to the peak area of the internal standard, divided by the DW of the extracted sample calculated relative response values.
Statistical Analyses
All parameters and calculated variables were tested for differences between transformants and control plants, using the Student's t test incorporated into Microsoft Excel 7.0 (Microsoft Corp.). Differences are described as significant, when a value P < 0.05 (marked with a) and P < 0.01 (b), respectively, was obtained. Significant expression values detected via transcriptional profiling and ratios of metabolite pools were further subjected to hierarchical cluster analyses using Cluster ver.3 and visualized via the TreeView software (http://otl.stanford.edu/industry/resources/rts.html; Stanford University).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Structure of the p35S-MtsucS1-nosT antisense construct in the binary vector MF2.
Supplemental Figure S2. Changes of initiation (A) and appearance (B) of leaves over the time on the main axis and the first branches B0 to B4 (as defined by Moreau et al., 2006) for MtSucS1-reduced transformants and control lines.
Supplemental Figure S3. Photochemical yield (ΦPSII) of leaves under ambient CO2 concentration and photon flux density of 120 μmol quanta m−2 s−1.
Supplemental Figure S4. Quantification of root morphology in MtSucS1-reduced lines.
Supplemental Figure S5. Development of the RSR of aeroponically grown plants in a time course between 16 and 26 dpi.
Supplemental Figure S6. Root development in MtSucS1-reduced and control lines at time point 47 dpi.
Supplemental Figure S7. Development of acetylen reduction capacity in MtSucS1-antisense and control lines.
Supplemental Table S1. Primer sequences used for real-time RT-PCR.
Supplemental Table S2. Expression profiles of selected plant and S. meliloti genes in MtSucS1-reduced lines in comparison to control lines.
Supplemental Table S3. Response ratios of selected metabolite levels in root nodules of MtSucS1-antisense and control plants at time points 21 and 26 dpi, respectively.
Supplementary Material
Acknowledgments
We thank Tom Steinlein (Department of Experimental and Systems Ecology, Bielefeld University, Germany) and his coworkers for the help in elemental and root system analysis. Florian Frugier (Institut des Sciences du Végétal [ISV], Centre National de la Recherche Scientifique-Gif sur Yvette, France) is acknowledged for the supply of binary vector MF2 and Ton Bisseling (Wageningen University, The Netherlands) for the supply of S. meliloti Sm2011-mRFP. We are grateful to Susanna Galvez (ISV) as well as Martin Crespi (ISV) for their advice in plant transformation.
This work was supported by the Deutsche Forschungsgemeinschaft Priority Programme 1084 “MolMyk: Molecular Basics of Mycorrhizal Symbioses” and by the International Graduate School in Bioinformatics and Genome Research (Center for Biotechnology, Bielefeld University, Germany; to A.B., N.H., and H.K.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Helge Küster (helge.kuester@genetik.uni-bielefeld.de).
The online version of this article contains Web-only data.
References
- Akazawa T, Okamoto K (1980) Biosynthesis and metabolism of sucrose. In PK Stumpf, EE Conn, eds, The Biochemistry of Plants, Vol 3. Academic Press, New York, pp 199–220
- Albrecht G, Mustroph A (2003) Localization of sucrose synthase in wheat roots: increased in situ activity of sucrose synthase correlates with cell wall thickening by cellulose deposition under hypoxia. Planta 217 252–260 [DOI] [PubMed] [Google Scholar]
- Allona I, Collada C, Casado R, Aragoncillo C (1994) 2S arginine-rich proteins from Pinus pinaster seeds. Tree Physiol 14 211–218 [DOI] [PubMed] [Google Scholar]
- Amor Y, Haigler CH, Johnson S, Wainscott M, Delmer D (1995) A membrane-associated form of sucrose synthase and its potential role in synthesis of cellulose and callose in plants. Proc Natl Acad Sci USA 92 9353–9357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthon GE, Emmerich DW (1990) Developmental regulation of enzymes of sucrose and hexose metabolism in effective and ineffective soybean nodules. Plant Physiol 92 346–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avigad G (1982) Sucrose and other disaccharides. In FA Loewus, W Tanner, eds, Encyclopedia of Plant Physiology. Springer Verlag, Berlin, pp 217–347
- Barratt DHP, Barber L, Kruger NJ, Smith AM, Wang TL, Martin C (2001) Multiple, distinct isoforms of sucrose synthase in pea. Plant Physiol 127 655–664 [PMC free article] [PubMed] [Google Scholar]
- Barsch A, Tellström V, Patschkowsky T, Küster H, Niehaus K (2006) Metabolite profiles of nodulated alfalfa plants indicate that distinct stages of nodule organogenesis are accompanied by global physiological adaptations. Mol Plant Microbe Interact 19 998–1013 [DOI] [PubMed] [Google Scholar]
- Bernier G, Havelange A, Houssa C, Petitjean A, Lejeune P (1993) Physiological signals that induce flowering. Plant Cell 5 1147–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniawska Z, Barratt DHP, Garlick AP, Thole V, Kruger NJ, Martin C, Zrenner R, Smith AM (2007) Analysis of the sucrose synthase gene family in Arabidopsis. Plant J 49 810–828 [DOI] [PubMed] [Google Scholar]
- Bolton E, Higgisson B, Harrington A, O'Gara F (1986) Dicarboxylic acid transport in Rhizobium meliloti: isolation of mutants and cloning of dicarboxylic acid transport genes. Arch Microbiol 144 142–146 [Google Scholar]
- Burstin J, Marget P, Huart M, Moessner A, Mangin B, Duchene C, Desprez B, Munier-Jolain N, Duc G (2007) Developmental genes have pleiotropic effects on plant morphology and source capacity, eventually impacting on seed protein content and productivity in pea. Plant Physiol 144 768–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson SJ, Chourey PS (1996) Evidence of plasma membrane-associated forms of sucrose synthase in maize. Mol Gen Genet 252 303–310 [DOI] [PubMed] [Google Scholar]
- Carlson SJ, Chourey PS, Helentjaris T, Datta R (2002) Gene expression studies on developing kernels of maize sucrose synthase (SuSy) mutants show evidence for a third SuSy gene. Plant Mol Biol 49 15–29 [DOI] [PubMed] [Google Scholar]
- Carlsson G, Huss-Danell K (2003) Nitrogen fixation in perennial forage legume in the fields. Plant Soil 253 353–372 [Google Scholar]
- Casse F, Boucher D, Julliot JS, Michel M, Dénarié J (1979) Identification and characterisation of large plasmids in Rhizobium meliloti using agarose gel electrophorese. J Gen Microbiol 113 229–242 [Google Scholar]
- Chen YC, Chourey PS (1989) Spatial and temporal expression of the two sucrose synthase genes in maize: immunohistological evidence. Theor Appl Genet 78 553–559 [DOI] [PubMed] [Google Scholar]
- Chourey PS, Taliercio EW, Carlson SJ, Ruan YL (1998) Genetic evidence that the two isoenzymes of sucrose synthase present in developing maize endosperm are critical, one for cell wall integrity and the other for starch biosynthesis. Mol Gen Genet 259 88–96 [DOI] [PubMed] [Google Scholar]
- Colebatch G, Kloska S, Trevaskis B, Freund S, Altmann T, Udvardi MK (2002) Novel aspects of symbiotic nitrogen fixation uncovered by transcript profiling with cDNA arrays. Mol Plant Microbe Interact 15 411–420 [DOI] [PubMed] [Google Scholar]
- Craig J, Barratt DHP, Tatge H (1999) Mutations at the rug4 locus alter the carbon and nitrogen metabolism in pea plants through an effect on sucrose synthase. Plant J 17 353–362 [Google Scholar]
- de la Pena TC, Frugier F, McKhann HI, Bauer P, Brown S, Kondorosi A, Crespi M (1997) A carbonic anhydrase gene is induced in the nodule primordium and its cell-specific expression is controlled by the presence of Rhizobium during development. Plant J 11 407–420 [DOI] [PubMed] [Google Scholar]
- El Yahyaoui F, Küster H, Ben Amor B, Hohnjec N, Pühler A, Becker A, Gouzy G, Vernié T, Gough C, Niebel A, et al (2004) Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol 136 3159–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemetakis E, Efrose RC, Ott T, Stedel C, Aivalakis G, Udvardi MK, Katinakis P (2006) Spatial and temporal organization of sucrose metabolism in Lotus japonicus nitrogen-fixing nodules suggests a role for the elusive alkaline/neutral invertase. Plant Mol Biol 62 53–69 [DOI] [PubMed] [Google Scholar]
- Fu HY, Park WD (1995) Sink- and vasculature-associated sucrose synthase functions are encoded by different gene classes in potato. Plant Cell 7 1369–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gálvez L, González EM, Arrese-Igor C (2005) Evidence of carbon flux shortage and strong carbon/nitrogen interactions in pea nodules at early stages of water stress. J Exp Bot 56 2551–2561 [DOI] [PubMed] [Google Scholar]
- Gong ZY, He ZS, Zhu JB, Yu GQ, Zou HS (2006) Sinorhizobium meliloti nifA mutant induces different gene expression profile from wild type in alfalfa nodules. Cell Res 16 818–829 [DOI] [PubMed] [Google Scholar]
- Gordon AJ, James CL (1997) Enzymes of carbohydrate and amino acid metabolism in developing and mature nodules of white clover. J Exp Bot 48 895–903 [Google Scholar]
- Gordon AJ, Minchin FR, James CL, Komina O (1999) Sucrose synthase in legume nodules is essential for nitrogen fixation. Plant Physiol 120 867–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon AJ, Ryle GJA, Mitchell DF, Lowry KH, Powell CE (1986) The effect of defoliation on carbohydrate, protein and geghaemoglobin content of white clover nodules. Ann Bot (Lond) 58 141–154 [Google Scholar]
- Gordon AJ, Thomas BJ, Reynolds PHS (1992) Localization of sucrose synthase in soybean root nodules. New Phytol 122 35–44 [DOI] [PubMed] [Google Scholar]
- Hardarson G, Bliss FA, Cigales-Rivero MR, Henson RA, Kipe-Nolt JA, Longeri L, Manrique A, Peña-Cabriales JJ, Pereira PAA, Sanabria CA, et al (1993) Genotypic variation in biological nitrogen fixation by common bean. Plant Soil 152 59–70 [Google Scholar]
- Hoffmann B, Trinh TH, Leung J, Kondorosi A, Kondorosi E (1997) A new Medicago truncatula line with superior in vitro regeneration, transformation, and symbiotic properties isolated through cell culture selection. Mol Plant Microbe Interact 10 307–315 [Google Scholar]
- Hohnjec N, Becker JD, Pühler A, Perlick AM, Küster H (1999) Genomic organization and expression properties of the MtSucS1 gene, which encodes a nodule-enhanced sucrose synthase in the model legume Medicago truncatula. Mol Gen Genet 261 514–522 [DOI] [PubMed] [Google Scholar]
- Hohnjec N, Perlick AM, Pühler A, Küster H (2003) The Medicago truncatula sucrose synthase gene MtSucS1 is activated both in the infected region of root nodules and in the cortex of roots colonized by arbuscular mycorrhizal fungi. Mol Plant Microbe Interact 16 903–915 [DOI] [PubMed] [Google Scholar]
- Hong ZQ, Copeland L (1990) Pentose phosphate pathway enzymes in nitrogen-fixing leguminous root nodules. Phytochemistry 29 2437–2440 [Google Scholar]
- Hood EE, Gelvin SB, Melchers LS, Hoekema A (1993) New Agrobacterium helper plasmid for gene-transfer to plants. Transgenic Res 2 208–218 [Google Scholar]
- Horst I, Welham T, Kelly S, Kaneko T, Sato S, Tabata S, Parniske M, Wang T (2007) TILLING mutants of Lotus japonicus reveal that nitrogen assimilation and fixation can occur in the absence of nodule-enhanced sucrose synthase. Plant Physiol 144 806–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jording D, Pühler A (1993) The membrane topology of the Rhizobium meliloti C4-dicarboxylate permease (DctA) as derived from protein fusions with Escherichia coli K12 alkaline phosphatase (PhoA) and β-galactosidase (LacZ). Mol Gen Genet 241 106–114 [DOI] [PubMed] [Google Scholar]
- Jørgensen FV, Ledgard SF (1997) Contribution from stolons and roots to estimates of the total amount of N2 fixed by white clover (Trifolium repens L.). Ann Bot (Lond) 80 641–648 [Google Scholar]
- Journet EP, El-Gachtouli N, Vernaud V, de Billy F, Pichon M, Dedieu A, Arnould C, Morandi D, Barker DG, Gianinazzi-Pearson V (2001) Medicago truncatula ENOD11: a novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol Plant Microbe Interact 14 737–748 [DOI] [PubMed] [Google Scholar]
- Journet EP, van Tuinen D, Gouzy J, Crespeau H, Carreau V, Farmer MJ, Niebel A, Schiex T, Jaillon O, Chatagnier O, et al (2002) Exploring root symbiotic programs in the model legume Medicago truncatula using EST analysis. Nucleic Acids Res 30 5579–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch KE, Nolte KD, Duke ER, McCarty DR, Avigne WT (1992) Sugar levels modulate differential expression of maize sucrose synthase genes. Plant Cell 4 59–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmüller E, Dormann P, Weckwerth W, Gibon Y, Stitt M, et al (2005) GMD@CSB.DB: the golm metabolome database. Bioinformatics 21 1635–1638 [DOI] [PubMed] [Google Scholar]
- Küster H, Frühling M, Perlick AM, Pühler A (1993) The sucrose synthase gene is predominantly expressed in the root nodule tissue of Vicia faba. Mol Plant Microbe Interact 6 507–514 [DOI] [PubMed] [Google Scholar]
- Larrainzar E, Wienkoop S, Weckwerth W, Ladera R, Arrese-Igor C, González M (2007) Medicago truncatula root nodule proteome analysis reveals differential plant and bacteroids responses to drought stress. Plant Physiol 144 1495–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodwig EM, Hosie AHF, Bourdès A, Findlay K, Allaway D, Karunakaran R, Downie JA, Poole PS (2003) Amino-acid cycling drives nitrogen fixation in the legume-Rhizobium symbiosis. Nature 422 722–726 [DOI] [PubMed] [Google Scholar]
- Manthey K, Krajinski F, Hohnjec N, Firnhaber C, Pühler A, Perlick AM, Küster H (2004) Transcriptome profiling in root nodules and arbuscular mycorrhiza identifies a collection of novel genes induced during Medicago truncatula root endosymbiosis. Mol Plant Microbe Interact 17 1063–1077 [DOI] [PubMed] [Google Scholar]
- Marschner H (1995) Mineral Nutrition of Higher Plants, Ed 2. Academic Press, London
- Mathis R, Gamas P, Meyer Y, Cullimore JV (2000) The presence of GSI-like genes in higher plants: support for the paralogous evolution of GSI and GSII genes. J Mol Evol 50 116–122 [DOI] [PubMed] [Google Scholar]
- McDonald AJS, Lohammar T, Ingestad T (1992) Net assimilation rate and shoot area development in birch (Betula pendula Roth.) at different steady-state values of nutrition and photon flux density. Trees (Berl) 6 1–6 [Google Scholar]
- Meade HM, Singer ER (1977) Genetic mapping of Rhizobium meliloti. Proc Natl Acad Sci USA 74 2076–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SS, Driscoll BT, Gregerson RG, Gantt JS, Vance CP (1998) Alfalfa malate dehydrogenase (MDH): molecular cloning and characterization of five different forms reveals a unique nodule-enhanced MDH. Plant J 15 173–184 [DOI] [PubMed] [Google Scholar]
- Moreau D, Salon C, Munier-Jolain N (2006) Using standard framework for the phenotypic analysis of Medicago truncatula: an effective method for characterizing the plant material used for functional genomics approaches. Plant Cell Environ 29 1087–1098 [DOI] [PubMed] [Google Scholar]
- Mustart PJ, Cowling RM (1992) Seed size: phylogeny and adaptation in two closely related Proteaceae species-pairs. Oecologia 91 292–295 [DOI] [PubMed] [Google Scholar]
- Nolte KD, Koch KE (1993) Companion-cell specific localization of sucrose synthase in zones of phloem loading and unloading. Plant Physiol 101 899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiter E, Schubert S (2003) Sugar uptake and proton release by protoplasts from the infected zone of Vicia faba L. nodules: evidence against apoplastic sugar supply of infected cells. J Exp Bot 54 1691–1700 [DOI] [PubMed] [Google Scholar]
- Richner W, Liedgens M, Bürgi H, Soldati A, Stamp P (2000) Root image analysis and interpretation. In AL Smit, AG Bengough, C Engels, M van Noordwijk, S Pellgrin, SC van de Geijn, eds, Root Methods: A Handbook. Springer Verlag, Berlin, pp 305–341
- Rolletschek H, Borisjuk L, Koschorreck M, Wobus U, Weber H (2002) Legume embryos develop in a hypoxic environment. J Exp Bot 53 1099–1107 [DOI] [PubMed] [Google Scholar]
- Rosendahl L, Vance CP, Pedersen WB (1990) Products of dark CO2 fixation in pea root nodules support bacteroid metabolism. Plant Physiol 93 12–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross HA, Davies H (1992) Purification and characterization of sucrose synthase from the cotyledons of Vicia faba L. Plant Physiol 100 1008–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan YL, Chourey PS, Delmer DP, Perez-Grau L (1997) The differential expression of sucrose synthase in relation to diverse patterns of carbon partitioning in developing cotton seed. Plant Physiol 115 375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan YL, Llewellyn DJ, Furbank RT (2003) Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation, and seed development. Plant Cell 15 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salnikow VV, Grimson MJ, Delmer DP, Haigler CH (2001) Sucrose synthase localizes to cellulose synthesis sites in tracheary elements. Phytochemistry 57 823–833 [DOI] [PubMed] [Google Scholar]
- Schreiber U, Bilger W, Neubauer C (1994) Chlorophyll fluorescence as a nonintrusive indicator for rapid assessment of in vivo photosynthesis. In ED Schulze, MM Caldwell, eds, Ecophysiology of Photosynthesis 100. Springer Verlag, Berlin, pp 49–70
- Smit P, Raedts J, Portyanko V, Debelle F, Gough C, Bisseling T, Geurts R (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308 1789–1791 [DOI] [PubMed] [Google Scholar]
- Somasegaran P, Hoben HJ (1985) Methods in Legume-Rhizobium Technology. NifTAL Project, Paia Maui, Hawaii
- Tesfaye M, Samac DA, Vance CP (2006) Insights into symbiotic nitrogen fixation in Medicago truncatula. Mol Plant Microbe Interact 19 330–341 [DOI] [PubMed] [Google Scholar]
- Trieu AT, Burleigh SH, Kardailsky IV, Maldonado-Mendoza IE, Versaw WK, Blaylock LA, Shin H, Chiou TJ, Katagi H, Dewbre GR, et al (2000) Technical advance: transformation of Medicago truncatula via infiltration of seedlings or flowering plants with Agrobacterium. Plant J 22 531–541 [DOI] [PubMed] [Google Scholar]
- Udvardi MK, Day DA (1997) Metabolite transport across symbiotic membranes of legume nodules. Annu Rev Plant Physiol Plant Mol Biol 48 493–523 [DOI] [PubMed] [Google Scholar]
- Vancanneyt G, Schmidt R, O'Connor-Sanchez A, Willmitzer L, Rocha-Sosa M (1990) Construction of an intron-containing marker gene. Mol Gen Genet 220 245–250 [DOI] [PubMed] [Google Scholar]
- Vance CP, Gregerson RG, Robinson DL, Miller SS, Gantt JS (1994) Primary assimilation of nitrogen in alfalfa nodules: molecular features of the enzymes involved. Plant Sci 101 51–64 [Google Scholar]
- Wang TL, Hadavizideh A, Harwood A, Welham TJ, Harwood WA, Faulks R, Hedley CL (1990) An analysis of seed development in Pisum sativum: the chemical induction of storage product mutants. Plant Breed 105 311–320 [Google Scholar]
- Wang TL, Hedley CL (1993) Seed mutants in Pisum. Pisum Genet 25 64–70 [Google Scholar]
- Warembourg FR, Roumet C (1989) Why and how to estimate the cost of symbiotic N2 fixation? A progressive approach based on the use of 14C and 15N isotopes. Plant Soil 115 167–177 [Google Scholar]
- Waring RH, Landsberg JJ, Williams M (1998) Net primary production of forests: a constant fraction of gross primary production? Tree Physiol 18 129–134 [DOI] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Heim U, Buchner P, Wobus U (1995) Seed coat-associated invertases of fava bean control both unloading and storage functions: cloning of cDNAs and cell type-specific expression. Plant Cell 7 1835–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckwerth W, Loureiro ME, Wenzel K, Fiehn O (2004) Differential metabolic networks unravel the effects of silent plant phenotypes. Proc Natl Acad Sci USA 101 7809–7814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter H, Huber JL, Huber SC (1997) Membrane association of sucrose synthase: changes during the graviresponse and possible control by protein phosphorylation. FEBS Lett 430 151–155 [DOI] [PubMed] [Google Scholar]
- Winter H, Huber JL, Huber SC (1998) Identification of sucrose synthase as an actin-binding protein. FEBS Lett 430 205–208 [DOI] [PubMed] [Google Scholar]
- Winter H, Huber SC (2000) Regulation of sucrose metabolism in higher plants: localization and regulation of activity of key enzymes. Crit Rev Plant Sci 19 31–67 [DOI] [PubMed] [Google Scholar]
- Witty JF, Minchin FR (1988) Measurement of nitrogen fixation by the acetylene reduction assay; myths and mysteries. In DP Beck, LA Materon, eds, Nitrogen Fixation by Legumes in Mediterranean Agriculture. Martinus Nijhoff Publ., Dordrecht, The Netherlands, pp 331–344
- Yu WP, Wang AY, Juang RH, Sung HY, Su JC (1992) Isolation and sequences of rice sucrose synthase cDNA and genomic DNA. Plant Mol Biol 18 139–142 [DOI] [PubMed] [Google Scholar]
- Zahran HH (1999) Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in arid climate. Microbiol Mol Biol Rev 63 968–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Wu Y, Avigne WT, Koch KE (1999) Rapid repression of maize invertase by low oxygen: invertase/sucrose synthase balance, sugar signalling potential, and seedling survival. Plant Physiol 121 599–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrenner R, Salanoubat M, Willmitzer L, Sonnewald U (1995) Evidence of the crucial role of sucrose synthase for sink strength using transgenic potato plants (Solanum tuberosum L.). Plant J 7 97–107 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.