Abstract
Although flowering regulatory mechanisms have been extensively studied in Arabidopsis (Arabidopsis thaliana), those in other species have not been well elucidated. Here, we investigated the role of OsMADS51, a type I MADS-box gene in the short-day (SD) promotion pathway in rice (Oryza sativa). In SDs OsMADS51 null mutants flowered 2 weeks later than normal, whereas in long days loss of OsMADS51 had little effect on flowering. Transcript levels of three flowering regulators—Ehd1, OsMADS14, and Hd3a—were decreased in these mutants, whereas those of OsGI and Hd1 were unchanged. Ectopic expression of OsMADS51 caused flowering to occur about 7 d earlier only in SDs. In ectopic expression lines, transcript levels of Ehd1, OsMADS14, and Hd3a were increased, but those of OsGI and Hd1 remained the same. These results indicate that OsMADS51 is a flowering promoter, particularly in SDs, and that this gene functions upstream of Ehd1, OsMADS14, and Hd3a. To further investigate the relationship with other flowering promoters, we generated transgenic plants in which expression of Ehd1 or OsGI was suppressed. In Ehd1 RNA interference plants, OsMADS51 expression was not affected, supporting our conclusion that the MADS-box gene functions upstream of Ehd1. However, in OsGI antisense plants, the OsMADS51 transcript level was reduced. In addition, the circadian expression pattern for this MADS-box gene was similar to that for OsGI. These results demonstrate that OsMADS51 functions downstream of OsGI. In summary, OsMADS51 is a novel flowering promoter that transmits a SD promotion signal from OsGI to Ehd1.
The ability to adjust the timing of flowering is an important factor when trying to adapt plants to regional environments and control their harvest time. Regulatory mechanisms that control flowering have been studied extensively in Arabidopsis (Arabidopsis thaliana). In this long-day (LD) species, flowering is promoted by several regulatory genes, including GIGANTEA (GI), CONSTANS (CO), FLOWERING LOCUS T (FT), SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1), and APETALA1 (AP1).
GI encodes a nuclear protein that defines the proper amplitude and period length of circadian rhythms (Park et al., 1999; Mizoguchi et al., 2005). In Arabidopsis, mutations in GI delay flowering in LDs, but have little or no effect in short days (SDs). GI expression is regulated by the circadian clock, with a peak in transcript levels at 8 to 10 h after dawn. Timing, height, and duration of this peak are influenced by daylength (Fowler et al., 1999). The GI protein acts between the circadian oscillator and CO to promote flowering by increasing CO and FT mRNA abundance (Mizoguchi et al., 2005). CO is a putative transcription factor containing two B-box zinc fingers and a C-terminal CCT domain (Putterill et al., 1995). CO promotes flowering in response to LDs but not SDs (Putterill et al., 1995). CO protein activates the expression of several early target genes (e.g. SOC1 and FT [Samach et al., 2000; Suarez-Lopez et al., 2001]). FT, which encodes a protein similar to phosphatidylethanolamine-binding proteins and Raf kinase inhibitor proteins, also promotes flowering in LDs (Kardailsky et al., 1999; Kobayashi et al., 1999). FT is primarily expressed in vascular tissues, whereas its interactor, FD, is predominantly expressed at the shoot apex (Abe et al., 2005; Wigge et al., 2005). SOC1 integrates vernalization and GA signals for flowering (Moon et al., 2005). Its expression is affected by the loss of FT, whereas activation-tagged FT mutants have been shown to induce SOC1 up-regulation (Schmid et al., 2003; Moon et al., 2005). In contrast, FT expression remains unaffected by either loss or gain of SOC1 expression. In response to FT-mediated floral induction, AP1 contributes to the inflorescence meristem becoming committed to flowering (Huang et al., 2005).
In contrast to Arabidopsis, rice (Oryza sativa) is a SD plant in which flowering is inhibited during early developmental stages, but induced when the daylength shortens in the fall. Although the roles of genes involved in the photoperiod pathway are generally conserved between rice and Arabidopsis, some differences exist. OsGI, a GI ortholog, regulates flowering time in response to photoperiodic conditions. Mutations in this gene delay flowering in inductive SDs, whereas flowering is marginally induced under LDs (Hayama et al., 2003). In comparison, overexpression of OsGI in transgenic rice causes late flowering in both SDs and LDs. Expression of Hd1, the ortholog of CO, is affected in these plants, which indicates that OsGI functions to regulate Hd1, a gene that also shows circadian rhythm (Hayama et al., 2003). However, its expression pattern differs from that of OsGI. Hd1 transcript levels are lowest at around midday and become high during the night (Izawa et al., 2002; Kojima et al., 2002; Hayama et al., 2003). In contrast to CO, Hd1 has a double role in regulating downstream flowering activators—repression in LDs and promotion in SDs (Yano et al., 2000). One of the downstream targets is Hd3a, a FT ortholog, which acts preferentially in inductive SDs (Kojima et al., 2002). In addition to this photoperiod-controlled pathway, rice contains an alternative inductive pathway that appears to function independently of Hd1. Ehd1, encoding a B-type response regulator, promotes flowering by inducing the expressions of Hd3a, FTL1, and OsMADS14 (Doi et al., 2004). These results indicate that two different signaling pathways are intrinsically involved in the photoperiodic flowering pathway in rice.
The relationship among AP1 group MADS-box genes seems to differ between rice and Arabidopsis. For example, AP1 and its related genes determine floral meristem identity downstream of FT. However, in rice, OsMASD14, a member of the AP1 group, apparently functions upstream of Hd3a as a flowering activator. Its ectopic expression causes significant early flowering in tissue culture (Jeon et al., 2000), and transcription of Hd3a is up-regulated in OsMADS14-overexpressing cultured cells (S. Lee and G. An, unpublished data).
Most plants eventually flower, even under nonpermissive conditions. This autonomous pathway is regulated mainly by SOC1 in Arabidopsis. The gene acts as a flowering inducer, a result of converging photoperiod, autonomous, vernalization, and GA pathways (Araki, 2001; Mouradov et al., 2002; Simpson and Dean, 2002). OsMASD50, the Arabidopsis SOC1 homolog, acts as a flowering activator similar to SOC1. Ectopic expression causes dramatic early flowering at the regenerating callus stage, and OsMASD50 mutants flower about a month late (Lee et al., 2004; Lee and An, 2007).
Most MADS-box genes that regulate flowering time belong to type II, a group that contains the conserved MEF2-like MADS-box and K regions. In addition, intermediate (I) and C-terminal (C) regions are present. Therefore, they are termed MIKC-type genes (Munster et al., 1997). The possible roles of the other group, type I MADS-box genes in flowering time, are not well understood. Type I group proteins contain a conserved SRF-like MADS domain, but the C-terminal region is not clearly defined. Furthermore, type I proteins do not carry the conserved K-box region (Alvarez-Buylla et al., 2000). Although overexpression of a type I MADS-box gene, AGAMOUS-LIKE28 (AGL28), promotes flowering within the autonomous pathway in Arabidopsis (Yoo et al., 2006), the roles of most MADS-box genes of that type remain unknown.
Here, we have elucidated the functional role of a type I MADS-box gene, OsMADS51, in rice. This gene was originally identified through large-scale cDNA analysis within the rice genome research program (Shinozuka et al., 1999). OsMADS51 is unique to rice; a homologous gene is lacking in Arabidopsis. However, homologous genes do occur, with 74% identity in wheat (Triticum aestivum; accession no. ABF57941) and maize (Zea mays; accession no. CAD23412). Nevertheless, their roles have not yet been elucidated. OsMADS51 is composed of 164 amino acids and contains an SRF-like MADS domain. However, like other type I MADS-box genes, the K-box region is not conserved and the C-terminal region is short. Expression is ubiquitous in all organs (Shinozuka et al., 1999). In this study, we have investigated the function of OsMADS51 with regard to flowering time.
RESULTS
Isolation of T-DNA Insertional Mutants in OsMADS51
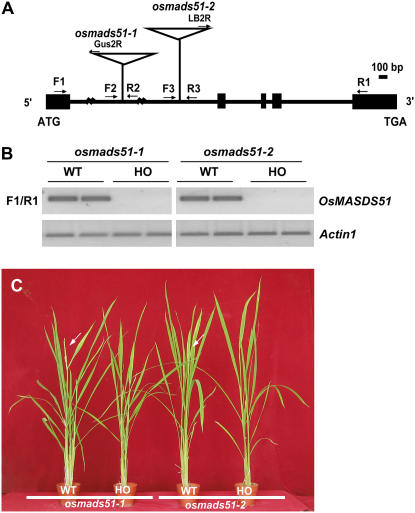
We have previously reported the generation of a T-DNA insertional mutant population in japonica rice (Jeon and An, 2001; Jeong et al., 2002, 2006; An et al., 2003, 2005b; Ryu et al., 2004). The sequence database is open to the public (www.postech.ac.kr/life/pfg/risd) and seeds are available to the scientific community. From that database, we have identified two independent T-DNA insertional mutant alleles, osmads51-1 and osmads51-2, in which T-DNAs are inserted into the first intron in both alleles, at 9,606 and 15,927 bp from the start codon in osmads51-1 and osmads51-2, respectively (Fig. 1A). Homozygous (osmads51/osmads51) plants did not accumulate OsMADS51 transcript (Fig. 1B). Under SDs, flowering in the homozygous progeny of both alleles was about 2 weeks later than in wild-type segregants (Fig. 1C; Table I). In contrast, little change in flowering time was noted under LDs (Table I). These results indicate that OsMADS51 is a flowering promoter that functions preferentially in SDs.
Figure 1.
OsMADS51 genomic structure, positions of T-DNA insertional mutant alleles, and osmads51 mutant phenotypes. A, Schematic diagram of T-DNA insertional position on OsMADS51. This gene consists of five exons (black boxes) and four introns (lines between black boxes). T-DNA is located at 9,606 bp (osmads51-1) and 15,927 bp (osmads51-2) from translation start site. F1 and R1 are primers used for RT-PCR and real-time PCR analyses. F2, R2, and Gus2R are primers for genotyping osmads51-1. F3, R2, and LB2R are primers for genotyping osmads51-2. B, Semiquantitative RT-PCR of OsMADS51 transcript in osmads51 mutants (HO) and wild-type segregants (WT). Actin1 was used for normalization of cDNA quantity. PCR cycles numbered 28 in OsMADS51 and 25 in Actin1. C, Phenotype of OsMADS51 mutants (HO) compared with wild-type segregants (WT). Plants were cultivated in a growth room under SD conditions (10 h light/14 h dark). Photograph was taken at 75 DAG. Arrows indicate panicles emerging at heading stage.
Table I.
Days to heading in osmads51
Days to heading was scored when the first panicle appeared. WT, Wild type; HO, osmads51/osmads51 homozygotes.
| Alleles | Genotypes | Days to Heading under SD | Days to Heading under LD |
|---|---|---|---|
| osmads51-1 | WT | 75 ± 2.2 | 96 ± 3.8 |
| HO | 89 ± 5.2 | 100 ± 3.2 | |
| osmads51-2 | WT | 75 ± 2.2 | 97 ± 2.5 |
| HO | 90 ± 3.8 | 101 ± 4.0 |
Ectopic Expression of OsMADS51 Causes Early Flowering
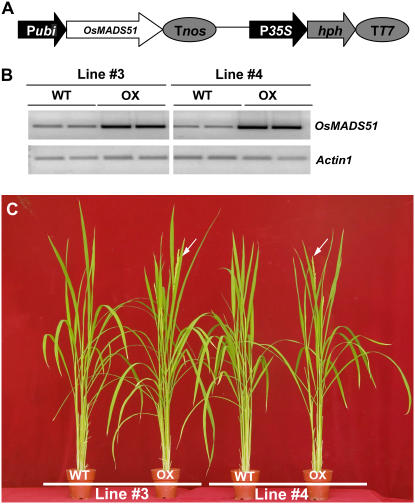
To further elucidate the functional roles of OsMADS51, we constructed a chimeric binary vector (Ubi:OsMADS51) by placing a OsMADS51 full-length cDNA under the maize ubiquitin (Ubi) promoter (Fig. 2A). This construct was introduced into Agrobacterium and transferred into plant cells via cocultivation (An et al., 1985; Hiei et al., 1997). Seven primary (T1) transgenic plants were obtained and planted in a paddy field. All showed early flowering, albeit to varying degrees. For example, two lines flowered 2 weeks early, three lines about 10 d sooner, and two lines by just about 3 d compared with our control transgenic plants. The extent of early flowering was correlated with OsMADS51 expression levels when confirmed by northern-blot analyses (data not shown).We then chose two lines (3 and 4) that manifested early flowering by 10 d and selected homozygous plants and segregating wild type using hygromycin selection. T3 progeny were reared in a growth room in either SDs (10 h light) or LDs (14 h light) to elucidate the daylength-dependent roles of OsMADS51. Homozygous plants exhibited early flowering by 9 d (line 3) and 7 d (line 4) in SDs. However, no significant phenotypic difference was observed between homozygous transgenic lines and wild-type plants grown in LDs (Table II). These results support our conclusion that OsMADS51 is a SD flowering activator.
Figure 2.
OsMADS51 overexpression. A, Schematic diagram of Ubi∷OsMADS51 construct. OsMADS51 full-length cDNA clone was placed between Ubi promoter (Pubi) and nos terminator (Tnos). B, Semiquantitative RT-PCR analyses of Ubi∷OsMADS51 transgenic (OX) lines 3 and 4 and their segregating wild-type controls (WT). Actin1 was used for normalization of cDNA quantity. RT-PCR was performed over 28 cycles for OsMADS51 and 25 cycles for Actin1. C, Phenotype Ubi∷OsMADS51 transgenic plants (OX) and their segregating wild-type controls (WT) cultivated for 62 d under SD conditions. Arrows indicate heading panicles.
Table II.
Days to heading in OsMADS51-overexpressing plants
Days to heading was scored when the first panicle appeared. WT, Wild type; OX, homozygous Ubi∷OsMADS51.
| Transgenic Plants | Genotypes | Days to Heading under SD | Days to Heading under LD |
|---|---|---|---|
| Line 3 | WT | 72 ± 3.8 | 93 ± 3.1 |
| OX | 63 ± 4.0 | 91 ± 2.4 | |
| Line 4 | WT | 73 ± 1.7 | 94 ± 2.4 |
| OX | 66 ± 2.3 | 93 ± 3.3 |
Relationship of OsMADS51 with Various Flowering-Time Regulators
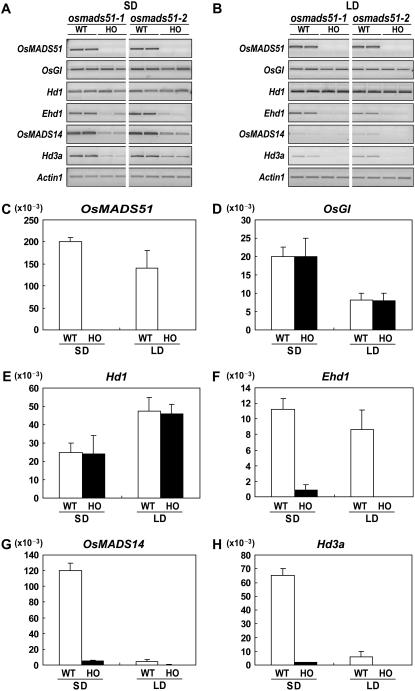
To evaluate the relationship between OsMADS51 and previously identified flowering regulators, we investigated the expression of OsGI, Hd1, Ehd1, OsMADS14, and Hd3a in the osmads51 mutants and their wild-type segregants grown in continuous SDs or LDs. At 55 d after germination (DAG), leaf blades were harvested and transcript levels were measured by reverse transcription (RT)-PCR and quantitative real-time PCR (Fig. 3; Supplemental Fig. S1). Expression of OsGI and Hd1 was unchanged in the OsMADS51 mutants under both lighting conditions (Fig. 3, A, B, D, and E). However, transcript levels of Ehd1, OsMADS14, and Hd3a were greatly reduced in the osmads51 mutants in SDs and LDs (Fig. 3, A, B, F, G, and H). These results indicate that Ehd1, Hd3a, and OsMADS14 probably function downstream of OsMADS51. As expected, the transcript levels of OsMADS51, OsGI, Ehd1, OsMADS14, and Hd3a were higher in SD-grown plants. Interestingly, this expression difference between SDs and LDs was greatest for OsMADS14 and Hd3a. Therefore, it appears that LD-specific inhibitors coordinately regulate these downstream genes. Hd1 is one candidate because it has been proposed to be a negative regulator in LDs (Yano et al., 2000), and we find its transcript level is higher in LDs compared with SDs.
Figure 3.
Expression profiles for various flowering regulators in OsMADS51 mutants. Shown are semiquantitative RT-PCR analyses of OsMADS51, OsGI, Hd1, Ehd1, OsMADS14, and Hd3a in wild-type (WT) and OsMADS51 mutant plants (HO) grown for 55 d under SD (A) or LD (B) conditions. Actin1 was used for normalization of cDNA quantity. RT-PCR cycles numbered 28 for all flowering regulators and 25 for Actin1. From at least three independent experiments, the most reproducible data are presented. Real-time PCR analyses of OsMADS51 (C), OsGI (D), Hd1 (E), Ehd1 (F), OsMADS14 (G), and Hd3a (H) in WT and HO plants grown for 55 d under SD or LD conditions. RNA was prepared from leaf blades harvested at 0 h after light was turned on. Each data point is the average of two or more independent experiments. y axis, Relative values between transcript levels of regulatory genes and Ubi; vertical bars, standard deviation.
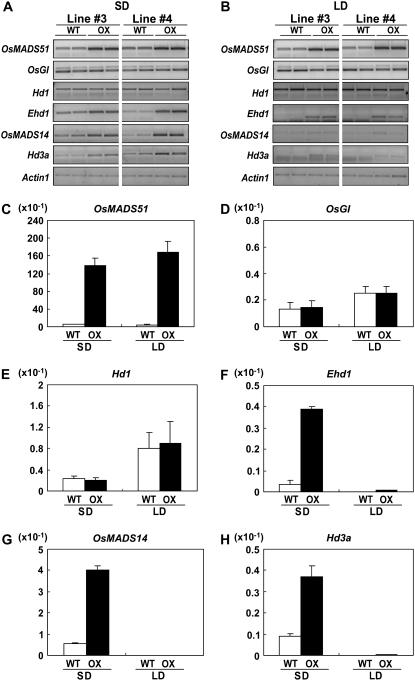
Transcript levels were also examined from these five genes in overexpressing OsMADS51 transgenic plants (Fig. 4; Supplemental Fig. S2). We used 35-DAG plants because they had flowered early and, therefore, their regulatory genes were expressed earlier. In wild-type control plants, transcript levels of Ehd1, OsMADS14, and Hd3a were very low at 35 DAG in LDs (Fig. 4), whereas transcripts were detected at 55 DAG (Fig. 3), showing age-dependent up-regulation patterns. As expected from the knockout experiment, Ehd1, Hd3a, and OsMADS14 transcript levels increased (Fig. 4, A, B, F, G, and H) under SD conditions, whereas those of OsGI and Hd1 were not significantly changed in the Ubi∷OsMADS51 transgenic plants in either daylength (Fig. 4, A, B, D, and E). These results support our conclusion that OsMADS51 functions upstream of Ehd1, Hd3a, and OsMADS14.
Figure 4.
Expression profiles for various flowering regulators in OsMADS51-overexpressing lines. Shown are semiquantitative RT-PCR analyses of OsMADS51, OsGI, Hd1, Ehd1, OsMADS14, and Hd3a in WT and Ubi∷OsMADS51 plants (OX) grown for 35 d under SD (A) or LD (B) conditions. Actin1 was used for normalization of cDNA quantity. RT-PCR cycles numbered 28 for all flowering regulators under SD; 32 for Ehd1, OsMADS14, and Hd3a; and 28 for OsMADS51, OsGI, and Hd1 under LD. For Actin1, 25 cycles were performed. From at least three independent experiments, the most reproducible data are presented. Real-time PCR analyses of OsMADS51 (C), OsGI (D), Hd1 (E), Ehd1 (F), OsMADS14 (G), and Hd3a (H) in WT and OsMADS51 mutant plants (HO) grown for 35 d under SD or LD conditions. RNA was prepared from leaf blades harvested at 0 h after light was turned on. Each data point is the average of two or more independent experiments. y axis, Relative values between transcript levels of regulatory genes and Ubi; vertical bars, standard deviation.
Expression Analyses of OsMADS51 in OsGI Antisense Plants
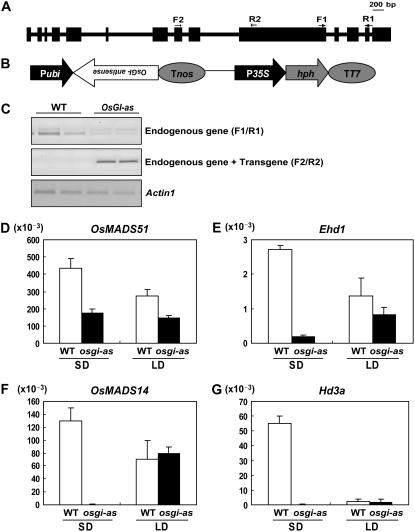
In osmads51 mutants and Ubi∷OsMADS51 transgenic plants, transcript levels of OsGI were unchanged (Figs. 3 and 4). Therefore, we believe that OsGI either functions upstream of OsMADS51 or is part of an independent pathway. Hayama et al. (2003) have previously reported that OsGI RNAi plants show delayed flowering in SDs, and we observed the same late-flowering phenotype in our OsGI antisense plants. Therefore, this SD-specific effect suggests that OsGI and OsMADS51 may function within the same pathway. To investigate the relationship between these two genes, we generated OsGI antisense plants and found that the OsMADS51 transcript level was reduced in plants grown under either SDs or LDs (Fig. 5A). As controls, we also examined expression levels from other flowering regulators—Ehd1, OsMADS14, and Hd3a—that had been demonstrated to function downstream of OsGI (Hayama et al., 2003; Doi et al., 2004). As expected, their transcript levels were reduced in our OsGI antisense plants (Fig. 5, E–G). These results indicate that OsGI does function in the same pathway as OsMADS51, with OsGI occurring upstream.
Figure 5.
Expression level analyses of flowering regulators in OsGi antisense plants. A, Schematic diagram of OsGi. F1, F2, R1, and R2 are primers used for RT-PCR and real-time PCR analyses. B, Schematic diagram of OsGi antisense construct. C, RT-PCR analysis of OsGi antisense (OsGi-as) plants and their wild-type segregants (WT). OsGi-as plants were grown for 55 d under SDs or LDs. RNA was prepared from leaf blades harvested at 0 h after light was turned on. Endogenous OsGI transcript was amplified with F1 and R1 primer sets, and both OsGI-as and endogenous transcripts were amplified with F2 and R2 primers. PCR cycles numbered 28 for OsGI and 25 for Actin1. Actin1 was used for normalization of cDNA quantity. Transcript amounts for OsMADS51 (D), Ehd1 (E), OsMADS14 (F), and Hd3a (G) were analyzed by quantitative real-time PCR. Each data point is the average of two or more independent experiments. y axis, Relative values between transcript levels of regulatory genes and Ubi; vertical bars, standard deviation.
Expression Analyses of OsMADS51 in Ehd1-RNAi Plants
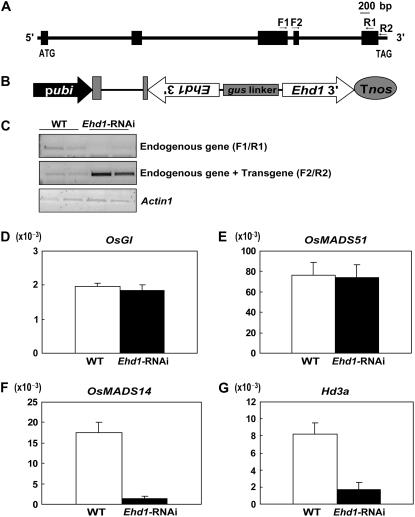
In osmads51 plants, the Ehd1 transcript level was significantly reduced (Fig. 3, A and F), whereas it was notably increased in the Ubi∷OsMADS51 plants (Fig. 4, A and F). Therefore, it is likely that Ehd1 is located downstream of OsMADS51. To confirm this hypothesis, we generated transgenic plants carrying the Ehd1-RNA interference (RNAi) construct (Fig. 6B). In all, 30 independently transformed plants were obtained, among which 20 flowered later than the control. We observed that Ehd1 transcript levels were reduced in those transgenics that were delayed. Two RNAi plants were selected that showed the most significant change in their flowering time. Under our SD conditions, they flowered about 7 d later than controls. Ehd1 transcript levels were reduced in those late-flowering transgenic plants in which a significant amount of RNAi transcript accumulated (Fig. 6C). Because the primers used for detecting the RNAi transcript amplified the endogenous Ehd1 transcript as well, we also found a PCR band in the wild type.
Figure 6.
Expression level analysis of flowering regulators in Ehd1-RNAi plants. A, Schematic diagram of Ehd1. F1, F2, R1, and R2 are primers used for RT-PCR and real-time PCR analyses. B, Schematic diagram of Ehd1-RNAi construct. C, RT-PCR analysis of Ehd1-RNAi plants and their wild-type segregants (WT). Endogenous Ehd1 transcript was amplified with F1 and R1 primer sets and both Ehd1-RNAi and endogenous transcripts were amplified with F2 and R2 primers. PCR cycles numbered 28 for Ehd1 and 25 for Actin1. Actin1 was used for normalization of cDNA quantity. D to G, Real-time PCR analysis of OsMADS51 (D), OsGI (E), OsMADS14 (F), and Hd3a (G) in Ehd1-RNAi plants. Samples for RT-PCR and real-time PCR were harvested at 0 h after light was turned on. Each data point is the average of two or more independent experiments. y axis, Relative values between transcript levels of regulatory genes and Ubi; vertical bars, standard deviation.
In the Ehd1-RNAi plants, OsGI and OsMADS51 transcript levels were not changed (Fig. 6, D and E), confirming that they function upstream of Ehd1. As previously reported (Doi et al., 2004), expression of Hd3a was significantly reduced in our Ehd1-RNAi plants (Fig. 6G). The OsMADS14 transcript level was also reduced in the RNAi plants, confirming the possibility that these flowering regulators are downstream of OsGI (Fig. 6F).
Diurnal Rhythms of OsMADS51 and OsGI
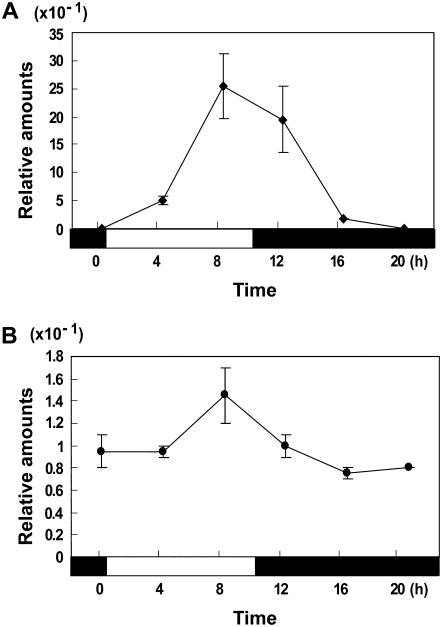
Because OsMAD51 functions between OsGI and Ehd1, the transcript level of a MADS-box gene may oscillate as do OsGI and Ehd1. Therefore, we measured OsMADS51 transcription in leaf blades from 55-DAG plants grown under SDs or LDs (Fig. 7). As reported previously (Hayama et al., 2003), the OsGI transcript level was low at the beginning of the day (Fig. 7A), but increased under light and reached a maximum at the end of the day. In darkness, that level was continuously reduced to a minimal amount. A similar rhythm was observed in LDs (data not shown). Under SDs, a similar induction curve was observed for OsMADS51 (Fig. 7B), increasing under light and decreasing in the dark. However, the change in transcript amounts was not as great as that for OsGI. Under LD, no such rhythm was found (data not shown).
Figure 7.
Diurnal rhythms of OsMADS51 (A) and OsGI (B) under SD conditions. Plants were grown under SDs of 10 h light (0–10 h, white box) and 14 h dark (10–24 h, black box). RNA was prepared from leaf blades of 50-DAG plants at 0, 4, 8, 12, 16, and 20 h after light was turned on. Each data point is the average of two or more independent experiments. y axis, Relative values between transcript levels of regulatory genes and Ubi; vertical bars, standard deviation.
Relationship between OsMADS51 and OsMADS50
The relationship between OsMADS51 and OsMADS50 was also investigated. We have previously reported that osmads50 plants show a late-flowering phenotype (delay of about 1 month), whereas overexpressors of OsMADS50 display an extreme early-flowering phenotype (Lee et al., 2004). Here, our mutations down-regulated the expression of OsMADS14 and Hd3a, suggesting that OsMADS50 functions upstream of OsMADS14. To evaluate whether OsMADS50 is another flowering activator that may influence OsMADS51, we examined whether OsMADS51 expression was altered in the osmads50 plants. However, that proved not to be the case (Supplemental Fig. S3). We also found that levels of OsMADS50 were unchanged in the osmads51 plants (Supplemental Fig. S3). Our results indicate that these two MADS-box genes function independently to control flowering time.
Analysis of Transcript Levels of OsMADS51 According to Developmental Stage
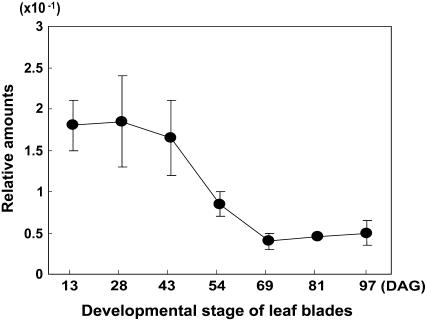
Because OsMADS51 is an early-acting activator, we measured its transcript levels at various developmental stages to see whether they would remain constant or change as plants matured. At 0 h (immediately after the light was turned on), we sampled leaf blades from plants grown in SDs. Real-time PCR analyses showed that transcription was high during the early developmental stages, up to day 43, but then declined and remained low over the remaining period (Fig. 8). This indicates that OsMADS51 functions early in the first 2 months of growth in rice.
Figure 8.
Changes in OsMADS51 transcript levels during growth phase. Plants were reared under SD conditions (10 h light/14 h dark). RNA was prepared from leaf blades of 13-, 28-, 43-, 54-, 69-, 81-, and 97-DAG plants at 0 h after light was turned on. Each data point is the average of two or more independent experiments. y axis, Relative values between transcript levels of regulatory genes and Ubi; vertical bars, standard deviation.
DISCUSSION
Intensive molecular genetics studies on the regulatory mechanisms that control flowering time have revealed that some major players are well conserved between rice and Arabidopsis (Izawa et al., 2003). However, some regulatory factors are found in just one of these model systems. For example, FLC is present only in Arabidopsis, whereas Ehd1 is exclusive to rice (Michaels and Amasino, 1999; Doi et al., 2004). Therefore, understanding the mechanisms that regulate flowering involves focusing on those species-specific genes to gain essential information that will elucidate the details of this diversification between species. In fact, the existence of the FLC family in Arabidopsis makes it possible to explain the vernalization pathway in crucifers, which does not exist in rice (Michaels and Amasino, 1999).
We, therefore, speculated that some MADS-box genes have been diversified to control gene-specific flowering mechanisms. OsMADS51 was selected for investigation in this regard because MADS-box genes of this group are found only in grass species. OsMADS51 was first identified via differential analysis of transcripts from mature spikelets (Ciaffi et al., 2005). Although the expression pattern for OsMADS51 has already been studied (Shinozuka et al., 1999), our report concerns the functional roles for this gene.
Our transgenic analyses showed that OsMADS51 is an SD-dependent flowering activator. Under SDs, OsMADS51 mutants flower about 2 weeks later compared with the control, whereas transformants that overexpress OsMADS51 flower 1 to 2 weeks early. Under LDs, these effects are milder, with OsMADS51 mutants flowering 3 to 4 d later and the overexpressors flowering no more than 1 to 2 d early. In rice, two SD promotion pathways have been proposed—one mediated by Hd1 and the other by Ehd1 (Yano et al., 2000; Doi et al., 2004). These pathways converge to induce Hd3a and the resultant protein product is moved to the shoot apical meristem to induce flowering (Tamaki et al., 2007). To investigate the relationship of these regulators with OsMADS51, we studied their expression patterns in OsMADS51 mutants. Transcript levels from OsMADS14 also were examined because this gene is down-regulated in the Ehd1 mutant background (Fig. 3) and its ectopic expression causes significant early flowering (Fig. 4).
RT-PCR and quantitative real-time PCR analyses demonstrated that Ehd1 expression is down-regulated in OsMADS51 mutants but up-regulated in Ubi:OsMADS51 plants. This suggests that OsMADS51 acts upstream of Ehd1. To further explore this relationship between OsMADS51 and Ehd1, we generated Ehd1 RNAi plants and measured OsMADS51 mRNA levels in RNAi plants. The previously identified Ehd1 mutant allele has one amino acid change that reduces DNA-binding ability and reduces Ehd1 function (Doi et al., 2004). Our RNAi results further support the role of Ehd1 as a flowering activator. In the Ehd1 RNAi plants, OsMADS51 mRNA levels are similar between RNAi and control plants, whereas OsMADS14 and Hd3a expression are affected by the mutation. Therefore, combined with the results from our OsMADS51 mutants, we conclude that OsMADS51 transmits a flowering signal through Ehd1, which then turns on OsMADS14 and Hd3a. OsMADS51 regulates Ehd1, OsMADS14, and Hd3a not only in SDs but also in LDs. However, the absolute amounts of these flowering activators are quite low in LDs, which is probably not sufficient for inducing flowering signal.
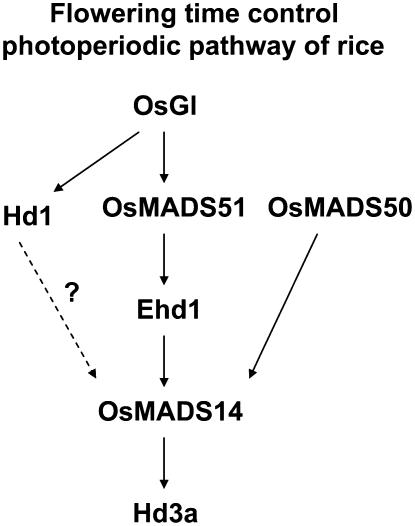
In contrast, levels of Hd1 and OsGI are not notably changed in OsMADS51 mutants or overexpressing plants, thereby implying that OsMADS51 acts either downstream of these genes or independently. To dissect these possibilities, we applied antisense technology to evaluate transgenic plants that suppress OsGI. Not only the level of Hd3a, a known downstream target of OsGI, but also the levels of OsMADS51 and Ehd1 are down-regulated in the OsGI antisense plants. These results suggest that OsMADS51 acts downstream of OsGI. The daily rhythm observed with OsMADS51 further supports this hypothesis because that rhythm is similar to the one for OsGI, showing a peak in the middle of the day (Fig. 7). Therefore, the SD promotion pathway mediated by OsGI can be divided into two routes. One is the well-known Hd1-mediated pathway, whereas the other is the OsMADS51-Ehd1-mediated pathway. Our finding is consistent with a previous report that Hd1 works regardless of the function of Ehd1 (Doi et al., 2004). Furthermore, our study confirms that Hd1 and Ehd1 operate independently and it provides additional evidence that OsGI is a common upstream regulator of Hd1 and Ehd1 (Fig. 9).
Figure 9.
Model of flowering-signal pathways in rice in which photoperiod signal is first translated to OsGI, then divided into two independent pathways. One is mediated by Hd1; the other by OsMADS51 and Ehd1. OsMADS50 generates an independent signal that turns on OsMADS14. All signals finally activate Hd3a, which produces a putative florigen.
Both Ehd1 and OsMADS51 occur as orphan genes within their family in the rice genome and no putative orthologs have been found in the Arabidopsis genome. Therefore, these unique genes probably function to differentiate SD-inducible rice from LD-inducible Arabidopsis. Thus, we can conclude that OsMADS51 plays an important role in transmitting the flowering signal from OsGI to Ehd1 in the SD promotion pathway and that this route is independent from previously reported pathways mediated by Hd1 or OsMADS50 (Fig. 9).
MATERIALS AND METHODS
Plant Materials and Growth
T-DNA-tagging lines were generated from rice (Oryza sativa) var. japonica ‘Dongjin’ and ‘Hwayoung’ (Jeon et al., 2000; Jeong et al., 2002). Insertion sites were determined via inverse PCR (An et al., 2003; Ryu et al., 2004; Jeong et al., 2006). T-DNA-tagged mutants were searched for in our flanking sequence database (www.postech.ac.kr/life/pfg/risd), which is also linked to the Salk Institute Genomic Analysis Laboratory (http://signal.salk.edu/cgi-bin/RiceGE; An et al., 2005a, 2005b). Plants were reared in growth rooms under either continuous SDs (10 h light/14 h dark) or LDs (14 h light/10 h dark).
Screening of OsMADS51 Mutants from T-DNA-Tagging Lines
The osmads51 homozygous progeny were screened from T1 seeds of the mutants. Genotyping was performed with 35 cycles of 95°C for 15 s, 55°C for 30 s, and 72°C for 40 s, using a combination of specific and T-DNA border primer sets as follows. The primer sets for osmads51-1 included F2 primer (5′-CAACAACTGACGAGGAGCAC-3′), R2 primer (5′-TAGATCATGACCTCCAGTAC-3′), and Gus2R primer in T-DNA (5′-TTGGGGTTTCTACAGGACGTAAC-3′). The osmads51-2 primer sets were F3 primer (5′-CCTCCCATGTTGTGTTCCTT-3′), R3 primer (5′-AAATGGCAAACCACTCGAAC-3′), and LB2R primer in T-DNA (5′-ACGTCCGCAATGTGTTATTAAG-3′). The Gus2R and LB2R primers were used to obtain the T-DNA flanking sequences by inverse PCR (An et al., 2003; Jeong et al., 2006).
Vector Construction and Transformation
The full-length cDNA clone of OsMADS51 (GenBank accession no. AB003327; Shinozuka et al., 1999) was amplified by PCR using a forward primer (5′-aagcttggggagctagggtttttcg-3′) and a reverse primer (5′-ggtaccaaacttatgcacaggacacc-3′). These primers contained one of two restriction enzyme sites, HindIII or Asp-718 (underlined in the primer sequences), for subsequent cloning. The PCR product was first cloned into pBluescript SK− (Stratagene). Afterward, cDNA was subcloned into the pGA1611 binary vector (Lee et al., 1999; Kim et al., 2003) between the maize (Zea mays) Ubi promoter and the nopaline synthase (nos) terminator to generate the Ubi∷OsMADS51 construct. To create the Ehd1-RNAi construct, we PCR amplified the 346-bp Ehd1 cDNA fragment (724–1,069 bp from the translation initiation site of the Ehd1 cDNA clone; GenBank accession no.AB092506), using a forward primer (5′-gttgccagtcatctgcag-3′) and a reverse primer (5′-ggatgtggatcatgagacat-3′). The PCR product was first cloned into pGA2960, a Gateway vector modified from pENTR 1A (Invitrogen). Afterward, the product was subcloned into the pANDA (p2K-1) binary vector by a clonase reaction (Miki and Shimamoto, 2004). Our OsGI antisense construct was generated by inserting a partial cDNA clone into the binary vector pGA1611 in the antisense orientation. The cDNA clone of OsGI (GenBank accession no. AJ133787mRNA) was amplified by PCR using a forward primer (5′-agacagcaaatttgactgcggcag-3′) and a reverse primer (5′-gacatgctgagtgcacggacatgcg-3′). The PCR product was first cloned into a pGEM-T vector (Promega). This OsGI cDNA product was then digested with restriction enzymes XhoI (991 bp from start codon) and XbaI (2,374 bp from start codon). The digested product was cloned into pBluescript SK− and subcloned into the pGA1611 binary vector using KpnI and SacI. Rice transformation was performed according to Agrobacterium-mediated cocultivation methods (Lee et al., 1999). All transgenic rice plants were grown in the greenhouse and then transferred to a confined paddy field.
RNA Isolation and Semiquantitative RT-PCR
Total RNA was isolated with Tri Reagent (MRC). First-strand cDNA was synthesized from 2 μg of total RNA, using Moloney murine leukemia virus reverse transcriptase (Promega), with 10 ng of the oligo(dT)15 primer and 2.5 mm deoxyriboneucleotide triphosphate, as described by Han et al. (2006). Actin1 was used for normalization of the cDNA quantity. Primers included 5′-GTTTGCTCTGCTCCTACTC-3′ and 5′-ACTCCTCCTCCAGCATTGAA-3′ for OsMADS51; 5′-TGGAGAAAGGTTGTGGATGC-3′ and 5′-CATAGACGGCACTTCAGCAGAT-3′ for OsGI; 5′-GTTGCCAGTCATCTGCAGAA-3′ and 5′-GGATGTGGATCATGAGACAT-3′ for Ehd1; 5′-TTCTCCTCTCCAAAGATTCC-3′ and 5′-CATACGCCTTTCTTGTTTCA-3′ for Hd1; 5′-TCCTATGCAGAAAAGGTCCTT-3′ and 5′-GGACGAAGCCAAAATATACAC-3′ for OsMADS14; 5′-ATGGCCGGAAGTGGCAGGGAC-3′ and 5′-ATCGATCGGGATCATCGTTAG-3′ for Hd3a; and 5′-GTATCCATGAGACTACATACATACAACT-3′ and 5′-TACTCAGCCTTGGCAATCCACA-3′ for Actin1. The reactions included an initial 5 min of denaturation at 95°C, followed by 95°C for 15 s, 55°C for 30 s, and 72°C for 40 s. PCR cycles numbered 28 for the flowering-regulator genes and 25 for Actin1.
Quantitative Real-Time RT-PCR
Real-time PCR was performed using a Roche LightCycler II as previously described (Han et al., 2006). cDNA was prepared from the leaf blades of plants at 35 or 55 DAG. The Ubi mRNA level was used to normalize the expression ratio for each gene. Primers included 5′-AACCAAGATCGGCAGTATGG-3′ and 5′-GATTGATTGCTCCAGCAGGT-3′ for Hd1; 5′-GCCAACTAATGCTCGAGTCC-3′ and 5′-CAGCTGCTGCAGGGTAGTTA-3′ for OsMADS14; 5′-AGCCCAAGTGACCCTAACCT-3′ and 5′-GTTGTAGAGCTCGGCGAAGT-3′ for Hd3a; and 5′-CACGGTTCAACAACATCCAG-3′ and 5′-TGAAGACCCTGACTGGGAAG-3′ for Ubi. Changes in gene expression were calculated via the ΔΔCt method.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AB003327, AB092506, and AJ133787.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression profiles for various flowering regulators in 55-DAG osmads51-2 plants.
Supplemental Figure S2. Expression profiles for various flowering regulators in 35-DAG Ubi∷osmads51 plants of line 4.
Supplemental Figure S3. Analyses of relationship between OsMADS51 and OsMADS50.
Supplementary Material
Acknowledgments
We express our thanks to Ko Shimamoto for providing the pANDA (p2K-1) binary vector, and Chung-Hwan Ryu and Dong-Hoon Jung for valuable discussions. We also acknowledge Priscilla Licht for critical reading of the manuscript.
This work was supported in part by the Crop Functional Genomic Center, the 21st Century Frontier Program (grant no. CG1111); the Biogreen 21 Program, Rural Development Administration; the Korea Science and Engineering Foundation (KOSEF) through the National Research Laboratory Program funded by the Ministry of Science and Technology (grant no. M10600000270–06J0000–27010); and the Korean government (KOSEF grant no. R15–2004–033–05002–0).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Gynheung An (genean@postech.ac.kr).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309 1052–1056 [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla ER, Pelaz S, Liljegren SJ, Gold SE, Burgeff C, Ditta GS, Ribas de Pouplana L, Martinez-Castilla L, Yanofsky MF (2000) An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc Natl Acad Sci USA 97 5328–5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G, Jeong DH, Jung KH, Lee S (2005. a) Reverse genetic approaches for functional genomics of rice. Plant Mol Biol 59 111–123 [DOI] [PubMed] [Google Scholar]
- An G, Lee S, Kim SH, Kim SR (2005. b) Molecular genetics using T-DNA in rice. Plant Cell Physiol 46 14–22 [DOI] [PubMed] [Google Scholar]
- An G, Watson BD, Stachel S, Gordon MP, Nester EW (1985) New cloning vehicles for transformation of higher plants. EMBO J 4 277–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S, Park S, Jeong DH, Lee DY, Kang HG, Yu JH, Hur J, Kim SR, Kim YH, Lee M, et al (2003) Generation and analysis of end sequence database for T-DNA tagging lines in rice. Plant Physiol 133 2040–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki T (2001) Transition from vegetative to reproductive phase. Curr Opin Plant Biol 4 63–68 [DOI] [PubMed] [Google Scholar]
- Ciaffi M, Paolacci AR, D'Aloisio E, Tanzarella OA, Porceddu E (2005) Identification and characterization of gene sequences expressed in wheat spikelets at the heading stage. Gene 346 221–230 [DOI] [PubMed] [Google Scholar]
- Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A (2004) Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev 18 926–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J (1999) GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J 18 4679–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MJ, Jung KH, Yi G, Lee DY, An G (2006) Rice Immature Pollen 1 (RIP1) is a regulator of late pollen development. Plant Cell Physiol 47 1457–1472 [DOI] [PubMed] [Google Scholar]
- Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K (2003) Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422 719–722 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Komari T, Kubo T (1997) Transformation of rice mediated by Agrobacterium tumefaciens. Plant Mol Biol 35 205–218 [PubMed] [Google Scholar]
- Huang T, Bohlenius H, Eriksson S, Parcy F, Nilsson O (2005) The mRNA of the Arabidopsis gene FT moves from leaf to shoot apex and induces flowering. Science 309 1694–1696 [DOI] [PubMed] [Google Scholar]
- Izawa T, Oikawa T, Sugiyama N, Tanisaka T, Yano M, Shimamoto K (2002) Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev 16 2006–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T, Takahashi Y, Yano M (2003) Comparative biology comes into bloom: genomic and genetic comparison of flowering pathways in rice and Arabidopsis. Curr Opin Plant Biol 6 113–120 [DOI] [PubMed] [Google Scholar]
- Jeon J, An G (2001) Gene tagging in rice: a high throughput system for functional genomics. Plant Sci 161 211–219 [DOI] [PubMed] [Google Scholar]
- Jeon JS, Lee S, Jung KH, Jun SH, Jeong DH, Lee J, Kim C, Jang S, Yang K, Nam J, et al (2000) T-DNA insertional mutagenesis for functional genomics in rice. Plant J 22 561–570 [DOI] [PubMed] [Google Scholar]
- Jeon JS, Lee S, Jung KH, Yang WS, Yi GH, Oh BG, An GH (2000) Production of transgenic rice plants showing reduced heading date and plant height by ectopic expression of rice MADS-box genes. Mol Breed 6 581–592 [Google Scholar]
- Jeong DH, An S, Kang HG, Moon S, Han JJ, Park S, Lee HS, An K, An G (2002) T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol 130 1636–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong DH, An S, Park S, Kang HG, Park GG, Kim SR, Sim J, Kim YO, Kim MK, Kim SR, et al (2006) Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J 45 123–132 [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kim SR, Lee S, Kang HG, Jeon JS, Kim KM, An G (2003) A complete sequence of the pGA1611 binary vector. J PIant Biol 16 211–214 [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286 1960–1962 [DOI] [PubMed] [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M (2002) Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol 43 1096–1105 [DOI] [PubMed] [Google Scholar]
- Lee S, An G (2007) Diversified mechanisms for regulating flowering time in a short-day plant rice. J Plant Biol 50 241–248 [Google Scholar]
- Lee S, Jeon JS, Jung KH, An G (1999) Binary vectors for efficient transformation of rice. J Plant Biol 42 310–316 [Google Scholar]
- Lee S, Kim J, Han JJ, Han MJ, An G (2004) Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in rice. Plant J 38 754–764 [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki D, Shimamoto K (2004) Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol 45 490–495 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Wright L, Fujiwara S, Cremer F, Lee K, Onouchi H, Mouradov A, Fowler S, Kamada H, Putterill J, et al (2005) Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17 2255–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Lee H, Kim M, Lee I (2005) Analysis of flowering pathway integrators in Arabidopsis. Plant Cell Physiol 46 292–299 [DOI] [PubMed] [Google Scholar]
- Mouradov A, Cremer F, Coupland G (2002) Control of flowering time: interacting pathways as a basis for diversity. Plant Cell (Suppl) 14 S111–S130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster T, Pahnke J, Di Rosa A, Kim JT, Martin W, Saedler H, Theissen G (1997) Floral homeotic genes were recruited from homologous MADS-box genes preexisting in the common ancestor of ferns and seed plants. Proc Natl Acad Sci USA 94 2415–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG (1999) Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285 1579–1582 [DOI] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80 847–857 [DOI] [PubMed] [Google Scholar]
- Ryu CH, You JH, Kang HG, Hur J, Kim YH, Han MJ, An K, Chung BC, Lee CH, An G (2004) Generation of T-DNA tagging lines with a bidirectional gene trap vector and the establishment of an insertion-site database. Plant Mol Biol 54 489–502 [DOI] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288 1613–1616 [DOI] [PubMed] [Google Scholar]
- Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU (2003) Dissection of floral induction pathways using global expression analysis. Development 130 6001–6012 [DOI] [PubMed] [Google Scholar]
- Shinozuka Y, Kojima S, Shomura A, Ichimura H, Yano M, Yamamoto K, Sasaki T (1999) Isolation and characterization of rice MADS box gene homologues and their RFLP mapping. DNA Res 6 123–129 [DOI] [PubMed] [Google Scholar]
- Simpson GG, Dean C (2002) Arabidopsis, the Rosetta stone of flowering time? Science 296 285–289 [DOI] [PubMed] [Google Scholar]
- Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410 1116–1120 [DOI] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science 316 1033–1036 [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309 1056–1059 [DOI] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, et al (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12 2473–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SK, Lee JS, Ahn JH (2006) Overexpression of AGAMOUS-LIKE 28 (AGL28) promotes flowering by upregulating expression of floral promoters within the autonomous pathway. Biochem Biophys Res Commun 348 929–936 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.