Abstract
Soil salinity is one of the most significant abiotic stresses for crop plants, including legumes. These plants can establish root symbioses with nitrogen-fixing soil bacteria and are able to grow in nitrogen-poor soils. Medicago truncatula varieties show diverse adaptive responses to environmental conditions, such as saline soils. We have compared the differential root growth of two genotypes of M. truncatula (108-R and Jemalong A17) in response to salt stress. Jemalong A17 is more tolerant to salt stress than 108-R, regarding both root and nodulation responses independently of the nitrogen status of the media. A dedicated macroarray containing 384 genes linked to stress responses was used to compare root gene expression during salt stress in these genotypes. Several genes potentially associated with the contrasting cellular responses of these plants to salt stress were identified as expressed in the more tolerant genotype even in the absence of stress. Among them, a homolog of the abiotic stress-related COLD-REGULATEDA1 gene and a TFIIIA-related transcription factor (TF), MtZpt2-1, known to regulate the former gene. Two MtZpt2 TFs (MtZpt2-1 and MtZpt2-2) were found in Jemalong A17 plants and showed increased expression in roots when compared to 108-R. Overexpression of these TFs in the sensitive genotype 108-R, but not in Jemalong A17, led to increased root growth under salt stress, suggesting a role for this pathway in the adaptive response to salt stress of these M. truncatula genotypes.
Maintaining crop yields under adverse stress environmental conditions is a major challenge in modern agriculture. To meet this goal, it is necessary to understand the contrasting adaptations of plants to growth in stressed conditions. Salinity is one of the major abiotic stresses that affects crop productivity and quality and has been described as one of the most serious threats to agriculture and the natural status of the environment (Chinnusamy et al., 2005). Increased salinization of arable land is expected to have devastating global effects, resulting in a 30% land loss within the next 25 years and up to 50% by the year 2050 (Wang et al., 2003).
Plant responses to salt stress are diverse and include modifications of root system architecture, activation of stress-induced transcriptional programs, and biochemical adaptations, as well as plant growth inhibition. Salinity imposes ionic, osmotic, and secondary stresses, such as nutritional disorders and oxidative stress (Zhu, 2001). Legumes, like most crop plants, are susceptible to salinity (Duzan et al., 2004; Chinnusamy et al., 2005). These plants are widely grown for grain and forage purposes, their world-wide economic importance being second only to grasses (Graham and Vance, 2003). In addition, legumes can establish root symbioses with nitrogen-fixing soil bacteria, enabling the plants to grow in nitrogen-poor soils. This ability to colonize soils where other plants cannot thrive makes the study of legumes and their symbioses important for agriculture. The establishment of successful symbiosis involves an elaborate exchange of molecular signals (Limpens and Bisseling, 2003). In the plant host, root nodule organogenesis is regulated by diverse hormonal, metabolic, and environmental conditions (Crespi and Gálvez, 2000), and this interaction is specifically affected in saline soils (Arrese-Igor et al., 1999; Zahran, 1999).
The problem of salinity has been approached through better management practices and the introduction of salt-tolerant varieties in the affected areas. Unfortunately, these approaches are generally uneconomical and difficult to implement on a large scale. However, major progress could be achieved through genetic improvement (Walia et al., 2005). Various legumes, such as the model legume Medicago truncatula, show a large diversity of varieties adapted to varying environmental conditions, including saline soils (http://www.noble.org/medicago/ecotypes.html). In recent years, M. truncatula has been recognized as an excellent legume model in view of its small, diploid genome, self fertility, and short life cycle, as well as availability of various genomic and genetic tools (Barker et al., 1990; Bell et al., 2001; Young et al., 2005). All these traits make M. truncatula suitable for identifying genes that could improve agronomic performances such as abiotic stress resistance (Cook, 1999).
Stress responses involve alterations in gene expression (Tester and Davenport, 2003), suppressive subtractive hybridizations, and array technologies using cDNAs or oligonucleotides are increasingly being used to monitor global gene expression changes in various plants in response to abiotic stresses (Ozturk et al., 2002; Seki et al., 2002; Bartels and Sunkar, 2005). However, few reports analyzed differences in gene expression between salt-sensitive and salt-tolerant genotypes in plants (Walia et al., 2005). Although such genes are useful in the dissection of genotype-specific regulatory pathways and mechanisms of salt tolerance, they usually represent only a fraction of all salt-regulated genes, and isolating them is a challenging task. The mechanisms underlying the genotype-dependent difference in expression of such genes are largely unknown.
Two genotypes (108-R and Jemalong A17) were shown to have a different adaptation to salt stress (Merchan et al., 2003). We have developed, in the M. truncatula 108-R genotype, a dedicated macroarray containing genes linked to salt stress and recovery responses in roots (Merchan et al., 2007). Here, we aimed to identify genes involved in genotype-specific regulatory pathways and mechanisms of salt acclimation. A comparison of molecular and physiological responses to salt stress in sensitive and tolerant M. truncatula genotypes (108-R and Jemalong A17, respectively) was performed. Expression analysis using the dedicated macroarray revealed several genes potentially associated with the contrasting responses to salt stress in these plants. Among them, we identified a homolog of the COLD-REGULATEDA1 (CorA1; MtCorA1; Laberge et al., 1993) gene, which is a putative target of MtZpt2 transcription factors (TFs; Merchan et al., 2007). Accordingly, these TFs showed higher expression levels in the tolerant variety. Overexpression of MtZpt2-1 or MtZpt2-2 in roots of the salt-sensitive variety allowed significant increase in root growth specifically under salt stress conditions, suggesting a role of these pathways in the differential adaptive response to salt stress of these M. truncatula genotypes.
RESULTS
Evaluation of Salt Stress Growth Responses of Two M. truncatula Genotypes
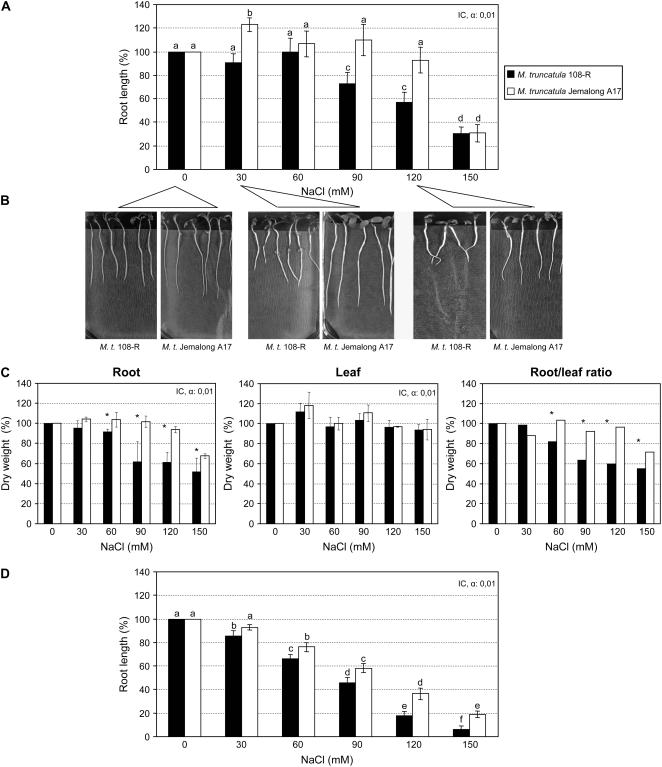
We have examined root growth and dry weight biomass in two genotypes of the model legume M. truncatula (108-R and Jemalong A17) in response to different salt stress conditions. Root length was measured after 5 d of growth on a rich medium (Fahräeus; Truchet et al., 1985) containing various concentrations of NaCl (Fig. 1). Root length of M. truncatula 108-R was significantly reduced after NaCl treatments in the range of 90 to 150 mm, as compared with plants grown without salt (Kruskal and Wallis test, P < 0.01; n = 20; Fig. 1A). However, root length in Jemalong A17 was negatively affected by salinity only at 150 mm (Kruskal and Wallis test, P < 0.01; n = 20; Fig. 1A). Altogether, we could identify a differential response to salt stress between the 108-R and Jemalong A17 genotypes. As shown in Figure 1B, M. truncatula Jemalong A17 grew well at high salinity levels (120 mm NaCl), while 108-R plants grew more slowly under these salt conditions. Similarly, salt treatment reduced the root dry weight more noticeably for the variety 108-R already after a 60 mm NaCl treatment, whereas it was only significantly affected at 150 mm in Jemalong A17 (Kruskal and Wallis test, P < 0.01; n = 20; Fig. 1C, left). No significant differences were, however, found for leaf dry weight biomass of each variety and in all treatments, suggesting greater salt sensitivity of the root than of the aerial part at this early postgermination stage (Fig. 1C, middle). The ratio of root dry weight to leaf dry weight (as percentage of control) reveals the relative effects of increasing NaCl concentration for each genotype (Fig. 1C, right). As expected, ratio values for M. truncatula 108-R were significantly reduced with increased salt treatments (60–150 mm NaCl; Kruskal and Wallis test, P < 0.01; n = 20). In contrast, this ratio was constant for the tolerant genotype, at least up to 120 mm NaCl. These results suggest that root dry weight is significantly different between the two genotypes at medium and high salt concentrations (60–150 mm).
Figure 1.
Effect of NaCl on root growth and dry weight of two M. truncatula genotypes. Root length and dry weight biomass under different salt stress conditions were evaluated in an in vitro system. Germinated seedlings were grown on vertical Fahräeus or “i” medium plates for 5 d in the presence of different NaCl concentrations (0, 30, 60, 90, 120, and 150 mm). A, Relative root length of each variety at 5 d.a.g. in in vitro conditions with Fahräeus medium is shown as percentage of control root growth without salt. B, Representative pictures taken 5 d after transfer of the seedlings to 0, 30, and 120 mm NaCl on Fahräeus medium. C, Relative dry weights of roots and leaves from M. truncatula 108-R and Jemalong A17 after 15 d of growth in Fahräeus medium submitted to different salt stress treatments. Results are shown as percentage of control without salt. Ratio (right) represents root dry weight/leaf dry weight to evaluate the effect of salinity on dry weight biomass of the plants. D, Relative root growth (%) of the two M. truncatula varieties grown under nonstressed and salt-stressed conditions in “i” medium for 5 d. Values indicated with different letters indicate statistically significant differences (P < 0.01), whereas those marked with the same letters show statistically similar values. IC, Interval of confidence (α = 0.01). Columns labeled with an asterisk are significantly different (P < 0.01) within a given salinity level. In both cases, the Kruskal and Wallis test has been used. A representative example out of two biological experiments is shown.
Root growth performance was also assessed on a low-nitrogen medium (“i”; Blondon, 1964) to examine the impact of the medium on the range of variability for salinity tolerance between these two genotypes of M. truncatula. At the same salinity concentration, root length of 108-R was even more significantly affected by salt treatment when compared with the results obtained in the rich medium. This difference is likely to be a consequence of minimal nutrient capacity of the “i” medium that may potentiate salt stress effects on growth. Statistically significant differences between 108-R and Jemalong A17 genotypes were found for all assayed salinity conditions (Fig. 1D). M. truncatula 108-R growth was already reduced by more than 50% at 90 mm of salt, and an NaCl concentration of 150 mm nearly abolished root growth. This negative effect of salt on root growth was less pronounced in Jemalong A17, in agreement with the results obtained on rich (Fahräeus) medium (Fig. 1A).
These various parameters allowed us to monitor the effects of salt treatments on plant growth and further revealed differential root growth responses between the two genotypes. Plants grown in low-nitrogen medium were more affected by salinity even though, independently of the growth medium, the Jemalong A17 genotype had a greater ability to tolerate salt stress conditions than 108-R.
Effects of Salt Stress on the Rhizobial Symbiotic Interaction in M. truncatula 108-R and Jemalong A17 Genotypes
The formation of nitrogen-fixing nodules results from the symbiotic interaction between legumes and rhizobia, a process sensitive to salinity at several stages (initiation, development, and function of nodules; Garg and Gupta, 2000). To study the effect of salinity on the nodulation capacity in these two Medicago genotypes, plants were grown in the presence of different NaCl concentrations and subsequently inoculated with Sinorhizobium meliloti strain 2011 (able to grow up to 300 mm NaCl; Rüberg et al., 2003). Three weeks after inoculation, these plants were assessed to determine their nodule number. A negative effect on nodule formation was clearly observed in both varieties at high salt concentration in the medium (Fig. 2A; Kruskal and Wallis test, P < 0.05; n = 20). Salinity decreased nodule number in the 108-R genotype at 60 mm NaCl by more than 50% versus only 20% in Jemalong A17. In contrast, the latter genotype reaches a similar reduction level only at 120 mm NaCl. These results correlated with the differential root growth sensitivity to salt stress of these genotypes. A drastic reduction in the total number of nodules was observed at 150 mm NaCl although to a lesser extent for Jemalong A17 (Fig. 2A). In addition, salinity reduced nodule size and differentiation in both varieties, as evidenced by the appearance of white nodules without high leghemoglobin content (data not shown). These perturbations of the nodulation process could explain the decrease in plant growth under saline conditions.
Figure 2.
Effect of salt stress on nodulation in M. truncatula 108-R and Jemalong A17. Seeds of the two genotypes were germinated on water agar plates, and seedlings were grown in the greenhouse in the presence of different NaCl concentrations (0, 30, 60, 90, 120, and 150 mm). The inoculation with S. meliloti strain 2011 was made 9 d.a.g.. Nodule number and root and leaf dry weights were determined at 30 d.a.g. (21 d.p.i.). A, The effect of salt concentration on nodulation of M. truncatula 108-R and Jemalong A17 by S. meliloti strain 2011 is measured as percentage of the total nodule number 21 d.p.i. observed in control (without salt) conditions. B, Relative dry weights (%) of root system and aerial part of M. truncatula 108-R and Jemalong A17 genotypes at 30 d.a.g. under different salt stress conditions. IC, Interval of confidence (α = 0.05). Columns labeled with an asterisk are significant differences (P < 0.05) between genotypes within a given salinity level. Statistical comparisons were performed using the Kruskal and Wallis test. A representative example out of two biological experiments is shown.
The effects of salinity on the dry weight of both 108-R roots and leaves are very significant already at 60 mm NaCl in contrast to what is observed for the Jemalong A17 variety (Fig. 2B). In nodulated plants, a major effect of salt in the aerial part was observed in contrast to recently germinated seedlings. The effect in root and leaf dry weights correlated with the reduction in nodule number under salt stress, with 108-R being significantly more affected than Jemalong A17. Perturbations in the nodulation process and in nodule functionality could explain the decrease in plant growth under saline conditions. The contrasting root physiological responses of the two genotypes to salt treatments may be partially responsible for their different ability to interact with Rhizobium under stress conditions. Nevertheless, independently of the nitrogen status (either from added combined nitrogen or through symbiotic fixation), the Jemalong A17 genotype is more tolerant to salt stress than 108-R.
Gene Expression Profiles of M. truncatula 108-R and Jemalong A17
Transcriptional profiling of selected salt stress-related genes is useful for the assessment and comparison of gene expression on a comprehensive scale across genotypes. To reveal molecular mechanisms that may sustain the contrasting salt tolerance responses in roots of both genotypes, we performed a comparative analysis of expression profiles using a salt stress dedicated macroarray containing 384 genes (Merchan et al., 2007). Gene expression analysis was performed using samples from roots of 108-R and Jemalong A17 after 0 or 4 d of salt stress (150 mm NaCl on “i” medium, referred to as “4i” and “4n,” respectively). This concentration and time was previously used for the analysis of recovery responses in M. truncatula (Merchan et al., 2003, 2007) and represent a condition that does not permanently affect root growth. Both varieties under these conditions can fully recover their root growth after changing into a normal medium. We directed our experimental design to detect genes differentially regulated between the two genotypes in control and stressed conditions (Fig. 3A). Four different treatments were used: untreated 108-R plants (R0), 108-R plants treated with 150 mm NaCl for 4 d (R4), untreated Jemalong A17 plants (J0), and Jemalong A17 plants treated with 150 mm NaCl for 4 d (J4). To identify statistically significant differentially expressed genes between genotypes during salt treatments, we used a Student's t test based on two biological replicates and four technical replicates (e.g. quadruplicated probes on macroarray) and retained genes with P values < 0.01.
Figure 3.
Gene expression profiles of M. truncatula 108-R and Jemalong A17. A, Experimental design is based on series of pairwise comparisons. Four d.a.g. seedlings from different genotypes (R for 108-R and J for Jemalong A17) were grown for 4 d without (0 mm; R0 and J0) or with (R4 and J4) 150 mm NaCl. Two biological and four technical replicates were available for each gene, for each physiological condition, and for each genotype. B, The histogram shows the total number of transcripts up- or down-regulated in 108-R (left bars) and Jemalong A17 (right bars) in response to salinity stress at a level of P < 0.01. C, Venn diagrams illustrating the number of genes up-regulated or down-regulated under salinity stress in either or both genotypes of M. truncatula (108-R, left, and Jemalong A17, right). D, Distribution of differentially expressed genes into functional categories according to BLASTN hits (based on Journet et al., 2002; Merchan et al., 2007). Percentages were calculated from the total number of differentially expressed genes from Jemalong A17 (112 genes).
Most of the genes were similarly regulated in these genotypes, because approximately one-half of the spots on the dedicated macroarray (e.g. about 150 spots) showed similar detectable signals for 108-R and Jemalong A17 hybridization, validating the use of the array for both genotypes. These included internal constitutive controls for gene expression analysis such as tubulin, elongation factor, and actin genes used for array normalization (Merchan et al., 2007). In Jemalong A17, expression of 14 genes was altered after a salt stress: four genes (1.4% of the 280 stress-related genes present in the array; Merchan et al., 2007) induced and 10 (3.6%) down-regulated (J0 versus J4; Fig. 3B). In contrast, in the salt-sensitive genotype, transcript levels of 63 genes were altered by the salt treatment. Among them, 23 genes (8.2%) were up-regulated and 40 (14.3%) down-regulated at 4n (R0 versus R4; Fig. 3B). Surprisingly, no gene was induced in common between 108-R and Jemalong A17 after a salt stress but three genes were down-regulated in both genotypes (Fig. 3C).
We can conclude that the tolerant and sensitive varieties have a different regulation of their transcriptome in response to salt stress.
Differences in Gene Regulation between the Salt-Sensitive and Salt-Tolerant M. truncatula Genotypes
The differential expression patterns that may lead to the increased salt tolerance of Jemalong A17 plants in comparison to 108-R were tested in two ways. First, a comparison of expression profiles under normal growth conditions between the salt-sensitive and the salt-tolerant varieties (J0 versus R0) revealed 38 genes up-regulated in Jemalong A17 roots relative to 108-R roots (Supplemental Table S1). In contrast, only five genes were more expressed in the sensitive genotype in the same conditions. This large gene activation observed in Jemalong A17 under control growth conditions may account for its relatively lower levels of gene regulation by salt stress when compared to 108-R. We found six salt-inducible genes in 108-R that were common with those up-regulated in Jemalong A17 in control conditions (Supplemental Table S1, genes marked in yellow), confirming that abiotic stress-related genes for the 108-R genotype are expressed at higher levels in the tolerant genotype in the absence of stress.
Second, intrinsic differences between the two genotypes after a salt stress were searched (J4 versus R4). Direct comparison of gene induction/repression between the two varieties after 4 d of salt stress detected large differences in expression patterns between the individual genotypes. This may be particularly relevant for genes whose levels of expression in control conditions are not the same for each genotype. This comparison allowed the detection of 118 differentially expressed genes that were classified into two groups: (1) those genes that are up-regulated in 108-R at 4n of salt (six genes); and (2) those genes that are up-regulated in Jemalong A17 at 4n (112 genes; Supplemental Table S2). These latter genes could be of interest in determining the differential tolerance to salt stress of these M. truncatula varieties. For this reason, these genes were further characterized. Sorting of the genes up-regulated at 4n in Jemalong A17 into functional categories (according to Journet et al., 2002) revealed that four categories were overrepresented: signal transduction and posttranslational regulation (16.9%), abiotic stimuli and development (11.6%), protein synthesis and processing (11.6%), and hormone and secondary metabolism (11.6%), whereas a significant proportion could not be assigned (unknown function, 24.1%; Fig. 3D). Among those classes underrepresented, we found genes linked to the cell cycle (0%) and to DNA synthesis and chromatin (0.9%) in correlation with the reduction of meristem activity induced by salt stress.
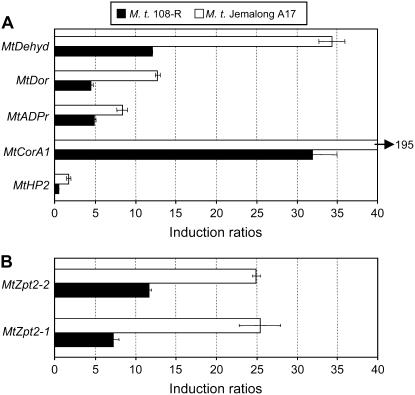
Among the 112 differentially expressed genes up-regulated in Jemalong A17 roots under salt stress, five genes showing different levels of induction in the tolerant variety were randomly selected to validate the changes detected on macroarrays using real-time reverse transcription (RT)-PCR on independent biological samples (Supplemental Table S2, genes indicated in yellow). One codes for a homolog to a ADP-ribosylation factor (MtADPr, TC67888); one has homology to a dormancy-associated protein (MtDor, TC85295), another codes for a cold-inducible CORA protein (MtCorA1, TC68022), a fourth matches the sequence of a dehydrin-related protein (MtDehyd, TC76699), and the fifth is MtHP2, a His-containing phosphotransfer protein potentially related to cytokinin signaling (González-Rizzo et al., 2006). The expression patterns revealed using real-time RT-PCR followed the same trends seen in macroarrays (Fig. 4A). Interestingly, we previously relate that a homolog of the CorA1 (Laberge et al., 1993) gene could be a target of TFIIIA-like C2H2 zinc finger TF, MtZpt2-1 and MtZpt2-2 (Merchan et al., 2007; Supplemental Fig. S1). Indeed, the other potential target gene, Fpf1 (Kania et al., 1997), as well as the MtZpt2-1 TF, were found to be up-regulated in the tolerant variety (Supplemental Table S2, genes indicated in light blue). Hence, we tested whether the MtZpt2 TF may be differentially regulated between these genotypes. Real-time RT-PCR showed that both TFs were more expressed in the salt-tolerant Jemalong A17 genotype than in 108-R (Fig. 4B). These regulatory genes could then be involved in the differential response to salt stress between these Medicago genotypes.
Figure 4.
Real-time RT-PCR analysis of selected genes differentially expressed between genotypes in response to salt stress. Specific gene expression in the salt-tolerant (Jemalong A17) versus salt-sensitive (108-R) genotypes were analyzed in control and salt stress conditions (4 d at 150 mm NaCl). Induction ratios were calculated between the salt-treated and nontreated samples. A representative example out of two biological experiments is shown, and error bars represent sd of three technical replicates. Numbers on the x axis indicate the fold-induction of gene expression in relation to the nonsalt stress condition. A, Real-time RT-PCR of five randomly selected differentially expressed genes between genotypes: MtHP2, a His-containing phosphotransfer protein homolog gene (a gene involved in cytokinin signal transduction); MtCorA1, a cold-and drought-regulated CORA protein homolog gene; MtADPr, a gene encoding an ADP-ribosylation factor homolog protein; MtDor, a gene coding for a dormancy-associated protein; and MtDehyd, a gene encoding a dehydrin-related protein. B, Real-time RT-PCR analysis of two genes that encode putative TFIIIA-type TFs (MtZpt2-1 and MtZpt2-2).
Role of the MtZpt Pathway in M. truncatula 108-R and Jemalong A17 Genotypes
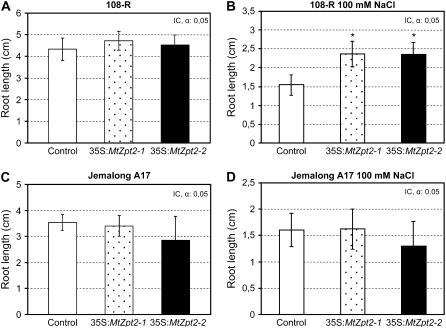
Our previous work (Merchan et al., 2003, 2007) demonstrated a potential role of MtZpt2 TFs in the adaptation of M. truncatula 108-R to salt stress, based either on stable transgenic antisense plants or on Agrobacterium rhizogenes-transformed roots overexpressing MtZpt2-1 (referred as composite plants; Boisson-Dernier et al., 2001). To analyze a potential physiological role of the MtZpt2 genes in root adaptation of M. truncatula Jemalong A17 to salt stress, we comparatively overexpressed the MtZpt2-1 or MtZpt2-2 genes from Jemalong A17 under the control of the constitutive 2× 35S cauliflower mosaic virus promoter in several independent transgenic roots of 108-R or Jemalong A17 genotypes to assay their salt responses. After 3 weeks of growth in control medium, the composite plants were transferred to a salt-containing (100 mm NaCl) or control medium and root length was measured 8 and 6 d after transfer in 108-R and Jemalong A17 plants, respectively (Fig. 5). In nonsaline conditions, the growth of MtZpt2-1 or MtZpt2-2 overexpressing 108-R roots, as well as control roots expressing the GUS reporter gene, were not significantly different (Fig. 5A). However, we detected for both TF-overexpressing 108-R roots a significant increase (Student's t test, P < 0.01 in two independent biological experiments; n > 25) in primary root growth compared to control roots 1 week after transfer on salt medium (Fig. 5B). These results agreed for MtZpt2-1 with our previous experiments (Merchan et al., 2007). In contrast, Jemalong A17 roots overexpressing MtZpt2-1 or MtZpt2-2 were not significantly affected in their growth either in the absence or presence of salt stress (Fig. 5, C and D, respectively), even though these roots were effectively overexpressing the corresponding MtZpt2 transgene (Supplemental Fig. S2).
Figure 5.
Evaluation of root growth in A. rhizogenes-transformed M. truncatula roots overexpressing MtZpt2-1 or MtZpt2-2. Composite plants were prepared as described in Boisson-Dernier et al. (2001) using control empty vector and MtZpt2-1 or MtZpt2-2 overexpressing constructs. After kanamycin selection of transgenic roots, composite plants were transferred to salt-containing or control media. A representative example out of two biological experiments is shown in all cases. *, Statistically significant differences (P < 0.01; n > 25). A and B, Growth of transgenic roots overexpressing either MtZpt2-1 or MtZpt2-2 in M. truncatula 108-R was monitored 1 week after transfer into control (A) or a salt-containing medium (100 mm NaCl; B). The initial position of the root apex after transfer was monitored to determine the degree of root elongation during the week. C and D, Idem as A and B for Jemalong A17 indicating transgenic root growth in control (C) or salt-containing medium (100 mm NaCl; D).
We also examined whether the overexpression of MtZpt2-1 or MtZpt2-2 TFs in M. truncatula 108-R and Jemalong A17 affected the nodulation capacity of these plants under salt stress conditions. We assayed nodulation capacity in control and salt stress conditions of several independent transgenic roots overexpressing these genes in these two genotypes. After 3 weeks of growth in control medium, composite plants were transferred to a salt-containing medium (100 mm NaCl) and inoculated with S. meliloti 2011. We determined the total number of nodules per plant 21 d postinoculation (d.p.i.). Overexpression of either one or the other MtZpt TFs did not alter significantly the nodulation capacity of these two genotypes under salt stress (108-R and Jemalong A17; Supplemental Fig. S3, A and B, respectively). Moreover, no differences in nodulation were observed even in the absence of salt, although the genotype-dependent differential inhibitory effect of salt stress on the symbiotic interaction was detected (Supplemental Fig. S3, A and B).
Hence, even though the MtZpt pathway could not be related to the different nodulation capacity of the genotypes either in the presence or absence of salt stress, their differential root growth response to salt stress may be partially linked to the higher expression levels of the two MtZpt2 TFs in the tolerant variety. These data suggest that the MtZpt2 pathway is activated in Jemalong A17.
DISCUSSION
Soil salinity is one of the major abiotic stresses reducing agricultural productivity. The direct selection of superior salt-tolerant genotypes under field conditions is hindered by the considerable influence that environmental factors have on the response of plants to salinity (Richards, 1996). Salt tolerance is a complex trait involving the function of many genes (Flowers, 2004; Foolad, 2004). In fact, the exploitation of natural genetic variations and the generation of transgenic plants introducing novel genes or altering expression levels of the existing genes are being used to improve salt tolerance (Austin, 1993; Jain and Selvaraj, 1997; Yeo, 1998; Hasegawa et al., 2000; Park et al., 2001; Xiong and Yang, 2003; Guo et al., 2004; Davletova et al., 2005; Dana et al., 2006; Hong and Hwang, 2006). In legumes, salt stress significantly limits productivity because of its adverse effects on the growth of the host plant and its symbiotic interactions. Root growth, nodule development, and nitrogen-fixation efficiency are particularly affected (Rai, 1992; Cardovilla et al., 1994). In this work, we analyze the differential adaptation to salt stress of different M. truncatula genotypes (108-R and Jemalong A17) with regard to root growth and nodulation responses. Analysis of gene expression patterns between these genotypes allowed us to propose the implication of the TFIIIA-type TF pathway in the increased tolerance of Jemalong A17 to salt stress.
Arrest of plant growth during stress conditions depends largely on the severity of the stress (Westgate and Boyer, 1985; Sharp et al., 1988; Spollen et al., 1993; Munns et al., 2000; Bartels and Sunkar, 2005). However, continuation of root growth under drought stress is an adaptive mechanism that facilitates water uptake from deeper soil layers (Sharp et al., 1988). Similarly, continued root growth under salt stress may provide additional surfaces for sequestration of toxic ions, leading to lower salt concentration, and enables reaching soil areas with lower salt concentrations. For example, salt tolerance of barley (Hordeum vulgare) was correlated with better root growth rates coupled with fast growth and early flowering (Munns et al., 2000; Bartels and Sunkar, 2005). In this study, specific experiments using two M. truncatula genotypes (108-R and Jemalong A17) revealed that increased primary root growth after germination may be linked to their different tolerance to salt stress. The magnitude of root growth inhibition was mainly dependent upon the genotype rather than on growth media. The poor “i” medium is probably more similar to the conditions of the rhizosphere than is the relatively rich Fahräeus medium and was chosen for macroarray analysis because good phenotypic variability in response to salinity was observed between these M. truncatula varieties.
The aerial organs also contribute to the various mechanisms aimed at postponing adverse saline effects or tolerating salt stress. These include reduction of water loss by increased stomatal resistance and accumulating sizeable amounts of Na+ in the vacuole (Apse et al., 1999; Steudle, 2000; Assmann and Wang, 2001; Zhang and Blumwald, 2001; Horié and Schroender, 2004). Nevertheless, in Arabidopsis (Arabidopsis thaliana), most of the genes were induced by salt stress only in roots, not in leaves (Shisong et al., 2006). The reason for this might be that leaves, compared with roots, have a large sodium storage capacity, or it might be a consequence of the relatively high concentration of sodium ions in roots as they perceive the salt conditions from the soil environment (Kreps et al., 2002; Volkov et al., 2004; Shisong et al., 2006). Under the conditions imposed in this work, 108-R and Jemalong A17 did not differ significantly in leaf dry weight after 15 d of treatment. However, in nodulated plants, the effects of salt on the aerial part were more significant, suggesting a later effect of salinity in this case. In the initial stages of growth, the aerial part may be less sensitive to salt stress than is the root in legumes.
In legume-Rhizobium symbiosis, the adverse effects of stresses on nodule functioning vary in intensity, depending on aspects such as plant species, rhizobial strain, and duration and conditions of exposure to the stressful condition (Garg and Gupta, 2000; Bouhmouch et al., 2005; Morón et al., 2005). Furthermore, the number and total weight of nodules decreased in salinity treatments (Delgado et al., 1994; Merchan et al., 2003). We showed that total nodule number in the two M. truncatula genotypes was adversely affected by salt stress. The salt present in the growth medium may inhibit the absorption of calcium, reducing the emergence and growth of roots and root hairs and decreasing potential infection sites (Zahran and Sprent, 1986). Nodule growth has previously been reported to be affected by salt stress in Glycine max (Delgado et al., 1994; Gordon et al., 1997) and Phaseolus vulgaris (Delgado et al., 1994; Bouhmouch et al., 2005). It has been proposed that the inhibition of photosynthesis in plants subjected to salt stress leads to a restriction of photosynthate transport toward nodules and a reduction in their size (Bekki et al., 1987). However, González et al. (2001) suggested that nodule growth reduction is linked to the inhibition of enzymes associated with Suc degradation in nodules. In nodulated plants, both aerial and ground tissues are affected by salt, and complex interactions between them may explain the better nodulation of Jemalong A17 plants.
Array analysis to characterize gene expression profiles for large numbers of transcripts has been used to describe the response to environmental stresses in various species, including Arabidopsis for cold, drought, and salt (Kreps et al., 2002; Seki et al., 2002; Takahashi et al., 2004), M. truncatula for recovery from salt stress (Merchan et al., 2007), and rice (Oryza sativa) for response to salt stress (Kawasaki et al., 2001). The large number of stress-regulated genes detected in these studies indicates that comparative transcript profiling could be applied between genotypes of the same species (Taji et al., 2004; Moore et al., 2005; Walia et al., 2005; Weber et al., 2006). This comparative analysis might reveal the responses shared across genotypes and also identify differential responses. Comparison of EST sequences from Jemalong A17 and 108-R revealed a high DNA sequence identity (90%–95%) for the large majority of transcripts, indicating that the macroarray of 108-R genotype (Merchan et al., 2007) can be used for expression profiling of Jemalong A17 genes.
The constitutive high expression of certain stress response genes in plants able to thrive in a particular stress environment emerges as a possibly widespread adaptive mechanism. This would imply that rather than the expression of particular species-specific stress-tolerance genes, it is the altered regulation of conserved genes that enables certain plants to survive in harsh environments. For instance, in the salt cress, Thellungiella halophila, a salt-tolerant relative of the glycophyte Arabidopsis, it was shown by comparative transcriptome analysis that the orthologs of several well-known Arabidopsis stress response genes are highly expressed even under control conditions (Taji et al., 2004); similarly, the metal-tolerant Arabidopsis halleri was found to express a number of metal-homeostasis genes at much higher levels than Arabidopsis, independently of micronutrient status (Becher et al., 2004; Weber et al., 2004). Our analysis suggests that Jemalong A17 is more tolerant than 108-R to salt stress, because several stress-related genes are expressed under nonstressed conditions. Jemalong A17 does not induce major changes at the transcriptional level among the dedicated macroarray population in response to salinity conditions as compared with 108-R. Additionally, gene repression may be moderated in the tolerant genotype, allowing growth to be achieved in the adverse conditions. In fact, many different functions have been identified as being differentially regulated between the two genotypes in response to salt stress (more than 30% of the tested genes in the macroarray). As expected, a major class is linked to stress-related pathways, together with a large class of regulatory genes (posttranscriptional regulation and signal transduction). Interestingly, several response regulators, a cytokinin receptor, and ethylene-related and auxin-inducible genes were found (Supplemental Table S2). This further reinforces the hypothesis of a cross talk between hormone- and environment-related signaling pathways in the control of root growth under adverse conditions (Malamy, 2005; Merchan et al., 2007). In addition, among those showing the largest differences, we detected a dehydrin-related gene and a homolog of imbibition proteins probably linked to the well-known osmotic stress component of salt stress.
TFs are crucial elements for the regulation of development and adaptation to abiotic stresses in plants, and the overexpression of specific TFs leads to increased tolerance to abiotic stress, such as salt stresses (Kasuga et al., 1999; Winicov and Bastola, 1999; Winicov, 2000; Guo et al., 2004; Kim et al., 2004; Mukhopadhyay et al., 2004; Davletova et al., 2005). We have previously studied the MtZpt2 TFIIIA-related TFs (MtZpt2-1 and MtZpt2-2) in the 108-R genotype (Merchan et al., 2003, 2007). Gene expression patterns in antisense plants and A. rhizogenes-transformed roots overexpressing MtZpt2-1 together with transient assays revealed that MtZpt2-1 may be an activator of the MtCorA1 genes in 108-R (Merchan et al., 2007). In this work, we found increased expression of the MtCorA1 gene in the tolerant Jemalong A17 variety that correlated with the differential expression of these TFs. Overexpression of either one or the other MtZpt2 TFs enhanced root growth of 108-R plants but not of Jemalong A17 plants under salt stress conditions. We speculate that activation of the TFIIIA Krüppel-like pathway may be partially involved in the tolerance of Jemalong A17 to salt stress.
In conclusion, our results demonstrate the contrasting salt tolerance at physiological and transcriptional levels between two M. truncatula genotypes. This variability in salt sensitivity may be linked to the activation of MtZpt TFs in the tolerant Jemalong A17 genotype and could be a potential genetic resource for improving the salt tolerance of M. truncatula and legume crops in selective breeding programs.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Two genotypes of Medicago truncatula, 108-R (Hoffmann et al., 1997) and Jemalong A17 (Barker et al., 1990), were used in this work. Seeds of these varieties of M. truncatula were scarified in concentrated sulfuric acid for 8 min and rinsed three times with distilled water. The scarified seeds were surface sterilized for 20 min in bleach (12% [v/v] sodium hypochlorite). After washing with sterilized water, seeds were sown on 1% water-agar plates and stored for 2 d at 4°C before incubating overnight at 24°C in the dark to ensure uniform germination. Germinated seedlings were transferred to square plates or pots containing appropriate medium and treatment (see below) and grown vertically in a growth chamber or in a greenhouse, respectively.
In this work, the Fahräeus and “i” media were used. The poor “i” medium (Blondon, 1964) was used for root growth and nodulation assays of seedlings. The Fahräeus medium (Truchet et al., 1985) with added nitrogen (1 mm) was used to measure root growth and to prepare composite plants (see below). For nodulation experiments on composite plants, Fahräeus medium without nitrogen was used for optimal nodulation.
Treatment and Evaluation of Salinity Tolerance
Plants were grown under various salt stress conditions (0, 30, 60, 90, 120, and 150 mm of sodium chloride) to elucidate differences in the root growth, dry weight biomass, and nodulation capacity between the varieties studied. NaCl is a common salt that adversely affects plant growth under natural conditions, although a unique salt is generally not found in soils (Bernstein, 1962). Different levels of NaCl were used in this study to provide a range of root growth and nodulation responses from the control treatment and of selected varieties.
For root growth comparisons, seedlings were placed in petri dishes with the appropriate medium (poor “i” medium or richer Fahräeus medium), and the position of primary root tips was marked on the petri dish every 24 h. Root length was measured the 5th d for around 20 seedlings in petri dishes, and two replications of each treatment (20 plants/genotypes) were performed. The experiment was conducted in a growth chamber with mean temperature 24°C under a 16-/8-h photoperiod, and 70% of relative humidity.
For measuring the effect of salt concentration on nodulation of M. truncatula genotypes, 2-d-old seedlings were placed in plastic pots containing perlite:sand (3:1, v/v) as mixed substrate (five seedlings per pot) and irrigated with the appropriate medium (“i” medium; Blondon, 1964) in the greenhouse. Plants were grown in this medium until the 5th d after germination (d.a.g.) when salt stress treatments were initiated. The plant growth media with salt stress was identical to that of controls, except for the addition of NaCl at different concentrations (30, 60, 90, 120, and 150 mm). After four additional days, plants were infected with a stationary-phase culture of wild-type Sinorhizobium meliloti strain 2011. Nodules were counted 21 d after inoculation (30 d.a.g.). This experiment was conducted in a greenhouse, minimizing evaporation to keep the total water volume of the pots constant. Two biological replicates per salinity treatment were performed (20 plants/genotype).
Fifteen and 30 d after the start of salt treatment in both types of experiments (root growth comparisons and determination of nodulation capacity in these two varieties, respectively), individual plants of salt stress and nonstress treatments were harvested. Plants were separated into root system and aerial part. Plant materials were dried at 60°C for 48 h and plant dry weight (under salt stress and as a percentage of dry weight under the nonstress treatment) was determined for individual plants of each genotype.
RNA Extraction and DNA Array Construction
Roots were collected after 4 d of salinity treatment at 150 mm NaCl and immediately frozen in liquid nitrogen for RNA extraction (Merchan et al., 2007). For real-time quantitative RT-PCR experiments, total RNA was extracted using the Total RNA Isolation kit (Macherey-Nagel). Macroarray hybridizations were performed as described in the EMBO M. truncatula Practical Course manual (http://www.isv.cnrs-gif.fr/embo01/manuels/pdf/module5.pdf). DNA macroarrays were constructed as described previously in Merchan et al. (2007).
Macroarray Expression Analysis and Real-Time RT-PCR Measurement of Transcript Level of Selected Genes
cDNA probe labeling, hybridization of macroarrays, and quantification of hybridization signals were performed as described in http://www.isv.cnrs-gif.fr/embo01/manuels/pdf/module5.pdf (Merchan et al., 2007). Experiments were normalized relative to expression of six housekeeping genes, whose expression was between 0.5- and 1.5-fold that of the control in all tested conditions and genotypes. These constitutive controls were selected to normalize all signals obtained from macroarray hybridizations (Merchan et al., 2007).
For real-time RT-PCR, after DNase treatment (RQ1 RNase-free DNase, Promega), 1.5 μg of total RNA were retrotranscribed for 1 h at 42°C using the SUPERSCRIPT II first-strand synthesis system (Invitrogen) and subsequently denatured for 10 min at 75°C. One-tenth of the cDNAs were used as a template in 10-μL PCR reactions. PCR was performed with a Light Cycler apparatus and the LC FastStart DNA Master SYBR Green I (Roche Diagnostics) according to the manufacturer's instructions. Gene-specific PCR primers were designed according to the cDNA sequences using the PRIMER3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi; maximum size, 300 bp; melting temperature, 60°C). The following gene-specific primers were used (forward and reverse, respectively): 5′-GGTTGTTTGCGAAGAAGGAG-3′ and 5′-GTACCCCACGGTTTCAACAT-3′ for TC67888 (MtADPr); 5′-GGTGTCGTCGGAGTCACAG-3′ and 5′-TGGCCTTGAGAAGCTTAGGA-3′ for TC85295 (MtDor); 5′-TGTCTCAGCAGATGGCACAG-3′ and 5′-CGAGGAGGAAGTTGATGGAG-3′ for TC76699 (MtDehyd for 108-R); 5′-GAGCGAGGAGGAAGTTGATGG-3′ and 5′-TGGTGCTGGTGGAGTTGTTA-3′ for TC106659 (MtDehyd for Jemalong A17); 5′-GGCGGAGGTGGTTACAATGG-3′ and 5′-GGCAACAGATTCAGCAGCAC-3′ for TC68024 (MtCorA1); 5′-ATAGATGCGTGCCGCAGGTG-3′ and 5′-GCATCTCTACAGATCCACTC-3′ for TC74018 (MtHP2); 5′-AAGTCCGGAAAAGCCGGGAGG-3′ and 5′-GCACTTAACTCACCCACCACTGC-3′ for MtZpt2-1; 5′-GGCAACGGACTTTCTACCTC-3′ and 5′-CTCCTCCATCAGCCACCGTG-3′ for MtZpt2-2. Parallel reactions to amplify MtActin11 were used to normalize the amount of template cDNA. Synthesis of three independent cDNA preparations from the same RNA sample (technical duplicates) allowed us to monitor reproducibility of the assay. A representative example out of the two biological replicates performed is shown for each figure.
Statistical Analysis
Root length, dry weight, and number of nodules in the various treatments were tested for significant differences using a Kruskal and Wallis test (Georgin and Gouet, 2000) because of the low sample size (n = 20). Statistical analysis of the macroarray data was based on a moderated Student's t test and is described in detail in Merchan et al. (2007). Genes with adjusted P values < 0.01 were selected as statistically significant. For composite plants, we analyzed the significance of differences in root growth lengths or nodulation capacities using a Student's t test (P < 0.01).
Agrobacterium rhizogenes Root Transformation
Agrobacterium rhizogenes-transformed M. truncatula roots were prepared as described in Boisson-Dernier et al. (2001), and the binary plasmid used was described in Merchan et al. (2007). Two weeks after inoculation with A. rhizogenes, plants developed transgenic roots and were transferred to the same medium without kanamycin, containing a brown filter paper (recovered from a growth pouch, Mega International) to allow root growth on the surface of the paper (without penetration into the agar), and easy transfer to a new plate with salt-containing medium (100 mm NaCl in “i” medium). Root length was measured 8 and 6 d after transfer in 108-R and Jemalong A17 plants, respectively, from the initial position of the root apex at the time of transfer. Two biological replicates were realized (n > 25).
For nodulation assays, composite plants were transferred to Fahräeus medium without nitrogen containing the brown filter paper and 5 d later were inoculated with 10 mL of S. meliloti strain 2011 suspension (OD600 nm = 0.05) per plate for 1 h in vitro. Nodulation efficiency was first evaluated by counting nodules at 10 d.p.i.. Then, infected composite plants (containing the A. rhizogenes-transformed roots obtained in vitro) were transferred into the greenhouse support (perlite:sand [3:1, v/v] mixed substrate) in “i” medium. After recovery of the transfer (3–4 d) in high humidity conditions, plants were submitted to different stress media and further inoculated with symbiotic bacteria. Nodule number was determined 21 d.p.i. after this second inoculation.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Homology analysis between Zpt2 sequences.
Supplemental Figure S2. Expression levels of MtZpt2 TFs in composite 108-R and Jemalong A17 plants.
Supplemental Figure S3. Evaluation of nodulation capacity in A. rhizogenes-transformed M. truncatula roots overexpressing MtZpt2-1 or MtZpt2-2 in 108-R and Jemalong A17.
Supplemental Table S1. Genes differentially regulated between the salt-tolerant and salt-sensitive genotypes grown under normal conditions (J0 versus R0).
Supplemental Table S2. Genes up-regulated in the salt-tolerant genotype grown under salt stress conditions compared with the sensitive one in the same conditions (J4 versus R4).
Supplementary Material
Acknowledgments
We thank Nathalie Mansion for photographic work and Liliane Troussard for sequencing.
This work was supported by the Spanish Department of Education and Science (a university professor training grant to L.d.L. and a postdoctoral grant to F.M.), and by the “Grain Legumes” FP6 European Economic Community project.
The author responsible for distribution of material integral to the findings presented in this article in accord with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Martin Crespi (crespi@isv.cnrs-gif.fr).
The online version of this article contains Web-only data.
References
- Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285 1256–1258 [DOI] [PubMed] [Google Scholar]
- Arrese-Igor C, González E, Gordon A, Minchin F, Gálvez L, Royuela M, Cabrerizo P, Aparicio-Tejo P (1999) Sucrose synthase and nodule nitrogen fixation under drought and environmental stresses. Symbiosis 27 189–212 [Google Scholar]
- Assmann S, Wang X (2001) From milliseconds to millions of years: guard cells and environmental responses. Plant Biol 4 421–428 [DOI] [PubMed] [Google Scholar]
- Austin RB (1993) Augmenting yield-based selection. In MD Hayward, I Romagosa, eds, Plant Breeding Principles and Prospects. Chapman and Hall, London, pp 391–405
- Barker DG, Bianchi S, Blondon F, Dattée Y, Duc G, Essad S, Flament P, Gallusci P, Génier G, Guy P, et al (1990) Medicago truncatula, a model plant for studying the molecular genetics of the Rhizobium-legume symbiosis. Plant Mol Biol Rep 8 40–49 [Google Scholar]
- Bartels D, Sunkar R (2005) Drought and salt tolerance in plants. Crit Rev Plant Sci 24 23–58 [Google Scholar]
- Becher M, Talke IN, Krall L, Krämer U (2004) Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri. Plant J 37 251–268 [DOI] [PubMed] [Google Scholar]
- Bekki A, Trinchant JC, Rigaud J (1987) Nitrogen fixation (C2H2 reduction) by Medicago nodules and bacteroids under sodium chloride stress. Physiol Plant 71 61–67 [Google Scholar]
- Bell CJ, Dixon RA, Farmer AD, Flores R, Inman J, Gonzales RA, Harrison MJ, Paiva NL, Scott AD, Weller JW, et al (2001) The Medicago genome initiative: a model legume database. Nucleic Acids Res 29 114–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein L (1962) Salt-affected soils and plants. In The Problems of the Arid Zone: Proceedings of the Paris Symposium. UNESCO, Paris, pp 139–174
- Blondon F (1964) Contribution à l'étude du développement de graminées fourragères: ray-grass et dactyle. Rev Gen Bot 71 293–381 [Google Scholar]
- Boisson-Dernier A, Chabaud M, García F, Becard G, Rosenberg C, Barker DG (2001) Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact 14 695–700 [DOI] [PubMed] [Google Scholar]
- Bouhmouch I, Souad-Mouhsine B, Brhada F, Aurag J (2005) Influence of host cultivars and Rhizobium species on the growth and symbiotic performance of Phaseolus vulgaris under salt stress. J Plant Physiol 162 1103–1113 [DOI] [PubMed] [Google Scholar]
- Cardovilla MP, Ligero F, Lluch C (1994) The effect of salinity on N fixation and assimilation in Vicia faba. J Exp Bot 45 1483–1488 [Google Scholar]
- Chinnusamy V, Jagendorf A, Zhu JK (2005) Understanding and improving salt tolerance in plants. Crop Sci 45 437–448 [Google Scholar]
- Cook DR (1999) Medicago truncatula, a model in the making! Curr Opin Plant Biol 2 301–304 [DOI] [PubMed] [Google Scholar]
- Crespi M, Gálvez S (2000) Molecular mechanisms in root nodule development. J Plant Growth Regul 19 155–166 [DOI] [PubMed] [Google Scholar]
- Dana MM, Pintor-Toro JA, Cubero B (2006) Transgenic tobacco plants overexpressing chitinases of fungal origin show enhanced resistance to biotic and abiotic stress agent. Plant Physiol 142 722–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S, Schlauch K, Coutu J, Mittler R (2005) The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol 139 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado MJ, Garrido JM, Lluch C (1994) Effects of salt stress on growth and nitrogen fixation by pea, faba-bean, common bean and soybean plants. Soil Biol Biochem 26 371–376 [Google Scholar]
- Duzan HM, Zhou X, Souleimanov A, Smith DL (2004) Perception of Bradyrhizobium japonicum Nod factor by soybean [Glycine max (L.) Merr.] root hairs under abiotic stress conditions. J Exp Bot 55 2641–2646 [DOI] [PubMed] [Google Scholar]
- Flowers TJ (2004) Improving crop salt tolerance. J Exp Bot 55 307–319 [DOI] [PubMed] [Google Scholar]
- Foolad MR (2004) Recent advances in genetics of salt tolerance in tomato. Plant Cell Tissue Organ Cult 76 101–119 [Google Scholar]
- Garg BK, Gupta IC (2000) Nodulation and symbiotic nitrogen fixation under salt stress. Curr Agric 24 23–35 [Google Scholar]
- Georgin P, Gouet M (2000) Statistiques Avec Excell. Eyrolles, Paris, p 338
- González EM, Gálvez L, Royurla M, Aparicio-Tejo PM, Arresse-Igor C (2001) Insights into the regulation of nitrogen fixation in pea nodules: lessons from drought, abscisic acid and increased photoassimilate availability. Agronomie 21 607–613 [Google Scholar]
- González-Rizzo S, Crespi M, Frugier F (2006) The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18 2680–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon AJ, Minchin FR, Skot L, James CL (1997) Stress induced declines in soybean N2 fixation are related to nodule sucrose synthase activity. Plant Physiol 114 937–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham PH, Vance CP (2003) Legumes: importance and constraints to greater use. Plant Physiol 131 872–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo ZJ, Chen XJ, Wu XL, Ling JQ, Xu P (2004) Overexpression of the AP2/EREBP transcription factor OPBP1 enhances disease resistance and salt tolerance in tobacco. Plant Mol Biol 55 607–618 [DOI] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51 463–499 [DOI] [PubMed] [Google Scholar]
- Hoffmann B, Trinh TH, Leung J, Kondorosi A, Kondorosi E (1997) A new Medicago truncatula line with superior in vitro regeneration, transformation, and symbiotic properties isolated through cell culture selection. Mol Plant Microbe Interact 10 307–315 [Google Scholar]
- Hong JK, Hwang BK (2006) Promoter activation of pepper class II basic chitinase gene, CAChi2, and enhanced bacterial disease resistance and osmotic stress tolerance in the CAChi2-overexpressing Arabidopsis. Planta 223 433–448 [DOI] [PubMed] [Google Scholar]
- Horié T, Schroender J (2004) Sodium transporters in plants: diverse genes and physiological functions. Plant Physiol 136 2457–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain RK, Selvaraj G (1997) Molecular genetic improvement of salt tolerance in plants. Biotechnol Annu Rev 3 245–267 [Google Scholar]
- Journet EP, Van Tuinen D, Gouzy J, Crespeau H, Carreau V, Farmer MJ, Niebel A, Schiex T, Jaillon O, Chatagnier O, et al (2002) Exploring root symbiotic programs in the model legume Medicago truncatula using EST analysis. Nucleic Acids Res 30 5579–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania T, Russenberger D, Peng S, Apel K, Melzer S (1997) FPF1 promotes flowering in Arabidopsis. Plant Cell 9 1327–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17 287–291 [DOI] [PubMed] [Google Scholar]
- Kawasaki S, Borchert C, Deyholos M, Wang H, Brazille S, Kawai K, Galbraith D, Bohnert H (2001) Gene expression profiles during the initial phase of salt stress in rice. Plant Cell 13 889–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kang JY, Cho DI, Park JH, Kim SY (2004) ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J 40 75–87 [DOI] [PubMed] [Google Scholar]
- Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in responses to salt, osmotic, and cold stress. Plant Physiol 130 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laberge S, Castonguay Y, Vezina LP (1993) New cold- and drought-regulated gene from Medicago sativa. Plant Physiol 101 1411–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E, Bisseling T (2003) Signaling in symbiosis. Curr Opin Plant Biol 6 343–350 [DOI] [PubMed] [Google Scholar]
- Malamy JE (2005) Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ 28 67–77 [DOI] [PubMed] [Google Scholar]
- Merchan F, Breda C, Hormaeche JP, Sousa C, Kondorosi A, Aguilar OM, Megías M, Crespi M (2003) A krüppel-like transcription factor gene is involved in salt stress responses in Medicago spp. Plant Soil 257 1–9 [Google Scholar]
- Merchan F, de Lorenzo L, González-Rizzo S, Niebel A, Megías M, Frugier F, Sousa C, Crespi M (2007) Analysis of regulatory pathways involved in the reacquisition of root growth after salt stress in Medicago truncatula. Plant J 51 1–17 [DOI] [PubMed] [Google Scholar]
- Moore S, Payton P, Wright M, Tanksley S, Giovannoni J (2005) Utilization of tomato microarrays for comparative gene expression analysis in the Solanaceae. J Exp Bot 56 2885–2895 [DOI] [PubMed] [Google Scholar]
- Morón B, Soria-Díaz ME, Ault J, Verroios G, Noreen S, Rodríguez-Navarro DN, Gil-Serrano A, Thomas-Oates J, Megías M, Sousa C (2005) Low pH changes the profile of nodulation factors produced by Rhizobium tropici CIAT899. Chem Biol 12 1029–1040 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A, Vij S, Tyagi AK (2004) Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc Natl Acad Sci USA 101 6309–6314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R, Hare RA, James RA, Rebetzke GJ (2000) Genetic variation for improving the salt tolerance of durum wheat. Aust J Agric Res 51 69–74 [Google Scholar]
- Ozturk ZN, Talame V, Deyholos M, Michalowski CB, Galbraith DW, Gozukirmizi N, Tuberosa R, Bohnert HJ (2002) Monitoring large-scale changes in transcript abundance in drought- and salt-stressed barley. Plant Mol Biol 48 551–573 [DOI] [PubMed] [Google Scholar]
- Park JM, Park CJ, Lee SB, Ham BK, Shin R, Paek KH (2001) Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 13 1035–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R (1992) Effect of nitrogen levels and Rhizobium strains on symbiotic N2 fixation and grain yield of Phaseolus vulgaris L. genotypes in normal and saline-sodic soils. Biol Fertil Soils 14 293–299 [Google Scholar]
- Richards RA (1996) Defining selection criteria to improve yield under drought. Plant Growth Regul 20 157–166 [Google Scholar]
- Rüberg S, Tian Z-X, Krol E, Linke B, Meyer F, Wang Y, Püuhler A, Weidner S, Becker A (2003) Construction and validation of a Sinorhizobium meliloti whole genome DNA microarray: genome-wide profiling of osmoadaptive gene expression. J Biotechnol 106 255–268 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, et al (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold, and high-salinity stresses using a full-length cDNA microarray. Plant J 31 279–292 [DOI] [PubMed] [Google Scholar]
- Sharp RE, Hsiao TC, Silk WK (1988) Growth of the maize primary root at low water potentials. I. Spatial distribution of expansive growth. Plant Physiol 87 50–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shisong M, Gong Q, Bohnert HJ (2006) Dissecting salt stress pathways. J Exp Bot 57 1097–1107 [DOI] [PubMed] [Google Scholar]
- Spollen WG, Sharp RE, Saab IN, Wu Y (1993) Regulation of cell expansion in roots and shoots at low water potentials. In JAC Smith, H Griffiths, eds, Water Deficits: Plant Responses from Cell to Community. BIOS Scientific Publishers, Oxford, pp 37–52
- Steudle E (2000) Water uptake by roots: effects of water deficit. J Exp Bot 51 1531–1542 [DOI] [PubMed] [Google Scholar]
- Taji T, Seki M, Satou M, Sakurai T, Kobayashi M, Ishiyama K, Narusaka Y, Narusaka M, Zhu JK, Shinozaki K (2004) Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiol 135 1697–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Seki M, Ishida J, Satou M, Sakurai T, Narusaka M, Kamiya A, Nakajima M, Enju A, Akiyama K, et al (2004) Monitoring the expression profiles of genes induced by hyperosmotic, high salinity, and oxidative stress and abscisic acid treatment in Arabidopsis cell culture using a full-length cDNA microarray. Plant Mol Biol 56 29–55 [DOI] [PubMed] [Google Scholar]
- Tester M, Davenport RJ (2003) Na+ transport and Na+ tolerance in higher plants. Ann Bot (Lond) 91 503–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truchet G, Déebelle F, Vasse J, Terzaghi B, Garnerone AM, Rosenberg C, Batut J, Maillet F, Dénarié J (1985) Identification of a Rhizobium meliloti pSym2011 region controlling the host specificity of root hair curling and nodulation. J Bacteriol 164 1200–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov V, Wang B, Dominy PJ, Fricke W, Amtmann A (2004) Thellungiella halophila, a salt-tolerant relative of Arabidopsis thaliana, possesses effective mechanisms to discriminate between potassium and sodium. Plant Cell Environ 27 1–14 [Google Scholar]
- Walia H, Wilson C, Condamine P, Liu X, Ismail AM, Zeng L, Wanamaker S, Mandal J, Xu J, Cui X, et al (2005) Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiol 139 822–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Vinocur B, Altman A (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218 1–14 [DOI] [PubMed] [Google Scholar]
- Weber M, Harada E, Vess C, von Roepenack-Lahaye E, Clemens S (2004) Comparative microarray analysis of Arabidopsis thaliana and Arabidopsis halleri roots identifies nicotianamine synthase, a ZIP transporter and other genes as potential metal hyperaccumulation factors. Plant J 37 269–281 [DOI] [PubMed] [Google Scholar]
- Weber M, Trampczynska A, Clemens S (2006) Comparative transcriptome analysis of toxic metal responses in Arabidopsis thaliana and the Cd2+-hypertolerant facultative metallophyte Arabidopsis halleri. Plant Cell Environ 29 950–963 [DOI] [PubMed] [Google Scholar]
- Westgate ME, Boyer JS (1985) Osmotic adjustment and the inhibition of leaf, root, stem and silk growth at low water potentials in maize. Planta 164 540–549 [DOI] [PubMed] [Google Scholar]
- Winicov I (2000) Alfin1 transcription factor overexpression enhances plant root growth under normal and saline conditions and improves salt tolerance in alfalfa. Planta 210 416–422 [DOI] [PubMed] [Google Scholar]
- Winicov I, Bastola DR (1999) Transgenic overexpression of the transcription factor Alfin1 enhances expression of the endogenous MsPRP2 gene in alfalfa and improves salinity tolerance of the plants. Plant Physiol 120 473–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Yang Y (2003) Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell 15 745–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo AR (1998) Molecular biology of salt tolerance in the context of whole-plant physiology. J Exp Bot 49 915–929 [Google Scholar]
- Young ND, Cannon SB, Sato S, Kim DJ, Cook DR, Town CD, Roe BA, Tabata S (2005) Sequencing the genespaces of Medicago truncatula and Lotus japonicus. Plant Physiol 137 1174–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahran HH (1999) Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate. Microbiol Mol Biol Rev 63 968–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahran HH, Sprent JI (1986) Effects of sodium chloride and polyethylene glycol on root-hair infection and nodulation of Vicia faba L. plants by Rhizobium leguminosarum. Planta 167 303–309 [DOI] [PubMed] [Google Scholar]
- Zhang HX, Blumwald E (2001) Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol 19 765–768 [DOI] [PubMed] [Google Scholar]
- Zhu JK (2001) Cell signaling under salt, water and cold stresses. Curr Opin Plant Biol 4 401–406 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.