Abstract
The B-class genes APETALA3 (AP3) and PISTILLATA (PI) in Arabidopsis (Arabidopsis thaliana) and their orthologs in other species have been the focus of studies to elucidate the development of petals and stamens in angiosperm flowers. Evolutionary analysis indicates that B-class genes have undergone multiple gene duplication events in angiosperms. The resultant B-class lineages are characterized by short, conserved amino acid sequences at the extreme C-terminal end of the B-class proteins. AP3 is a member of the euAP3 lineage that contains both the euAP3 and PI-derived motifs at the C terminus. PI is a member of the PI lineage that contains the C-terminal PI motif at the C terminus. Despite conservation over a wide evolutionary distance, the function of C-terminal motifs is not well understood. In this study, we demonstrate that truncated forms of AP3 and PI, which lack the conserved C-terminal motifs, function to direct floral organ identity specification in Arabidopsis plants. By contrast, larger truncations, which remove the third putative amphipathic α-helix in the K domain of AP3 or PI, are nonfunctional. We conclude that the euAP3 and PI-derived motifs of AP3 and the PI motif of PI are not essential for floral organ identity function of AP3 and PI in Arabidopsis.
The dominant paradigm for floral organ identity specification in angiosperms is the well-established ABC model, which states that multiple activities are present in the flower and these activities function in a combinatorial manner to specify floral organ identity (Coen and Meyerowitz, 1991; Krizek and Fletcher, 2005). Genes that possess ABC activity have been extensively characterized in model species such as Arabidopsis (Arabidopsis thaliana), snapdragon (Antirrhinum majus), and petunia (Petunia hybrida), resulting in elucidation of many of the molecular mechanisms that underlie floral organ identity specification. In recent years, it has been demonstrated that variation in the function and expression of ABC genes contributes to the evolution of diverse floral morphologies of angiosperms. In particular, B-class genes (e.g. APETALA3 [AP3] and PISTILLATA [PI] in Arabidopsis) have been at the forefront of studies that seek to establish a relationship between sequence evolution and function (Kramer and Hall, 2005; Kramer et al., 2006).
The majority of ABC genes encode MADS transcription factors that function as sequence-specific DNA-binding proteins. In Arabidopsis, the MADS family is large and consists of over 100 members, most of which have been demonstrated to be involved in various aspects of plant development (Becker and Theissen, 2003; Parenicova et al., 2003; de Folter and Angenent, 2006). The MADS proteins involved in specifying floral organ identity possess a characteristic MIKC domain structure (Alvarez-Buylla et al., 2000). The highly conserved MADS (M) domain, located at the N terminus, possesses DNA-binding, dimerization, and nuclear localization activities (Riechmann et al., 1996a, 1996b). The K domain, a keratin-like coiled-coil domain, is located in the central region of MIKC MADS proteins (Ma et al., 1991). The K domain encodes three putative amphipathic α-helices that function in protein-protein interaction (Yang et al., 2003a). The intervening (I) domain, located between the M and K domains, plays a role in MADS protein dimerization and functional specificity (Riechmann et al., 1996a). The M, I, and K domains exhibit greater amino acid conservation than the C-terminal (C) domain. Consistent with this, the C domain possesses different functions in different MADS proteins. In a subset of MADS proteins, the C domain encodes a transcriptional activation function (Cho et al., 1999; Honma and Goto, 2001; de Folter et al., 2005), whereas in other proteins the C domain has been implicated in higher order MADS protein-protein interactions (Egea-Cortines et al., 1999; Honma and Goto, 2001).
Recent advances in the understanding of the function and evolution of MIKC MADS genes have emerged from studying the B-class genes AP3 and PI in Arabidopsis and their orthologs in other species. AP3 and PI are floral organ identity genes that are essential for proper petal and stamen identity. In strong mutants for either ap3 or pi, petals develop as sepals and stamens develop as carpels (Jack et al., 1992; Goto and Meyerowitz, 1994). Whereas most MADS proteins bind to DNA as homodimers or heterodimers, AP3 and PI function as an obligate heterodimer (Riechmann et al., 1996b; Hill et al., 1998; Tilly et al., 1998). All plant MIKC MADS proteins recognize and bind to similar consensus DNA sequences; therefore, the functional specificity of AP3 and PI is most likely mediated by interactions with other proteins and not by differential DNA sequence recognition (Riechmann and Meyerowitz, 1997). Considerable evidence supports a model that predicts that the AP3/PI heterodimer forms a higher order protein complex with additional MADS proteins allowing distinctive MADS complexes to specify floral organ identity in different floral whorls (Egea-Cortines et al., 1999; Honma and Goto, 2001; Theissen, 2001). Although higher order MADS complexes have been postulated, they have not yet been isolated from plant cells.
Phylogenic analysis of AP3 and PI orthologs from a wide range of plant species has allowed the evolutionary history of B-class genes to be reconstructed. AP3 and PI lineages are sister lineages that likely resulted from a gene duplication event of an ancestral B-class gene that occurred at the base of the angiosperms (Kramer et al., 1998; Stellari et al., 2004). In both the AP3 and PI lineages, the highest level of amino acid identity is in the M domain and the lowest level of overall sequence identity/similarity is in the C domain. Despite the low overall sequence conservation in the C domain, alignment of AP3 and PI orthologs from a wide range of plant species reveals small regions of high amino acid conservation at the extreme C-terminal end of the AP3 and PI orthologs (Kramer et al., 1998). The C-terminal end of PI orthologs contains the approximately 16-amino acid PI motif with the consensus sequence MPFxFRVQPxQPNLQE. AP3 orthologs contain a motif similar to the PI motif, called the PI-derived motif, with the consensus sequence FxFRLQPSQPNLH (Kramer et al., 1998). In higher eudicots, the AP3 lineage has undergone a second duplication event leading to two separate AP3 lineages: the TM6 lineage and the euAP3 lineage. The TM6 lineage possesses the PI-derived motif as well as the paleoAP3 motif, which has the consensus sequence YGxHDLRLA (Kramer et al., 1998). By contrast, the euAP3 lineage has replaced the paleoAP3 motif with the euAP3 motif, presumably by frameshift mutation (Vandenbussche et al., 2003; Kramer et al., 2006). The euAP3 motif has the consensus sequence SDLTTFALLE. The high level of amino acid conservation in the C-terminal motifs suggests that these motifs provide a critical function for the AP3 and PI proteins.
At present, the function of C-terminal motifs is unknown. Also, it remains unclear whether C-terminal motifs are necessary for the function of AP3- and PI-like genes. One previous study demonstrated that C-terminal motifs were essential for function because truncations of AP3 and PI that remove these motifs were unable to rescue ap3 or pi mutants when expressed in Arabidopsis (Lamb and Irish, 2003). By contrast, a second study demonstrated that a PI ortholog in pea (Pisum sativum) that does not encode the PI motif is still capable of rescuing pi mutants when expressed in Arabidopsis (Berbel et al., 2005). Evidence that C-terminal motifs have a function comes from domain-swap experiments. For example, when the paeloAP3 motif from the lower eudicot Dicentra eximia is substituted for the euAP3 motif in AP3, the chimeric AP3cPALEO gene can partially rescue the stamen defects of an ap3 mutant, but not the petal defects (Lamb and Irish, 2003). Surprisingly, a chimeric PIcAP3 construct, in which the PI-derived and euAP3 motifs of AP3 replaced the PI motif of PI, functioned as AP3 in mutant rescue experiments, suggesting that the C-terminal end of AP3 provides AP3 functional specificity. By contrast, a chimeric protein that replaces the AGAMOUS (AG) C domain with the AP3 C domain functions like AG, not AP3, in planta (Krizek and Meyerowitz, 1996a), demonstrating that the C domain of AP3 is not capable of providing AP3 functional specificity to all floral organ identity MADS proteins. The bottom line from these experiments is that no clear function has been established for the C-terminal motifs in AP3 and PI.
To clarify the functional relevance of the PI-derived and euAP3 motifs of AP3 and the PI motif of PI, we have constructed versions of AP3 and PI lacking these domains and tested their floral organ identity function in Arabidopsis plants. In contrast to an earlier study (Lamb and Irish, 2003), we find that the C-terminal motifs of AP3 and PI are not required for floral organ identity specification.
RESULTS
PI-Derived and euAP3 Motifs of AP3 Are Not Necessary for Floral Organ Identity Function in Planta
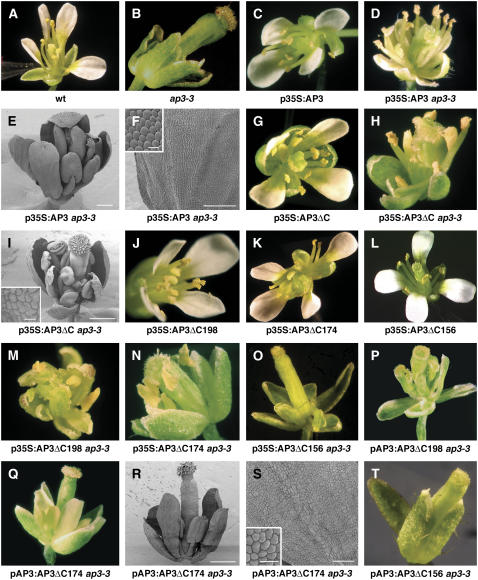
To better understand the functional significance of the PI-derived and euAP3 motifs of AP3, a nonsense mutation was introduced at amino acid 175 of AP3 (AP3ΔC). In AP3ΔC, most of the C domain, including the PI-derived and euAP3 motifs, are downstream of the engineered stop codon (Fig. 1). The construct was placed under the broadly expressed cauliflower mosaic virus (CaMV) 35S promoter and transformed into Arabidopsis. Transgenic plants that express full-length AP3 cDNA under the CaMV 35S promoter exhibit a homeotic conversion of carpels to stamens in the fourth whorl of the flower (Fig. 2C; compare with wild type in Fig. 2A; Jack et al., 1994). p35S:AP3ΔC transgenic flowers exhibit a phenotype similar to p35S:AP3 (Fig. 2G), suggesting that truncated AP3 possesses floral organ identity function in the fourth whorl of the flower. To test whether AP3ΔC could function in place of endogenous AP3, p35S:AP3ΔC plants were assayed in an ap3-3 mutant background. ap3-3 is a strong allele of AP3 that contains a premature stop codon at amino acid 18 (Jack et al., 1992). ap3-3 flowers have strong floral organ identity transformation of petals to sepals in the second whorl and stamens to carpels in the third whorl (Fig. 2B). p35S:AP3 ap3-3 exhibits strong rescue of third-whorl defects, but only partial rescue of second-whorl defects of ap3-3 flowers (Fig. 2, D–F; Supplemental Table S1). p35S:AP3 ap3-3 second-whorl organs develop as sepaloid petals that contain primarily small, rounded cells characteristic of petals (Fig. 2F, inset), but some cells are more elongated and irregularly shaped and thus are more similar to sepal cells. Second-whorl organs of p35S:AP3ΔC ap3-3 contain cells characteristic of both sepals and petals and thus exhibit less rescue than p35S:AP3 ap3-3 (Fig. 2I, inset). p35S:AP3ΔC ap3-3 flowers exhibit a degree of rescue similar to p35SAP3 ap3-3 in the third whorl, where stamens are strongly rescued (Fig. 2, H and I).
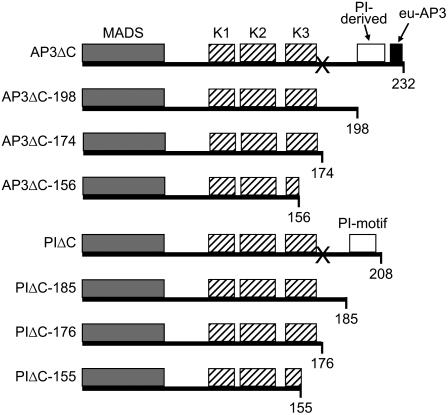
Figure 1.
Summary of constructs. Schematic representation of the constructs used in this study. The gray box indicates the DNA-binding M domain. Hatched boxes indicate the three putative amphipathic α-helices in the K domain (K1, K2, and K3). Positions of the PI-derived (white box) and euAP3 motif (black box) of AP3 and the PI motif (white box) of PI are indicated. Numbers at the bottom represent amino acid positions. “X” represents the position of the engineered stop codon.
Figure 2.
Phenotype of transgenic plants expressing AP3 variants. In planta assays of AP3 transgenes lacking the euAP3 and PI-derived motifs. A to D, G, H, J to Q, and T, Photographs of flowers. E, F, I, R, and S are scanning electron micrographs. A, Wild-type. B, ap3-3. C, p35S:AP3. D to F, p35S:AP3 ap3-3. G, p35S:AP3ΔC. H and I, p35S:AP3ΔC ap3-3. J, p35S:AP3ΔC198. K, p35S:AP3ΔC174. L, p35S:AP3ΔC156. M, p35S:AP3ΔC198 ap3-3. N, p35S:AP3ΔC174 ap3-3. O, p35S:AP3ΔC156 ap3-3. P, pAP3:AP3ΔC198 ap3-3. Q to S, pAP3:AP3ΔC174 ap3-3. T, pAP3:AP3ΔC156 ap3-3. Scale bars: 500 μm (R), 200 μm (E), 100 μm (F and S), 20 μm (I and R; inset), and 10 μm (F; inset).
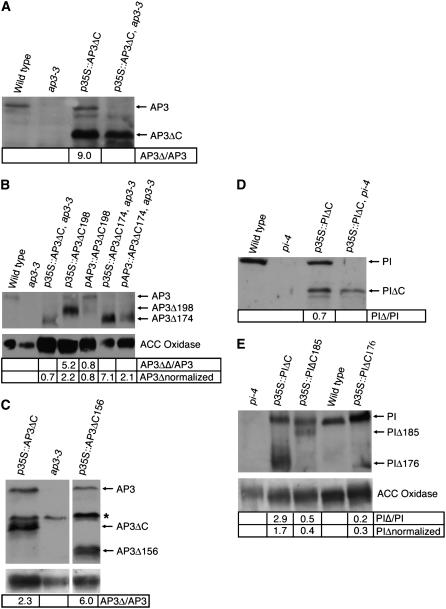
To demonstrate that rescue of the mutant phenotype is due to the AP3ΔC transgene producing a truncated AP3 protein and not full-length product, immunoblots were performed on p35S:AP3ΔC ap3-3 flowers utilizing AP3 antisera. Truncated AP3 protein was detected at the predicted molecular mass (19 kD) and no full-length (25 kD) AP3 protein was detected (Fig. 4A). These results suggest that the euAP3 and PI-derived motifs are not essential for the floral organ identity function of AP3.
Figure 4.
Detection of truncated AP3 and PI proteins via immunoblot. Immunoblots detect truncated AP3 and PI proteins in plant extracts. A to C, Immunoblots probed with α-AP3 sera. D to E, Immunoblots probed with α-PI sera. ACC oxidase loading control shown for B, C, and E. A, The top arrow indicates endogenous AP3 and the lower arrow indicates AP3ΔC. The quantitation compares the signal strength of AP3ΔC to endogenous AP3 within a single lane. B, The top arrow indicates endogenous AP3, the middle arrow points to AP3ΔC198, and the bottom arrow indicates AP3ΔC and AP3ΔC174. The AP3Δ/AP3 quantitation compares the signal strength of AP3ΔC to endogenous AP3 within a single lane. The AP3Δ normalized quantitation compares the ratio of signal strength of AP3Δ:ACC oxidase to AP3:ACC oxidase in lane 1. C, The top arrow points endogenous AP3, the middle arrow points to AP3ΔC, and the bottom arrow points to AP3ΔC156. Asterisk indicates a cross-reacting band. The AP3Δ/AP3 quantitation compares the signal strength of AP3ΔC to endogenous AP3 within a single lane. D, The top arrow points to endogenous PI and the lower arrow points to PIΔC. The quantitation compares the signal strength of PIΔC to endogenous PI within a single lane. E, The top arrow points to endogenous PI, the middle arrow points to PIΔC185, and the bottom arrow points to PIΔC and PIΔC176. The PIΔ/PI quantitation compares the signal strength of PIΔC to endogenous PI within a single lane. The PIΔ normalized quantitation compares the ratio of signal strength of PIΔ to ACC oxidase to the ratio of PI to ACC oxidase in lane 4.
Although immunoblots indicate that no full-length AP3 protein was produced in p35S:AP3ΔC ap3-3 flowers, it is possible that there is read-through of the engineered stop codon, resulting in a low level of full-length AP3 protein that is below the detection limit of our immunoblots. To eliminate this possibility, we made constructs that truncate AP3 after amino acids 198 and 174 (Fig. 1). The truncation point in the AP3ΔC198 construct was chosen because it mimicked the AP3 constructs that have been previously reported to be nonfunctional in Arabidopsis plants (Lamb and Irish, 2003). The AP3ΔC174 construct was chosen because it produces a protein that is the same length as the engineered stop codon AP3ΔC construct that we found to be functional in planta.
In the first series of experiments, truncated AP3 genes were placed under the control of the CaMV 35S promoter to look at the ability of these constructs to direct organ identity transformations in the fourth whorl of the flower. p35S:AP3ΔC198 transgenic plants exhibit strong transformation of fourth-whorl carpels to stamens, a phenotype similar to p35S:AP3 (Fig. 2J; Supplemental Table S1). Similarly, p35S:AP3ΔC174 plants exhibit carpel-to-stamen conversions, but the organ identity transformation is less complete (Fig. 2K). Immunoblots indicate that truncated AP3 proteins are detected at the predicted molecular mass for AP3ΔC198 and AP3ΔC174 (22 and 19 kD, respectively; Fig. 4B). The p35S:AP3 truncations were also analyzed in an ap3-3 mutant background to assay for rescue of the mutant phenotype. Both the p35S:AP3ΔC198 and the p35S:AP3ΔC174 constructs rescue the third-whorl stamen defects and partially rescue the second-whorl petal defects of ap3-3 mutants (Fig. 2, M and N). p35S:AP3, p35S:AP3ΔC, p35S:AP3ΔC198, and p35S:AP3ΔC174 all exhibit similar strong rescue of the third-whorl defects of ap3-3 flowers, but none of these lines demonstrates complete rescue of second-whorl defects and they instead develop organs that possess characteristics of both sepals and petals (Fig. 2, F, I, and S; Supplemental Table S1).
AP3ΔC198 and AP3ΔC174 truncations were also expressed under the control of the AP3 promoter, which directs expression primarily in the second and third whorls (Hill et al., 1998; Tilly et al., 1998). pAP3:AP3ΔC174 ap3-3 strongly rescues the third-whorl defects and rescue of second-whorl petals is as strong as full-length AP3 (Fig. 2, Q–S; Supplemental Table S1). In pAP3:AP3ΔC174, the second-whorl organs are indistinguishable from wild-type petals except for the slightly stunted appearance (Fig. 2, Q and R). Scanning electron microscopy reveals that the cells of the second whorl exhibit rounded morphology characteristic of petal cells, and irregularly shaped and elongated cells characteristic of sepals are not observed (Fig. 2S). Immunoblots reveal that the pAP3:AP3ΔC174 lines express truncated protein at levels equivalent to or exceeding the levels of endogenous AP3 (Fig. 4B). By contrast, all pAP3:AP3ΔC198 lines analyzed are unable to rescue the petal and stamen defects of ap3-3 mutant flowers (Fig. 2P). Analysis of protein expression in the pAP3:AP3ΔC198 lines reveals that truncated protein is expressed below endogenous levels, perhaps below the level required to provide AP3 function (Fig. 4B). In summary, because AP3ΔC174 can function to the same extent as wild-type AP3 in overexpression and mutant rescue assays, we conclude that the PI-derived motif and the euAP3 motif are not required for floral organ identity function of AP3.
The PI Motif of PI Is Not Required for Floral Organ Identity Function in Planta
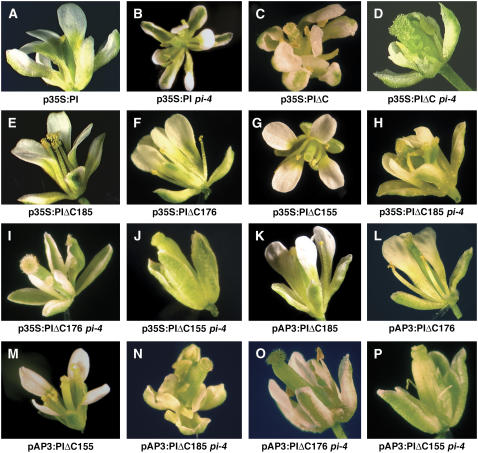
To test the functional necessity of the PI motif, a nonsense mutation was engineered at amino acid 177 of PI (PIΔC). In PIΔC, most of the C domain, including the PI motif, is downstream of the stop codon (Fig. 1). The PIΔC construct was placed under the control of the CaMV 35S promoter and transformed into Arabidopsis plants. Transgenic plants that express full-length wild-type PI under the CaMV 35S promoter exhibit a partial organ identity transformation of sepals to petals in the first whorl of the flower (Fig. 3A; Krizek and Meyerowitz, 1996b). p35S:PIΔC transgenic flowers, which lack the PI motif, also exhibit a partial homeotic conversion of sepals to petals in the first whorl of the flower (Fig. 3C). To test whether PIΔC could function to direct normal petal and stamen development, p35S:PIΔC plants were assayed in a pi-4 mutant background. pi-4 is a strong allele of PI that disrupts the 5′ splice site for intron 1 (Yang et al., 2003b) and results in organ identity transformations of petals to sepals in the second whorl and stamens to carpels in the third whorl. p35S:PIΔC pi-4 flowers exhibit petals in the second whorl and primarily stamens in the third whorl (Fig. 3D; Supplemental Table S2). Some third-whorl organs are chimeric organs possessing both stamen and carpel tissue, but the extent of third-whorl chimeric organs is similar to that observed in p35S:PI pi-4 (Fig. 3B; Supplemental Table S2). Immunoblots utilizing PI-specific antisera demonstrate that the p35S:PIΔC pi-4 flowers produce a truncated protein product at the predicted molecular mass (19 kD) and no full-length PI protein is detectable (Fig. 4D). Thus, these results suggest that the PI motif of PI is not necessary for PI floral organ identity function.
Figure 3.
Phenotype of transgenic plants expressing PI variants. In planta assays of PI transgenes lacking the PI motif. A, p35S:PI. B, p35S:PI pi-4. C, p35S:PIΔC. D, p35S:PIΔC pi-4. E, p35S:PIΔC185. F, p35S:PIΔC176. G, p35S:PIΔC155. H, p35S:PIΔC185 pi-4. I, p35S:PIΔC176 pi-4. J, p35S:PIΔC155 pi-4. K, pAP3:PIΔC185. L, pAP3:PIΔC176. M, pAP3:PIΔC155. N, pAP3:PIΔC185 pi-4. O, pAP3:PIΔC176 pi-4. P, pAP3:PIΔC155 pi-4.
To eliminate the possibility that a small amount of endogenous PI protein is produced due to read-through of the engineered stop codon, we made constructs that truncate PI after amino acids 185 and 176 (Fig. 1). The PIΔC185 construct produces a protein that terminates just past amino acid 185 and is similar to a construct previously reported to be nonfunctional in planta (Lamb and Irish, 2003). The PIΔC176 construct produces a protein that terminates immediately after amino acid 176 and produces the same size protein as the p35S:PIΔC construct that we found to be functional in planta.
The PIΔC176 and PIΔC185 truncations were expressed under CaMV 35S control and transformed into Arabidopsis. Flowers of both p35S:PIΔC185 and p35S:PIΔC176 plants exhibit floral organ conversion in the first whorl of the flower, causing sepals to develop as petals or petaloid organs, indicating that these truncated PI proteins are functional (Fig. 3, E and F; Supplemental Table S2). We also assayed p35S:PIΔC185 and p35S:PIΔC176 in a pi-4 background. Both p35S:PIΔC185 pi-4 and p35S:PIΔC176 pi-4 flowers exhibit first-whorl organ identity transformation of sepals to petals and exhibit restoration of petals in the second whorl (Fig. 3, H and I). Rescue of third-whorl stamens is variable between constructs; the p35S:PIΔC185 pi-4 third-whorl organs are primarily stamenoid with some carpeloid features, whereas the p35S:PIΔC176 pi-4 third-whorl organs are chimeric organs that clearly consist of a mixture of stamen and carpel tissue (Fig. 3, H and I; Supplemental Table S2). Immunoblots utilizing PI-specific antisera confirm that proteins of the expected molecular mass are produced in the p35S:PIΔC185 and p35S:PIΔC176 lines (Fig. 4E). Analysis of the levels of expression demonstrate that PI truncations that express at less than one-half of endogenous levels are still able to provide PI activity, thus ruling out the possibility that PI function is an artifact due to overexpression (Fig. 4E).
The PIΔC185 and the PIΔC176 truncations were also placed under the control of the AP3 promoter, which expresses primarily in the second and third floral whorls but also is active in a few cells at the base of the first-whorl sepals (Jack et al., 1994; Krizek and Meyerowitz, 1996b). PI transcript is normally expressed in the second, third, and fourth whorls of young flowers, but absent in the first whorl (Goto and Meyerowitz, 1994). PI expression in the fourth whorl of wild-type flowers is not maintained because AP3 is not expressed in the fourth whorl, resulting in a failure of AP3/PI autoregulation (Goto and Meyerowitz, 1994). When PI is expressed under the AP3 promoter, PI transcript is expressed together with AP3 at the base of the first whorl, and simultaneous expression of both AP3 and PI in the first whorl results in an expansion of B-class activity, leading to partial organ identity conversion of sepals to petals (Lamb and Irish, 2003). Both pAP3:PIΔC185 and pAP3:PIΔC176 display first-whorl sepal-to-petal transformations in an otherwise wild-type background (Fig. 3, K and L), suggesting that these truncations are functional. In addition, both pAP3:PIΔC185 pi-4 and pAP3:PIΔC176 pi-4 not only rescue the pi-4 phenotype in the second and third whorls, but also exhibit sepal-to-petal transformations in the first whorl (Fig. 3, N and O; Supplemental Table S2). Immunoblots using PI antisera confirm that no full-length PI protein is detectable in pAP3:PIΔC185 pi-4 and pAP3:PIΔC176 pi-4 (data not shown). In summary, these results clearly demonstrate that the PI motif is not required for the floral organ identity function of PI.
The K3 Domain Is Necessary for Function of AP3 or PI in Planta
The above results suggest that truncation of the C-terminal 30 amino acids of PI and the C-terminal 55 amino acids of AP3 does not affect the function of these proteins. To attempt to pinpoint the minimal functional length, we generated constructs that truncate PI and AP3 at amino acids 155 and 156, respectively. This location was chosen because it truncates the proteins between the putative K2 and K3 helices in the K domain.
p35S:AP3ΔC156 transgenic flowers do not exhibit floral organ identity transformations in the fourth whorl (Fig. 2L). In addition, p35S:AP3ΔC156 do not rescue the second- and third-whorl defects of ap3-3 mutants (Fig. 2O). Immunoblots using AP3 antisera detect a truncated protein at the correct molecular mass (15 kD), demonstrating that the failure to observe organ identity transformation is not due to a failure of protein accumulation (Fig. 4C). By contrast, although the AP3ΔC156 construct driven by the AP3 promoter was unable to rescue ap3-3 mutants (Fig. 2O), we were unable to detect truncated protein via immunoblot (data not shown).
As with AP3, PI truncations at amino acid 155 do not function to direct floral organ identity. Neither p35S:PIΔC155 nor pAP3:PIΔC155 plants exhibit floral organ identity transformations in the first whorl of the flower (Fig. 3, G and M). Also, neither line could rescue the petal and stamen defects of pi-4 mutants (Fig. 3, J and P). Expression of the truncated PIΔ155 protein could not be confirmed by immunoblot analysis using PI antisera; thus, the failure of this truncated protein to function may be due to low levels of expression or instability of the truncated protein. In summary, removal of the K3 α-helix together with the C domain clearly prevents proper floral organ identity specification by AP3 and may prevent specification by PI as well.
The PI Motif of PI Is Not Required for Higher Order Complex Formation in Yeast
AP3 and PI have been demonstrated to interact with one another in yeast two-hybrid assays and to interact with other MADS proteins such as SEPALLATA1 (SEP1) and SEP3 in yeast three-hybrid assays (Honma and Goto, 2001; Yang et al., 2003b; Yang and Jack, 2004). To test whether the PI motif of PI was necessary for AP3/PI/SEP1 ternary complex formation in yeast, we tested interactions of PI and SEP1 with three versions of PI: full-length PI (PI-MIKC), a MADS deleted form of PI (PI-IKC), and a C domain deleted form of PI (PIΔC). A strong interaction was observed when either PIΔC or PI-IKC was coexpressed together with both a MADS deleted form of AP3 and a full-length version of SEP1 (Fig. 5), demonstrating that the C domain of PI, including the PI motif, is not required for ternary complex formation in yeast. We also detected a weak interaction between PI-IKC and SEP1 in the absence of an AP3 intermediate (i.e. in a yeast two-hybrid assay); such an interaction is consistent with previous results (Yang and Jack, 2004). Interestingly, when PIΔC is tested with SEP1 in a yeast two-hybrid assay, interaction is weaker (Fig. 5), suggesting that the C domain of PI may play a role in mediating interaction between PI and SEP1.
Figure 5.
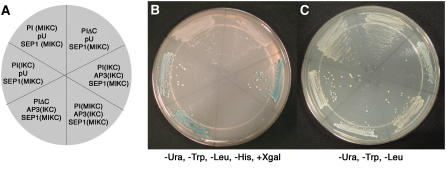
PIΔC is not defective in ternary complex interaction with AP3 and SEP1. Yeast three-hybrid interactions of AP3, PI, and SEP1. A to C, PI (MIKC) is full-length PI, PI (IKC) is a MADS deleted version of PI, PIΔC is PI lacking the C domain, AP3 (IKC) is AP3 lacking the M domain, and SEP1 (MIKC) is full-length SEP1. PI variants were fused to the GAL4 DNA-binding domain. Full-length SEP1 was fused to the GAL4 activation domain. An unfused MADS deleted version of AP3 was cloned into the pU vector. A, Schematic summarizing yeast genotypes streaked on the plates in B and C. B, Two microliters of yeast culture was plated on Sc-Ura-Leu-Trp-His + Xgal plates. C, Two microliters of yeast culture was plated on Sc-Ura-Leu-Trp plates.
DISCUSSION
In this study, we demonstrate that the euAP3 and PI-derived motifs are not required for the floral organ identity function of AP3. Similarly, the PI motif is not required for the floral organ identity function of PI. For AP3, transgenic plants lacking the euAP3 and PI-derived motifs exhibit fourth-whorl carpel-to-stamen floral organ identity conversions when expressed under the CaMV 35S promoter, indicating that AP3 proteins lacking the conserved C domains are functional in planta. In addition, when the AP3 C-terminal deleted proteins are expressed under the AP3 promoter at or above the levels of endogenous AP3, they are able to rescue the petal and stamen defects of ap3-3 mutants. Likewise, for PI, we demonstrate that transgenic plants lacking the PI motif exhibit a first-whorl sepal-to-petal floral organ identity transformation when expressed under both the CaMV 35S and AP3 promoters and were able to rescue the petal and stamen defects of pi-4 flowers.
Our demonstration that the C-terminal deleted motifs are not required for the floral organ identity function contradicts previous findings (Lamb and Irish, 2003). Lamb and Irish demonstrated that truncated forms of AP3 and PI, lacking the conserved C-terminal motifs, when expressed under either the CaMV 35S promoter or the AP3 promoter, did not exhibit floral organ identity function (Lamb and Irish, 2003). To address this discrepancy, we obtained the lines used in the Lamb and Irish study, prepared floral protein extracts, and performed immunoblots using our AP3 and PI antisera. We could not detect expression of the truncated AP3 and PI proteins in these lines (Supplemental Fig. 1). Based on this, it is likely that, in the Lamb and Irish study, low levels of expression of AP3 and PI, rather than the lack of C-terminal motifs, is the reason that truncated proteins failed to exhibit AP3 and PI activity.
In all AP3 and PI truncated lines that we analyzed, except PIΔC155, we could detect truncated protein, but the levels of protein varied widely among lines and constructs. For PI, both the PIΔC185 and PIΔC176 proteins exhibit floral organ identity function when expressed under the control of either the 35S or AP3 promoters. Analysis of protein expression levels demonstrates that some PI truncation lines clearly express at below endogenous PI levels. For example, some p35S:PIΔC176 lines express at only 20% to 30% of endogenous levels (Fig. 4E), but are still able to rescue the second- and third-whorl defects of pi-4 flowers. Thus, the ability of PI truncations to function, even when expressed below endogenous levels, rules out the possibility that organ identity function is an artifact of overexpression.
The situation for AP3 is slightly different. In several pAP3:AP3ΔC174 lines that rescue the petal and stamen identity in ap3 mutants, the AP3ΔC174 protein is expressed at or above endogenous levels. However, none of the pAP3:AP3ΔC198 lines that we analyzed was able to rescue petal and stamen identity in ap3 mutants. Whereas we could detect expression of the AP3ΔC198 protein, in all pAP3:AP3ΔC198 lines the level was no greater than 80% of endogenous AP3. Although the inability of AP3ΔC198 to function in organ identity specification could be interpreted as evidence for the necessity of the euAP3 and PI-derived motifs, we think the failure to function is due instead to low levels of expression. When the level of AP3ΔC198 protein is increased under the CaMV 35S promoter, AP3ΔC198 functions to direct floral organ identity. AP3 truncations, unlike PI truncations, appear to be sensitive to protein expression level. Thus, the lack of petal and stamen rescue in the pAP3:AP3ΔC198 ap3-3 flowers is most likely due to suboptimal levels of protein and not to the absence of the euAP3 and PI-derived motifs.
The euAP3 and PI-derived motifs of AP3 and the PI motif of PI are highly conserved, so it is surprising that the AP3 and PI organ identity function is not altered when these motifs are removed. However, other evidence in the literature implies that the conserved C-terminal motifs may not be essential for the floral organ identity functions of AP3 and PI. In vitro studies indicate that the C domain is not required to form DNA-binding AP3/PI heterodimers (Riechmann et al., 1996b; Winter et al., 2002). Two lines of in planta evidence indicate that the PI motif of PI orthologs and the euAP3 motif of AP3 orthologs may not be required for floral organ identity function. The first involves PsPI, the ortholog of PI in pea, which lacks the PI motif. Constitutive expression of PsPI in tobacco (Nicotiana tabacum) and Arabidopsis results in first-whorl organ identity transformations, demonstrating PI organ identity function. Similarly, p35S:PsPI rescues the petal and stamen defects of pi-1 mutants (Berbel et al., 2005). This demonstrates that a PI ortholog, lacking the conserved PI motif, is functional in Arabidopsis. The second involves Silky1, an AP3 ortholog in maize (Zea mays) that contains a paleoAP3 motif in place of the euAP3 motif. When Silky1 is expressed from the AP3 promoter in Arabidopsis, it rescues the petal and stamen defects of ap3-3 mutants (Whipple et al., 2004). This demonstrates that an AP3 ortholog that lacks the euAP3 motif is functional in Arabidopsis. This evidence, together with results presented in this study, indicates that conserved C-terminal motifs are not required for floral organ identity function.
Because C-terminal motifs are highly conserved, it would be surprising if they had no function. One possibility is that C-terminal motifs may have a function that is redundantly specified elsewhere in the protein. Under this scenario, C-terminal motifs may be dispensable under most circumstances, but if the redundant activities are compromised, then removal of C-terminal motifs is more critical. Evidence from the literature (Lamb and Irish, 2003; Tzeng et al., 2004), as well as our demonstration that the strength of protein-protein interaction of PI with SEP1 is reduced when the C domain is removed from PI, suggests that C-terminal motifs do possess a function in some circumstances. Therefore, we postulate that C-terminal motifs enhance activities that can occur in their absence but may occur more efficiently in their presence.
One surprising observation was that the function of AP3 truncations is much more sensitive to levels of protein expression compared to PI truncations. For example, pAP3:AP3ΔC174 lines that express AP3 protein at levels equivalent to endogenous levels rescue both second- and third-whorl defects of ap3-3 flowers. By contrast, pAP3:AP3ΔC174 lines that express below endogenous levels rescue third-whorl stamens but are less efficient at rescuing second-whorl petals (Supplemental Table S3). Moreover, all pAP3:AP3ΔC198 lines express below endogenous levels and none of the lines tested is able to rescue either the second- or third-whorl defects of ap3-3 flowers. When AP3ΔC198 and AP3ΔC174 are placed under the 35S promoter, they express at higher than endogenous levels and their organ identity transformations are more complete, demonstrating that these truncations can function to specify floral organ identity. At present, it is unclear why AP3 C-terminal truncations are more sensitive than PI C-terminal truncations to protein expression levels. If we assume that the C-terminal motifs provide a function that is redundantly encoded elsewhere in the protein, then one possibility is that, for AP3, these redundant activities function less well than they do in PI and thus AP3 C-terminal truncations are more sensitive to protein levels. When AP3 C-terminal truncations are expressed at endogenous levels, the lack of C-terminal motifs is not critical, but if protein levels fall too low, then the redundantly specified activity is insufficient.
In summary, although C-terminal motifs are not necessary for AP3 and PI organ identity function, there is experimental evidence that, in some cases, C-terminal motifs can affect functional specificity and protein-protein interaction ability (Lamb and Irish, 2003; Tzeng et al., 2004). Because C-terminal motifs can be removed without affecting organ identity function, essential activities like functional specificity and protein-protein interaction must be redundantly specified in these proteins. It has been previously demonstrated that protein-protein interaction is primarily mediated by the M, I, and K domains (Riechmann et al., 1996a; Yang et al., 2003a), whereas functional specificity is mainly encoded in the M and K domains of floral MADS proteins (Krizek and Meyerowitz, 1996a; Riechmann and Meyerowitz, 1997). Therefore, whereas the C-terminal motifs of B-class genes may redundantly contribute to protein-protein interaction ability, we have demonstrated that C-terminal motifs can be removed from both AP3 and PI without affecting their function in floral organ identity specification.
MATERIALS AND METHODS
Plant Growth
Arabidopsis (Arabidopsis thaliana) plants were grown in constant light (150 μmol m−2 s−1) at 22°C in a 3:1:1 mixture of promix:vermiculite:perlite.
Plasmid Construction and Transformations
Site-specific nonsense mutations and truncated forms of AP3 and PI were generated via PCR from genomic DNA, AP3 cDNA, or PI cDNA plasmids (see Supplemental Table S4 for primer sequences). AP3 and PI variants were cloned into the BamHI site of two different binary transformation vectors (Yang et al., 2003a): pD1954 (contains the AP3 promoter separated from the AP3 3′-untranslated region and 3′-nopaline synthase trailer sequence by a BamHI site) and pD1950 (contains the CaMV 35S promoter separated from the AP3 3′-untranslated region and 3′-nopaline synthase trailer sequence by a BamHI site). Mutations were confirmed by DNA sequencing from plant transformation vectors.
AP3 and PI plasmids were transformed into Agrobacterium tumefaciens strain ASE by heat shock and positive colonies were selected by antibiotic resistance and confirmed by amplification of the transgene via PCR. Wild-type flowers of Landsberg erecta or Columbia Arabidopsis plants were vacuum transformed or floral dipped with Agrobacterium. Transformed plants were selected on kanamycin Murashige and Skoog agar plates (Bechtold and Pelletier, 1998; Clough and Bent, 1998). Multiple independent transgenic lines were generated and analyzed for each construct (Supplemental Table S3).
Genotyping
DNA was isolated from antibiotic-resistant plants and the presence of the transgene was confirmed by PCR amplification. Transgenic plants were crossed to ap3-3 and F2 plants were genotyped by creating a cleaved amplified polymorphic sequence marker that engineers an RsaI site in ap3-3 but not the wild type (see Supplemental Table S4 for primer sequences). Transgenic plants were crossed to pi-4 and the F2 plants were genotyped using a cleaved amplified polymorphic sequence marker that detects an Mnl1 site that is present in pi-4 but is absent in the wild type (see Supplemental Table S4 for primer sequences).
Immunoblots
Inflorescence or leaf protein was isolated by grinding tissue in liquid nitrogen and adding 2× isolation buffer (50 mm Tris, pH 7.5, 150 mm NaCl, and 1% SDS [w/v]), then briefly vortexing and microcentrifuging for 15 min at 12,500 rcf at 4°C. For immunoblots, supernatants were boiled in loading dye for 5 min and equal amounts of total protein were loaded on a SDS-PAGE gel. Protein gels were transferred to polyvinylidene difluoride (Millipore IPVH00010) and blocked for 1 h at room temperature in 2% bovine serum albumin (BSA; [w/v]) in 1× 100 mm Tris base, 150 mm NaCl, and 0.05% Tween 20 (TBS-T; w/v). Both primary antibodies were incubated for 1 h at room temperature. Peroxidase-labeled anti-rabbit secondary antibody (Amersham NA934VS) was diluted to 1:20,000 in 1% BSA (w/v), 1× TBS-T, and blots incubated for 45 min at room temperature. Pierce SuperSignal West Femto maximum sensitivity substrate was used for detection (Pierce 34095).
Primary antibodies were generated in rabbits against MADS deleted forms of AP3 (Jack et al., 1994) and PI that were purified from Escherichia coli. AP3 antisera were used directly at 1:30,000 diluted in 1% BSA (w/v), 1× TBS-T. PI antisera were affinity purified on an AminoLink Plus immobilization column (Pierce 20394); peak fractions were combined and diluted to 1:3,000 in 1% BSA (w/v), 1× TBS-T.
Blot Quantitation
ImageJ software was used for all immunoblot quantifications. AP3ΔC/AP3 and PIΔC/PI values were calculated by measuring the pixel intensity of the truncated protein product and dividing it by the pixel intensity of the endogenous protein band in the same lane. For the normalized AP3Δ and PIΔ values in the aminocyclopropane carboxylic acid (ACC) oxidase blots, a control ratio was established by calculating the pixel intensity of the ACC oxidase band in each lane and dividing it by the pixel intensity of the ACC oxidase band in the wild-type lane (for Fig. 4, B and E). For the AP3Δ normalized quantification, two ratios were calculated: (1) the ratio of signal strengths of AP3Δ/PIΔ to ACC oxidase in a given lane; and (2) the ratio of signal strengths of AP3-wild type/PI-wild type to ACC oxidase in the wild-type lane were calculated. To calculate the normalized AP3Δ or PIΔ value, the AP3Δ/PIΔ:ACC oxidase ratio was divided by the AP3-wild type/PI-wild type:ACC oxidase ratio.
Yeast Three-Hybrid Assays
Yeast (Saccharomyces cerevisiae) three-hybrid assays were performed as previously described (Yang and Jack, 2004) with the following modifications. PI variants were expressed in the pGBDU vector fused to the GAL4 DNA-binding domain with Ura3 as a selectable marker. SEP1 was expressed in the pGAD vector fused to the GAL4 activation domain with Leu-2 as a selectable marker. AP3 was the expressed pU vector, a variation of the pGBDU vector with the GAL4 DNA-binding domain deleted. pU utilizes Trp-1 as a selectable maker. Transformations were grown on Sc-Leu-Trp-Ura plates and individual colonies were grown in Sc-Leu-Trp-Ura liquid medium. Two microliters of liquid culture was streaked on either Sc-Leu-Trp-Ura plates or Sc-Leu-Trp-Ura-His + Xgal. Photographs were taken after yeast colonies grew at 30°C for 4 d.
Scanning Electron Microscopy
Inflorescence tissue was fixed at room temperature for 48 h in 4% gluteraldehyde in 20 mm potassium phosphate buffer, pH 7.0. Fixative was removed and samples were coated in 1% osmium tetroxide in 20 mm potassium phosphate buffer. After 48 h, osmium was removed and samples washed several times in buffer. Samples were then taken through an ethanol series to 100% ethanol and subjected to critical point drying. Samples were dissected, mounted, and sputter coated in gold palladium. Images were taken at an accelerating voltage of 5 kV using a FEI XL-30 ESEM-FEG scanning electron microscope.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Immunoblots comparing expression of AP3 and PI transgenes in planta.
Supplemental Table S1. Phenotype of AP3ΔC lines.
Supplemental Table S2. Phenotype of PIΔC lines.
Supplemental Table S3. Transgenic lines generated.
Supplemental Table S4. Primer sequences.
Supplementary Material
Acknowledgments
We thank Rebecca Lamb and Vivian Irish for providing their truncated AP3 and PI lines, allowing us to test them for expression using our AP3 and PI antisera. We thank Stacey King and Anwesha Nag for comments on the manuscript, and Chuck Daghlian for help with scanning electron microscopy.
This work was supported by the National Science Foundation (grant no. MCB–0516736).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Thomas Jack (thomas.p.jack@dartmouth.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alvarez-Buylla ER, Pelaz S, Liljegren SJ, Gold SE, Burgeff C, Ditta GS, de Pouplana LR, Martinez-Castilla L, Yanofsky MF (2000) An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc Natl Acad Sci USA 97 5328–5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82 259–266 [DOI] [PubMed] [Google Scholar]
- Becker A, Theissen G (2003) The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol Phylogenet Evol 29 464–489 [DOI] [PubMed] [Google Scholar]
- Berbel A, Navarro C, Ferrandiz C, Canas LA, Beltran JP, Madueno F (2005) Functional conservation of PISTILLATA activity in a pea homolog lacking the PI motif. Plant Physiol 139 174–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Jang S, Chae S, Chung KM, Moon YH, An G, Jang SK (1999) Analysis of the C-terminal region of Arabidopsis thaliana APETALA1 as a transcription activation domain. Plant Mol Biol 40 419–429 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353 31–37 [DOI] [PubMed] [Google Scholar]
- de Folter S, Angenent GC (2006) trans meets cis in MADS science. Trends Plant Sci 11 224–231 [DOI] [PubMed] [Google Scholar]
- de Folter S, Immink RG, Kieffer M, Parenicova L, Henz SR, Weigel D, Busscher M, Kooiker M, Colombo L, Kater MM, et al (2005) Comprehensive interaction map of the Arabidopsis MADS box transcription factors. Plant Cell 17 1424–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea-Cortines M, Saedler H, Sommer H (1999) Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS, and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J 18 5370–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM (1994) Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev 8 1548–1560 [DOI] [PubMed] [Google Scholar]
- Hill TA, Day CD, Zondlo SC, Thackeray A, Irish VF (1998) Discrete spatial and temporal cis-acting elements regulate transcription of the Arabidopsis floral homeotic gene APETALA3. Development 125 1711–1721 [DOI] [PubMed] [Google Scholar]
- Honma T, Goto K (2001) Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409 525–529 [DOI] [PubMed] [Google Scholar]
- Jack T, Brockman LL, Meyerowitz EM (1992) The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68 683–697 [DOI] [PubMed] [Google Scholar]
- Jack T, Fox GL, Meyerowitz EM (1994) Arabidopsis homeotic gene APETALA3 ectopic expression: transcriptional and post-transcriptional regulation determine floral organ identity. Cell 76 703–716 [DOI] [PubMed] [Google Scholar]
- Kramer EM, Dorit RL, Irish VF (1998) Molecular evolution of genes controlling petal and stamen development: duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics 149 765–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM, Hall JC (2005) Evolutionary dynamics of genes controlling floral development. Curr Opin Plant Biol 8 13–18 [DOI] [PubMed] [Google Scholar]
- Kramer EM, Su HJ, Wu CC, Hu JM (2006) A simplified explanation for the frameshift mutation that created a novel C-terminal motif in the APETALA3 gene lineage. BMC Evol Biol 6 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA, Fletcher JC (2005) Molecular mechanisms of flower development: an armchair guide. Nat Rev Genet 6 688–698 [DOI] [PubMed] [Google Scholar]
- Krizek BA, Meyerowitz EM (1996. a) Mapping the protein regions responsible for the functional specificities of the Arabidopsis MADS domain organ-identity proteins. Proc Natl Acad Sci USA 93 4063–4070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA, Meyerowitz EM (1996. b) The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development 122 11–22 [DOI] [PubMed] [Google Scholar]
- Lamb RS, Irish VF (2003) Functional divergence within the APETALA3/PISTILLATA floral homeotic gene lineages. Proc Natl Acad Sci USA 100 6558–6563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Yanofsky MF, Meyerowitz EM (1991) AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev 5 484–495 [DOI] [PubMed] [Google Scholar]
- Parenicova L, de Folter S, Kieffer M, Horner DS, Favalli C, Kater MM, Davies B, Angenent GC, Colombo L (2003) Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Krizek BA, Meyerowitz EM (1996. a) Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc Natl Acad Sci USA 93 4793–4798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Meyerowitz EM (1997) Determination of floral organ identity by Arabidopsis MADS domain homeotic proteins AP1, AP3, PI, and AG is independent of their DNA-binding specificity. Mol Biol Cell 8 1243–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Wang M, Meyerowitz EM (1996. b) DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res 24 3134–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellari GM, Jaramiillo MA, Kramer EM (2004) Evolution of the APETALA3 and PISTILLATA lineages of MADS-box containing genes in basal angiosperms. Mol Biol Evol 21 506–519 [DOI] [PubMed] [Google Scholar]
- Theissen G (2001) Development of floral organ identity; stories from the MADS house. Curr Opin Plant Biol 4 75–85 [DOI] [PubMed] [Google Scholar]
- Tilly J, Allen DW, Jack T (1998) The CArG boxes in the promoter of the Arabidopsis floral organ identity gene APETALA3 mediate diverse regulatory effects. Development 125 1647–1657 [DOI] [PubMed] [Google Scholar]
- Tzeng TY, Liu HC, Yang CH (2004) The C-terminal sequence of LMADS1 is essential for the formation of homodimers for B function proteins. J Biol Chem 279 10747–10755 [DOI] [PubMed] [Google Scholar]
- Vandenbussche M, Theissen G, Van de Peer Y, Gerats T (2003) Structural diversification and neo-functionalization during floral MADS-box gene evolution by C-terminal frameshift mutations. Nucleic Acids Res 31 4401–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple CJ, Ciceri P, Padilla CM, Ambrose BA, Bandong SL, Schmidt RJ (2004) Conservation of B class floral homeotic gene function between maize and Arabidopsis. Development 131 6083–6091 [DOI] [PubMed] [Google Scholar]
- Winter K-U, Weiser C, Kaufmann K, Bohne A, Kirchner C, Kanno A, Saedler H, Theissen G (2002) Evolution of class B floral homeotic proteins: obligate heterodimerization originated from homodimerization. Mol Biol Evol 19 587–596 [DOI] [PubMed] [Google Scholar]
- Yang Y, Fanning L, Jack T (2003. a) The K domain mediates heterodimerization of the Arabidopsis floral organ identity proteins APETALA3 and PISTILLATA. Plant J 33 47–59 [DOI] [PubMed] [Google Scholar]
- Yang Y, Jack T (2004) Defining subdomains of the K domain important for protein-protein interactions of plant MADS proteins. Plant Mol Biol 55 45–59 [DOI] [PubMed] [Google Scholar]
- Yang Y, Xiang H, Jack T (2003. b) pistillata-5, an Arabidopsis B class mutant with strong defects in petal but not in stamen development. Plant J 33 177–188 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.