Abstract
Phosphate (Pi) is a macronutrient that is essential for plant growth and development. However, the low mobility of Pi impedes uptake, thus reducing availability. Accordingly, plants have developed physiological strategies to cope with low Pi availability. Here, we report that the characteristic Arabidopsis thaliana Pi starvation responses are in part dependent on the activity of the nuclear growth-repressing DELLA proteins (DELLAs), core components of the gibberellin (GA)-signaling pathway. We first show that multiple shoot and root Pi starvation responses can be repressed by exogenous GA or by mutations conferring a substantial reduction in DELLA function. In contrast, mutants having enhanced DELLA function exhibit enhanced Pi starvation responses. We also show that Pi deficiency promotes the accumulation of a green fluorescent protein-tagged DELLA (GFP-RGA [repressor of ga1-3]) in root cell nuclei. In further experiments, we show that Pi starvation causes a decrease in the level of bioactive GA and associated changes in the levels of gene transcripts encoding enzymes of GA metabolism. Finally, we show that the GA-DELLA system regulates the increased root hair length that is characteristic of Pi starvation. In conclusion, our results indicate that DELLA-mediated signaling contributes to the anthocyanin accumulation and root architecture changes characteristic of Pi starvation responses, but do not regulate Pi starvation-induced changes in Pi uptake efficiency or the accumulation of selected Pi starvation-responsive gene transcripts. Pi starvation causes a reduction in bioactive GA level, which, in turn, causes DELLA accumulation, thus modulating several adaptively significant plant Pi starvation responses.
Phosphate (Pi) is an essential plant macronutrient (Raghothama, 1999). Phosphorus forms insoluble compounds in acid soils or is unevenly distributed as Pi in alkaline soils (Holford, 1997). Thus, although the total amount of phosphorus is high in many soils, Pi availability is often a significant limiting factor for plant growth in both natural and agricultural systems (López-Bucio et al., 2000). To sustain growth in such limiting conditions, plants have evolved a number of developmental and metabolic responses to adapt both the internal Pi status in planta and the external soil Pi availability. These responses include changes in root morphology and architecture, accumulation of anthocyanin, and increases in the synthesis and secretion of organic acids into the rhizosphere (which enhance the utilization of Pi from insoluble inorganic compounds; del Pozo, 1999; Raghothama, 1999; López-Bucio et al., 2002). One characteristic of plant Pi starvation response is simultaneous reduction in shoot growth and increase in root proliferation. The outcome of this response is the formation of a highly branched root system (associated with reduced primary root length, increased lateral root number, and density) and increases in both frequency and length of root hairs. These changes enhance the exploratory capacity of roots to search for Pi-rich patches present in the soil (Raghothama, 1999; Lynch and Brown, 2001). Recently, LPR1 (for LOW PHOSPHATE ROOT1), a major quantitative trait locus with a large effect on primary root growth arrest in response to Pi starvation, has been isolated (Svistoonoff et al., 2007). Loss-of-function mutations of LPR1 and its close paralog, LPR2, both of which encode multicopper oxidases, reduce Pi starvation-induced inhibition of primary root growth. This suggests that LPR1 protein enables cells of the Arabidopsis (Arabidopsis thaliana) primary root cap to sense low-phosphate (LP) conditions, thus triggering root growth arrest (Svistoonoff et al., 2007).
The phytohormone auxin and the associated polar auxin transport mechanism are known to be essential for lateral root formation (Muday and Haworth, 1994; Reed et al., 1998; Casimiro et al., 2001). When plants are grown in LP conditions, exogenous auxin treatment dramatically inhibits the growth of primary roots and induces the formation of lateral roots. In contrast, a 10- to 100-fold higher auxin dose is necessary for seedlings grown in high-Pi (HP) conditions to develop a similar root architecture to that induced by Pi starvation (López-Bucio et al., 2002). In addition, auxin-resistant mutants axr2-1, axr3-1, and axr4-1 display normal responses to Pi deficiency, whereas the iaa28-1 mutant displays resistance to the stimulatory effects of LP on root hair and lateral root formation (López-Bucio et al., 2002). These observations suggest that Pi starvation increases the sensitivity of Arabidopsis roots to auxin (López-Bucio et al., 2002).
Additional phytohormones are also known to be involved in inhibiting primary root growth and in promoting the production of both lateral roots and root hairs of Pi-starved Arabidopsis seedlings (Gilbert et al., 2000; Skene and James, 2000; López-Bucio et al., 2002; Al-Ghazi et al., 2003). For example, exogenous cytokinin treatment reduces the expression of selected Pi starvation-responsive marker genes (e.g. At4, AtIPS1, AtPT1, and AtACP5; Muchhal et al., 1996; del Pozo et al., 1999; Martin et al., 2000). Recently, it has been shown that cytokinin modulates the level of meristem cell cycle activity and that this, in turn, influences the expression of Pi starvation-responsive genes (Lai, et al., 2007). Ethylene is also involved in primary root elongation and root hair formation of seedlings grown in Pi starvation conditions (Ma et al., 2003; He et al., 2005). However, analysis of the root architecture of ethylene-signaling mutants, such as etr1, ctr1, ein2, ein3, and hls1, and of plants treated with the ethylene precursor 1-aminocyclopropane-1-carboxylic acid show that ethylene does not promote the formation of lateral roots when Pi is limiting (López-Bucio et al., 2002).
The role of the phytohormone GA in Pi starvation responses remains largely unknown. Accordingly, we performed experiments to determine whether the GA-DELLA growth regulatory system contributes to Pi starvation plant growth responses. GA plays an important role in regulating plant growth and development throughout the life cycle, including seed germination, root elongation, hypocotyl growth, leaf expansion, floral initiation, and floral development (Hooley, 1994; Richards et al., 2001). GA-deficient mutant plants, such as the Arabidopsis ga1-3 mutant, are dwarfed, exhibit dark-green leaves, and are late flowering, whereas exogenous GA treatment can restore normal growth to these mutants (Koornneef and van der Veen, 1980). GA promotes plant growth by relieving the growth restraint imposed by a family of nuclear growth-repressing DELLA proteins (DELLAs; Peng et al., 1997; Silverstone et al., 1998; Dill and Sun, 2001; King et al., 2001; Lee et al., 2002; Wen and Chang, 2002; Fu and Harberd, 2003). In Arabidopsis, the DELLAs comprise a family of five distinct proteins (GAI, RGA [repressor of ga1-3], RGL1, RGL2, and RGL3; Lee et al., 2002). Binding of bioactive GA to the Arabidopsis GA receptors (AtGID1a, AtGID1b, and AtGID1c) promotes interaction between these GA receptors and DELLAs (Nakajima et al., 2006). The DELLAs are subsequently polyubiquitinated by the SCFSLY1/SLY2 E3 ubiquitin ligase and thus targeted for destruction in the 26S proteasome (Fu et al., 2002; McGinnis et al., 2003; Sasaki et al., 2003; Dill et al., 2004; Fu et al., 2004; Griffiths et al., 2006). Thus, DELLAs restrain plant growth, whereas GA stimulates growth by promoting destruction of the DELLAs.
Recent advances have revealed that the DELLAs play important roles in many aspects of the adaptation of plant growth and development in response to environmental variables (Fu et al., 2002; Lee et al., 2002; Fu and Harberd, 2003; Yu et al., 2004; Cao et al., 2005; Achard et al., 2006, 2007a, 2007b; Penfield et al., 2006). In this study, we systematically determined whether the GA-DELLA signaling pathway contributes to the control of plant Pi starvation responses. We found that exogenous GA overcomes several of the characteristic growth responses of Arabidopsis to Pi starvation and that mutants that are substantially DELLA deficient do not exhibit these Pi starvation growth responses. We further found that Pi starvation promotes the accumulation of a GFP-tagged DELLA (GFP-RGA) in root cell nuclei. Thus Pi starvation growth responses are, in part, determined by DELLA accumulation. However, we also found that Pi starvation does not alter the GA sensitivity of 26S proteasome-dependent DELLA destruction. This latter observation suggested that Pi starvation might cause the accumulation of DELLAs by reducing the levels of bioactive GA. Accordingly, we found that Pi starvation induced changes in the levels of transcripts encoding GA metabolism enzymes and decreases in bioactive GA levels. We additionally found that the GA deficiency ga1-3 mutation confers a significant reduction in root hair length in Pi-starved plants and that GA treatment can restore this root hair length to that of wild type. Thus, an appropriate bioactive GA level is necessary for the increased root hair growth that is characteristic of Pi-starved roots. We conclude that DELLA restraint is a component of the mechanism via which Pi starvation modulates growth. Essentially, Pi starvation reduces bioactive GA levels, thus causing the accumulation of DELLAs and, in turn, triggering multiple Pi starvation responses, including alteration of root architecture, reduction of shoot growth, and accumulation of anthocyanin.
RESULTS
Pi Starvation Regulates Root Growth and Architecture via a GA-Dependent Mechanism
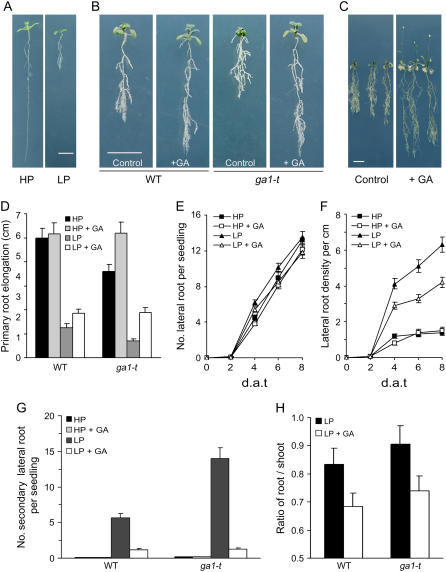
Inhibition of primary root growth is a characteristic plant Pi starvation response (Lynch and Brown, 2001; López-Bucio et al., 2003). For example, Arabidopsis seedling primary roots grow longer in HP (1 mm NaH2PO4) than in LP (10 μm NaH2PO4) conditions (Fig. 1A). Primary root length is also regulated by GA. The length of the primary seedling root of the GA-deficient ga1-3 mutant is shorter than that of wild type, whereas exogenous GA can restore ga1-3 primary roots to wild-type length (Fu and Harberd, 2003). To determine the effect of GA on the seedling primary root Pi starvation response, we compared the length of primary roots, numbers of lateral roots, and lateral root densities of wild-type (Columbia) and GA-deficient ga1-t mutant seedlings grown in HP versus LP conditions. We found that GA increases the primary root length of wild-type seedlings grown in LP (Fig. 1, B and C). In addition, LP-grown ga1-t primary root length was 2- to 3-fold reduced compared with that of wild type, whereas exogenous GA restored the LP-grown ga1-t primary root to wild-type (+GA) length (Fig. 1, B and D). There was no detectable difference in the root architectures of GA-treated and control wild-type seedlings grown in HP (Fig. 1D). Thus, GA increases the primary root length of wild-type seedlings grown in LP.
Figure 1.
GA promotes root growth and changes in root morphology in responses to Pi starvation. Four-day-old Arabidopsis Columbia wild-type (WT) and GA-deficient ga1-t mutant seedlings were transferred to LP or HP conditions (as described in “Materials and Methods”) in the presence or absence of exogenous 1 μm GA. A, Pi starvation affects primary root growth. Four-day-old wild-type seedlings were transferred to LP or HP medium for 6 d. B and C, Effects of GA treatment on root growth and morphology of seedlings grown in LP conditions for 8 d (B) and 28 d (C). D, Comparison of primary root elongation of 8-d-after-transfer (d.a.t.) wild-type and ga1-t mutant seedlings grown in LP and HP conditions in the presence or absence of GA. E and F, Kinetic analysis of lateral root number per each seedling (E) and lateral root density (F) of wild-type seedlings grown during Pi starvation. G, Effects of GA and Pi starvation on secondary lateral root development of 12-d-after-transfer wild-type and ga1-t seedlings. H, Root-to-shoot ratio of wild-type and ga1-t mutant seedlings grown in Pi starvation conditions. Results are presented as means with error bars representing se. Bar = 1 cm.
Pi starvation promotes the growth of lateral roots (López-Bucio et al., 2002; Fig. 1, A–C). We found that exogenous GA did not significantly alter the number of lateral roots formed by wild-type seedlings in HP or LP (Fig. 1E). However, due to GA-promoted increases in primary root length, exogenous GA caused a reduction in lateral root density in LP-grown wild-type seedlings (Fig. 1F). In HP, at 12 d following transfer, wild-type seedlings have a long primary root and relatively few lateral and secondary lateral roots. However, LP-grown wild-type seedlings produce a highly branched root system with abundant lateral and secondary lateral roots (Fig. 1G). Moreover, LP-grown ga1-t mutant seedlings produce even higher numbers of secondary lateral roots than do LP-grown wild-type seedlings (Fig. 1, B and G). Interestingly, we found that exogenous GA significantly reduced the number of secondary lateral roots (in both wild type and ga1-t) of seedlings grown in LP (Fig. 1G).
An increased root-to-shoot ratio is another characteristic plant Pi starvation response (Lynch and Brown, 2001; López-Bucio et al., 2003; Fig. 1H). We found that exogenous GA decreases the root-to-shoot ratio of LP-grown wild-type and ga1-t plants (Fig. 1H). Thus, exogenous GA promotes the growth of both roots and shoots in LP, suggesting the possibility that Pi starvation reduces growth via a reduction in bioactive GA levels.
Taken together, the above results indicate that GA contributes (at least partially) to regulating the alterations in root and shoot growth and architecture that are characteristic of plant Pi starvation responses and play a particularly prominent role in controlling the development of secondary lateral roots.
Pi Starvation Inhibits Plant Growth via the GA-DELLA Signaling Mechanism
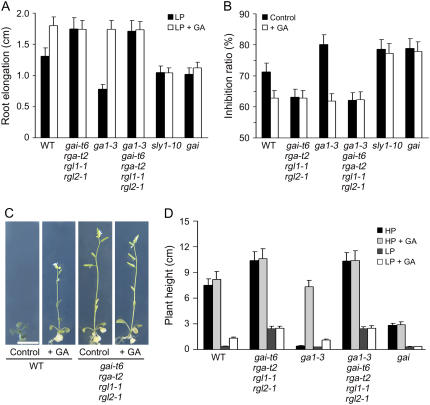
DELLAs act as plant growth repressors in GA signaling (Richards et al., 2001). GA promotes growth by targeting the growth-restraining DELLAs for destruction in the 26S proteasome (Fu and Harberd, 2003; McGinnis et al., 2003; Dill et al., 2004; Fu et al., 2004). To determine whether GA regulates plant Pi starvation responses via the GA-DELLA mechanism, we examined the Pi starvation responses of wild-type (Landsberg erecta), ga1-3, gai-t6 rga-t2 rgl1-1 rgl2-1, ga1-3 gai-t6 rga-t2 rgl1-1 rgl2-1, gai, and sly1-10 mutant seedlings. Mutants carrying the gai-t6, rga-t2, rgl1-1, and rgl2-1 mutations, respectively, lack the DELLAs GAI, RGA, RGL1, and RGL2 (although they retain RGL3) and are thus substantially DELLA deficient (Cheng et al., 2004; Cao et al., 2005; Achard et al., 2006). gai and sly1-10 confer GA insensitivity because they reduce the susceptibility of DELLAs to GA-promoted degradation, thus enhancing DELLA accumulation. We found that the primary root length of LP-grown ga1-3 gai-t6 rga-t2 rgl1-1 rgl2-1 mutant seedlings was approximately twice that of LP-grown control GA-deficient ga1-3 mutant seedlings (Fig. 2A). Furthermore, the primary root length of LP-grown ga1-3 gai-t6 rga-t2 rgl1-1 rgl2-1 mutant seedlings was identical to that of GA-treated LP-grown wild-type, ga1-3, and gai-t6 rga-t2 rgl1-1 rgl2-1 seedlings. Conversely, the LP-grown primary root lengths of the GA-insensitive gai and sly1-10 mutant lines were unaffected by exogenous GA (Fig. 2A). In addition, when wild-type seedlings were grown in LP, the root length was only 28% of that grown in HP and the LP-to-HP growth-inhibition ratio was therefore approximately 72% in the absence of GA (Fig. 2B). But GA treatment restored root growth in LP conditions and the wild-type LP-to-HP growth inhibition ratio was reduced to approximately 63% (Fig. 2B). These GA-promoted differentials were enhanced in ga1-3 and reduced in DELLA-deficient lines (gai-t6 rga-t2 rgl1-1 rgl2-1; ga1-3 gai-t6 rga-t2 rgl1-1 rgl2-1) and in GA-insensitive gai and sly1-10 mutant lines (Fig. 2B).
Figure 2.
Pi starvation inhibits plant growth via a DELLA-dependent mechanism. A, Comparison of root growth of wild-type and DELLA-associated mutant seedlings. Four-day-old seedlings were transferred to LP conditions in the presence or absence of 1 μm GA for 6 d. B, Primary root growth inhibition ratios (1-LP/HP elongation length) from 4-d-old seedlings transferred to appropriate Pi conditions in the presence or absence of 1 μm GA for 6 d. C and D, Comparison of the plant heights of 4-d-old seedlings transferred to HP or LP conditions in the presence or absence of GA for 21 d. Results are presented as means with error bars representing se. Bar = 1 cm.
We also found, similarly to what had been observed for root growth, that the effects of Pi starvation on shoot growth (plant height) are mediated in part via the GA-DELLA pathway. For example, either exogenous GA or substantial DELLA deficiency (conferred by gai-t6 rga-t2 rgl1-1 rgl2-1) increases the stem length of LP-grown wild-type plants (Fig. 2, C and D). GA increases the stem length of LP-grown ga1-3 plants, but has no effect on LP-grown ga1-3 plants that are substantially DELLA deficient (ga1-3 gai-t6 rga-t2 rgl1-1 rgl2-1) or on LP-grown gai plants (Fig. 2, C and D). Taken together, the above results indicate that Pi starvation affects both root and shoot via a mechanism that is (at least in part) dependent on the GA-DELLA signaling system.
Pi Starvation Induces Anthocyanin Accumulation via a DELLA-Dependent Mechanism
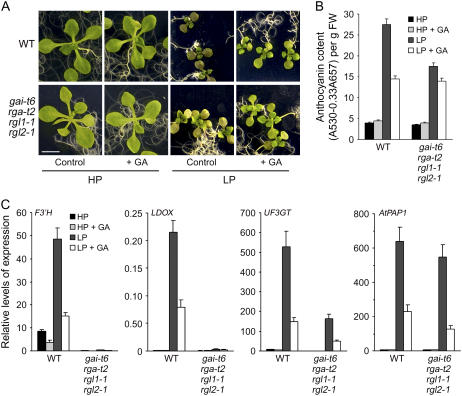
The visible accumulation of anthocyanin pigmentation is one of the characteristic responses of plants to Pi starvation. Anthocyanins and other polyphenolic compounds (e.g. flavonols and condensed tannins) have a wide range of functions in plants related to UV absorption, pathogen attack, and nutrient stress (Stewart et al., 2001; Kliebenstein, 2004). To gain insight into the relationship between DELLA function and anthocyanin accumulation, we compared the effects of Pi starvation on the anthocyanin content of wild-type and gai-t6 rga-t2 rgl1-1 rgl2-1 seedlings. In the absence of exogenous GA, leaves of LP-grown wild-type plants were visibly purple and accumulated higher levels of anthocyanin than did HP-grown controls (Fig. 3, A and B). In contrast, LP-grown gai-t6 rga-t2 rgl1-1 rgl2-1 plants were visibly less purple than LP-grown wild-type plants (Fig. 3A) and accumulated anthocyanin to a lower level (Fig. 3B). GA treatment caused a reduction in anthocyanin accumulation in wild-type plants grown in LP conditions, whereas anthocyanin accumulation of LP-grown gai-t6 rga-t2 rgl1-1 rgl2-1 plants was less responsive to GA-induced reductions (Fig. 3B). These results suggest that DELLA activity promotes the accumulation of anthocyanin during Pi starvation.
Figure 3.
DELLAs contribute to changes in anthocyanin accumulation and levels of transcripts encoding enzymes of anthocyanin metabolism in Pi starvation conditions. A, Effects of Pi availability and GA treatment on anthocyanin accumulation for 12-d-after-transfer Arabidopsis seedlings. B, Comparison of anthocyanin content of wild-type and gai-t6 rga-t2 rgl1-1 rgl2-1 mutant seedlings as shown in A. C, Levels of F3′H, PAP1, LDOX, and UF3GT gene transcripts in seedlings as shown in A and B (determined by real-time PCR). Results are displayed as the copy per 1,000 copy of 18S rRNA.
Anthocyanin is a branch for synthesis of flavonols via flavonol synthase in the flavonoid pathway. The enzymes of each step for anthocyanin synthesis are required: chalcone synthase, chalcone isomerase, flavone-3-β-hydroxylase, dihydroflavonol-4-reductase, leucoanthocyanidin dioxygenase (LDOX; Jaffe et al., 2006). Moreover, UDP-Glc-flavonoid 3-O-glucosyltransferase (UF3GT), UDP-glycose:UF3GT is a specific enzyme for anthocyanin synthesis (Jaffe et al., 2006). We next examined the effects of Pi starvation on the expression of genes encoding enzymes of anthocyanin metabolism. We found that wild-type plants grown in LP conditions accumulated relatively high levels of PAP1 (for PRODUCTION OF ANTHOCYANIN PIGMENT1, AtMYB75; Borevitz et al., 2000), F3′H (flavone 3′ hydroxylase), LDOX, and UF3GT transcripts, and that GA treatment dramatically reduced the levels of these transcripts (Fig. 3C). These observations indicate that the anthocyanin accumulation characteristic of Pi starvation is due to increases in PAP1, F3′H, LDOX, and UF3GT activity, whereas the GA-promoted reduction in anthocyanin level is the consequence of a reduction in PAP1, F3′H, LDOX, and UF3GT activity. Furthermore, LP-grown gai-t6 rga-t2 rgl1-1 rgl2-1 plants contained undetectable levels of F3′H and LDOX transcripts in both HP and LP conditions and had significantly lower than LP-grown wild-type plant levels of UF3GT transcripts (Fig. 3C). Interestingly, exogenous GA promoted a decrease in the levels of both UF3GT and PAP1 transcripts in both wild type and gai-t6 rga-t2 rgl1-1 rgl2-1, whereas there were no differences in levels of PAP1 transcripts between LP-grown wild-type and gai-t6 rga-t2 rgl1-1 rgl2-1 plants (regardless of the presence or absence of exogenous GA; Fig. 3C). These observations indicate that down-regulation of PAP1 transcript level is GA dependent, but not dependent on the function of GAI, RGA, RGL1, or RGL2.
In conclusion, the data in Figure 3 indicate that the accumulation of anthocyanin that is characteristic of Pi starvation is the consequence of increases in expression of genes of anthocyanin metabolism. For some of these genes (F3′H and LDOX), this up-regulated expression is GA-DELLA dependent. For others (UF3GT and PAP1), GA can overcome LP-induced increases in transcript level via a mechanism that is independent of the DELLAs GAI, RGA, RGL1, and RGL2 (and may therefore be dependent upon RGL3 activity).
DELLAs Do Not Detectably Regulate LP-Induced Changes in Pi Uptake Efficiency or Levels of Pi Starvation-Induced Transcripts
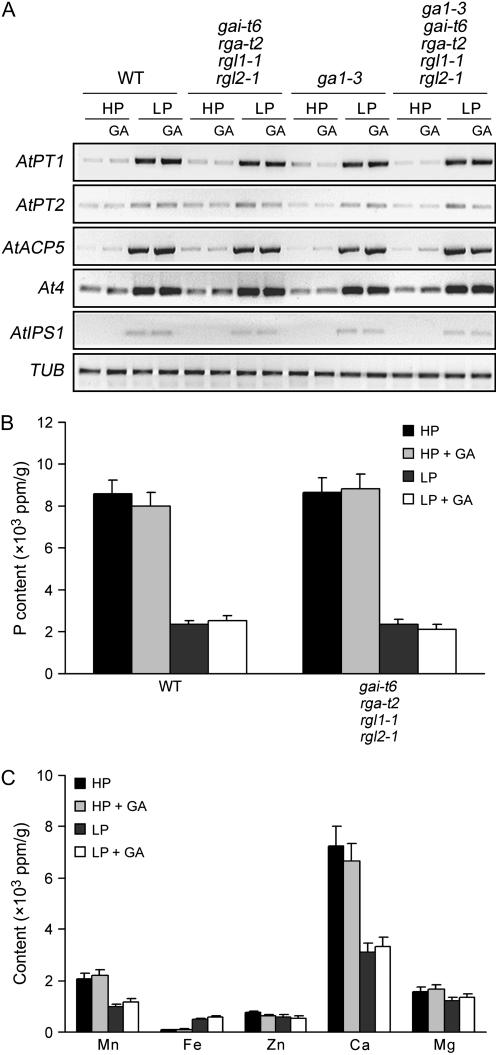
Plants have evolved numerous adaptive responses to Pi starvation, including changes in root architecture, accumulation of anthocyanin pigments, and improvement of Pi uptake efficiency (Raghothama, 1999). Activation of expression of plant Pi starvation marker genes, such as genes encoding Pi transporters, appears to be a universal response, suggesting the presence of a highly regulated molecular network controlling the expression of the genes involved (Rausch and Bucher, 2002). The above-described experiments had indicated a link between GA signaling and various aspects of the plant Pi starvation response. It remained possible that the growth promoted by GA under Pi starvation was due to increases in Pi transporter levels and consequent enhancement of Pi uptake. We therefore analyzed the effect of GA and DELLAs on the expression of Pi starvation-induced marker genes. As shown previously, we found that the expression of genes encoding Pi transporters (AtPT1 and AtPT2) and of additional Pi starvation-responsive marker genes (AtACP5, At4, and AtIPS1) was enhanced in LP conditions (Muchhal et al., 1996; del Pozo et al., 1999; Martin et al., 2000; Fig. 4A). However, GA treatment had no detectable effect on the levels of these marker gene transcripts in wild-type plants (Fig. 4A). Furthermore, mutant lines having altered GA-DELLA signaling functions (e.g. gai-t6 rga-t2 rgl1-1 rgl2-1 and ga1-3 gai-t6 rga-t2 rgl1-1 rgl2-1) also exhibited LP-induced accumulation of transcripts of the above Pi starvation-responsive marker genes, accumulations that were not changed by exogenous GA (Fig. 4A). Thus, the growth promotion of GA-treated LP-grown wild-type seedlings (or of LP-grown seedlings substantially deficient for DELLA function) is unlikely to be due to a DELLA-dependent further increase in Pi uptake efficiency (resulting from an increase in the levels of transcripts encoding the Pi transporters AtPT1 and AtPT2).
Figure 4.
DELLAs do not contribute to the expression of Pi starvation-induced marker genes or to the regulation of phosphorus absorption. A, Comparison of the levels of Pi starvation-induced marker gene transcripts (determined by RT-PCR). Plants were 6-d-after-transfer wild-type, gai-t6 rga-t2 rgl1-1 rgl2-1, ga1-3, and ga1-3 gai-t6 rga-t2 rgl1-1 rgl2-1 mutant seedlings grown with treatments as indicated. Tubulin (TUB) transcripts provided loading control. B and C, Comparison of the phosphorus and other micronutrient contents of 6-d-after-transfer wild-type, gai-t6 rga-t2 rgl1-1 rgl2-1, ga1-3, and ga1-3 gai-t6 rga-t2 rgl1-1 rgl2-1 mutant seedlings grown in the same conditions as in A. Results are presented as means with se bars.
We next compared the phosphorus content of wild-type and gai-t6 rga-t2 rgl1-1 rgl2-1 seedlings grown in HP versus LP conditions. Whereas plants grown in HP contained more phosphorus than plants grown in LP, we found no significant differences attributable to genotype (the phosphorus content of wild-type and gai-t6 rga-t2 rgl1-1 rgl2-1 seedlings were identical in HP and again in LP; Fig. 4B). In addition, GA treatment had no detectable effect on phosphorus content (Fig. 4B) or on the content of additional nutrients such as manganese, iron, zinc, calcium, and magnesium (Fig. 4C). Thus, DELLAs do not detectably alter changes in Pi absorption or the expression of Pi starvation-responsive marker genes.
Pi Starvation Regulates the Levels of Transcripts Encoding Enzymes of GA Metabolism
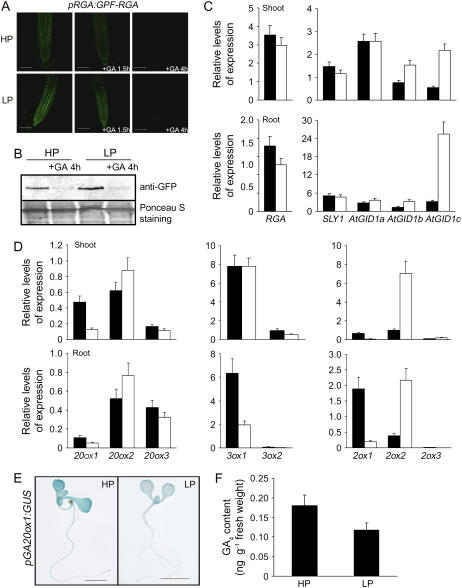
Because Pi starvation inhibits plant growth via a mechanism that is in part DELLA dependent (as shown in Figs. 1 and 2), we determined whether DELLA-dependent Pi starvation-induced inhibition of root growth is associated with DELLA accumulation. These experiments used a transgenic line expressing a pRGA:GFP-RGA construct (that encodes a fusion protein comprising the GFP and the DELLA protein RGA; Silverstone et al., 2001). We found that fluorescence attributable to GFP-RGA was more intense in root cell nuclei of pRGA:GFP-RGA seedlings grown in LP conditions than it was in HP-grown pRGA:GFP-RGA seedlings (Fig. 5A). Furthermore, immunologically detectable GFP-RGA (detected using an anti-GFP antibody) was more abundant in LP-grown roots than in HP-grown roots (Fig. 5B). These observations indicate that Pi starvation increases the accumulation of RGA in root cell nuclei. The GFP-RGA fusion protein is rapidly destroyed following GA treatment when pRGA:GFP-RGA seedlings are grown in HP conditions (Silverstone et al., 2001; Fu and Harberd, 2003; Fig. 5, A and B). In contrast, GFP-RGA was clearly detectable in root cell nuclei of LP-grown pRGA:GFP-RGA seedlings after 1.5 h GA treatment (and not after 1.5 h GA treatment of HP-grown pRGA:GFP-RGA seedlings [Fig. 5A]). However, GFP-RGA was not detectable in root cell nuclei of both LP- and HP-grown pRGA:GFP-RGA seedlings within 4 h of onset of GA treatment (Fig. 5, A and B). These observations suggest that Pi starvation enhances the accumulation of DELLAs, but does not change the vulnerability of DELLAs to GA-promoted destruction in the 26S proteasome.
Figure 5.
Pi starvation regulates the levels of GA biosynthesis gene transcripts. A, Pi starvation promotes accumulation of GFP-RGA protein in nuclei of 4-d-old pRGA:GFP-RGA seedlings grown on LP condition for 5 d, GFP-RGA fluorescence is still sensitive to GA response. B, Immunodetection of GFP-RGA in pRGA:GFP-RGA roots grown in HP or LP conditions in the presence or absence of GA. C, Levels of GA signaling RGA, SLY1, AtGID1a, AtGID1b, and AtGID1c gene transcripts (determined by real-time PCR) in 4-d-old seedlings grown on GM medium, then transferred to HP or LP conditions for 5 d. Results are displayed as the copy per 1,000 copy of 18S rRNA. D, Expression profiles of GA biosynthesis GA 20-oxidase, GA 3-oxidase, and GA 2-oxidase genes. Four-day-old wild-type seedlings were transferred to LP or HP conditions for 6 d, respectively, and then the samples were collected and analyzed using real-time PCR. Results are displayed as the ratio of expression to 18S rRNA. E, GUS staining of pGA20OX1:GUS seedling grown on GM medium, then transferred to LP condition for 5 d. F, Determination of GA4 content of 4-d-old seedlings transferred to LP or HP conditions for 5 d using the gas chromatography-mass spectrometry method.
We next determined whether the LP-promoted accumulation of GFP-RGA was associated with Pi starvation-induced increases in the levels of gene transcripts encoding DELLAs or decreases in the levels of gene transcripts encoding other GA-signaling components, such as the SLY1 components of the SCFSLY1/SLY2 E3 ubiquitin ligase (Dill et al., 2004; Fu et al., 2004) and the three Arabidopsis GA receptors (AtGID1a, AtGID1b, and AtGID1c; Griffiths et al., 2006). We found that levels of SLY1, RGA, GAI, RGL1, RGL2, and RGL3 transcripts were not affected by Pi status (Fig. 5C; data not shown). In contrast, whereas AtGID1a and AtGID1b transcript levels were not detectably different in LP or HP conditions, Pi starvation dramatically promoted AtGID1c transcript accumulation (Fig. 5C). Thus, whereas GFP-RGA accumulates in LP-grown pRGA:GFP-RGA root nuclei, this accumulation is unlikely to be due to increased accumulation of DELLA-encoding transcripts or to decreased accumulation of transcripts encoding the F-box (SLY1) and GA receptors (AtGID1a,b,c) of the GA-signaling pathway.
Whereas an increase in bioactive GA level causes a decrease in GFP-RGA accumulation, a reduction in bioactive GA level causes an increase in GFP-RGA accumulation (Silverstone et al., 2001; Fu and Harberd, 2003). We next analyzed whether the Pi starvation-induced accumulation of GFP-RGA might be the consequence of a decrease in bioactive GA level. Bioactive GA level is elevated by increases in the levels of transcripts encoding GA 20-oxidases (GA20ox) and GA 3-oxidases (GA3ox) or by decreases in the levels of transcripts encoding GA 2-oxidases (GA2ox; Chiang et al., 1995; Phillips et al., 1995; Thomas et al., 1999). We therefore analyzed the effects of Pi starvation on the levels of GA20ox, GA3ox, and GA2ox transcripts. We found evidence of reduced levels of GA20ox1 transcripts in LP-grown (compared with HP-grown) seedlings whether determined via real-time reverse transcription (RT)-PCR (Fig. 5D) or visualized as the relative level of GUS activity expressed from a pGA20ox1:GUS (promoter-GUS) fusion construct (Fig. 5E). Although there was no detectable difference in GA3ox1 transcript levels in shoots, roots were found to contain much lower levels of GA3ox1 transcripts when grown in LP conditions than were found in HP conditions (Fig. 5C). In contrast, both shoots and roots of LP-grown seedlings had relatively high levels of GA2ox2 transcripts (Fig. 5D).
Increases in the levels of GA2ox2 transcripts (that encode an enzyme that deactivates bioactive GAs) and decreases in the levels of GA20ox and GA3ox transcripts (both of which encode enzymes of bioactive GA biosynthesis) would be expected to reduce the in planta levels of bioactive GAs. To test this possibility, we determined the levels of GA4 (the principal bioactive GA species in Arabidopsis) in wild-type seedlings grown in LP versus HP conditions. We found that the level of GA4 in LP-grown seedlings was significantly less than was detected in HP-grown controls (Fig. 5F). Thus, Pi starvation causes a reduction in GA4 levels and this reduction, in turn, likely explains the Pi starvation-induced accumulation of GFP-RGA.
Pi Starvation Enhancement of Root Hair Elongation Is GA Dependent
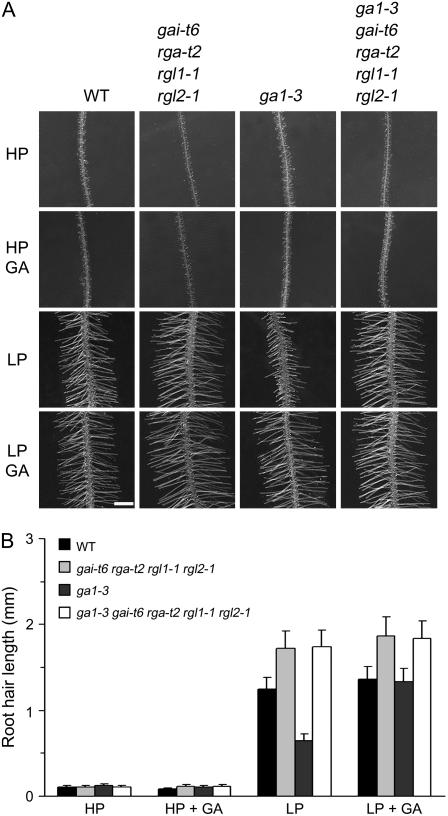
Growth of Arabidopsis seedlings in Pi-limiting conditions causes an increase in the length and frequency of root hairs, thus enlarging the root surface area and enhancing the ability of the roots to absorb phosphorus (Schikora and Schmidt, 2001; Ma et al., 2003; Fig. 3A). Recently, it has been reported that the phytohormones ethylene and auxin are involved in Pi starvation-induced root hair development in Arabidopsis (Schikora and Schmidt, 2001; He et al., 2005). To investigate the possible role of the GA-DELLA system in Pi deficiency-induced changes in the formation and growth of root hairs, we compared the roots of wild-type, gai-t6 rga-t2 rgl1-1 rgl2-1, ga1-3, and ga1-3 gai-t6 rga-t2 rgl1-1 rgl2-1 seedlings. We found no significant differences in root hair formation in GA-treated and control LP-grown wild-type seedlings (Fig. 6A; data not shown), suggesting that DELLAs do not contribute to LP-stimulated changes in epidermal cell fate. Whereas the root hairs of LP-grown GA-deficient ga1-3 mutants were mostly formed in the same position as in wild-type controls, the ga1-3 root hair density was somewhat high, and the lengths of individual ga1-3 root hairs were much shorter than those of wild type (Fig. 6A). However, the length of LP-grown ga1-3 root hairs could be restored to that of wild type by exogenous GA (Fig. 6, A and B). Thus, GA is required for Pi deficiency-induced root hair elongation. Pi starvation causes a reduction in GA4 levels (Fig. 5F) and an increase in root AtGID1c transcript levels (Fig. 5C). Perhaps Pi starvation-enhanced AtGID1c GA receptor function explains the Pi starvation-induced root hair elongation. Furthermore, we found that LP-grown gai-t6 rga-t2 rgl1-1 rgl2-1 root hairs are longer than those of LP-grown wild-type seedlings. The density of Pi-starved ga1-3 gai-t6 rga-t2 rgl1-1 rgl2-1 root hairs was lower than that of ga1-3 and slightly lower than that of the wild type (data not shown). These results indicate that GA-DELLA signaling contributes to the regulation of root hair length in Pi starvation conditions.
Figure 6.
Bioactive GAs are required for Pi starvation promotion of root hair growth. A, Effect of Pi availability and GA treatment on root hair growth for 9-d-after-transfer wild-type, gai-t6 rga-t2 rgl1-1 rgl2-1, ga1-3, and ga1-3 gai-t6 rga-t2 rgl1-1 rgl2-1 mutant seedlings. Bar = 1 mm. B, Comparison of root hair length in mature root hair zone of Arabidopsis seedlings shown in A; results are presented as means ± se.
DISCUSSION
It has recently become apparent that the GA-DELLA mechanism plays an important role in modulating plant growth via integration of both environmental and endogenous signals (Lee at al., 2002; Fu and Harberd, 2003; Alvey and Harberd, 2005; Achard et al., 2006, 2007a, 2007b; Penfield et al., 2006). The work described in this article shows that the plant growth and developmental effects of nutrient limitation, in particular of Pi starvation, are also mediated (at least in part) via the GA-DELLA mechanism. First, we have shown that GA is involved in regulating Pi starvation-induced changes in root and shoot architecture and, in particular, in promoting the development of secondary lateral roots. Second, we have shown that Pi starvation inhibits plant growth and promotes anthocyanin accumulation and root hair elongation via mechanisms that are DELLA dependent. Third, we have shown that Pi starvation results in DELLA accumulation (accumulation of GFP-RGA) and that this accumulation is associated with a reduction in the levels of bioactive GA.
The mechanisms underlying Pi starvation signaling are well understood in bacteria and yeast (Saccharomyces cerevisiae; Torriani, 1990; Lenburg and O'Shea, 1996). However, it is not currently clear how plant primary responses to Pi starvation are initiated. Our results indicate that the plant GA-DELLA mechanism does not regulate the changes in Pi uptake efficiency or levels of Pi starvation-induced transcripts that are characteristic of Pi starvation. Thus, the GA-DELLA mechanism likely contributes to Pi starvation responses as follows. Following perception and initial signaling of Pi starvation conditions (via unknown mechanisms), the levels of gene transcripts encoding enzymes that activate bioactive GAs are reduced, whereas those that deactivate GAs are increased. In consequence, bioactive GA levels fall and DELLAs accumulate. Accumulation of DELLAs, in turn, contributes to a range of characteristic Pi starvation growth and developmental responses, including changes in shoot and root architecture, accumulation of anthocyanins, and root hair elongation.
Whereas our observations identify GA-DELLA-dependent components of the plant Pi starvation response, we have also identified DELLA-independent components. Thus, although a significant contributor, the GA-DELLA mechanism is not the sole developmental regulator of Pi starvation response. Future experiments will determine whether the developmental effects of nutrient limitation, in general, are partially DELLA dependent or whether DELLA dependency is specifically restricted to the Pi starvation response.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The experiments used transgenic line pGA20ox1:GUS and ga1-t Arabidopsis (Arabidopsis thaliana) ecotype Columbia (laboratory strain genetic background). The Landsberg erecta laboratory strain, mutant lines of ga1-3, gai, gai-t6 rga-t2 rgl1-1 rgl2-1, ga1-3 gai-t6 rga-t2 rgl1-1 rgl2-1, and sly1-10 and transgenic line pRGA:GFP-RGA were as described previously (Reed et al., 1993; Whitelam et al., 1993; Cheng et al., 2004; Fu et al., 2004; Achard et al., 2006). All seeds were surface sterilized and placed on glucose minimal (GM) medium plates at 4°C for 4 d to synchronize germination as described previously (Fu and Harberd, 2003). Plates were then placed in vertical orientation in controlled-environment chambers (22°C, 16-h light). Four-day-old seedlings were transferred to LP medium (10 μm NaH2PO4) or HP medium (1 mm NaH2PO4), which was supplemented with 2.0 mm NH4NO3, 1.9 mm KNO3, 0.3 mm CaCl2·2H2O, 0.15 mm MgSO4·7H2O, 5 μm KI, 100 μm H3BO3, 100 μm MnSO4·H2O, 30 μm ZnSO4·7H2O, 1 μm Na2MoO4·2H2O, 0.1 μm CuSO4·5H2O, 0.1 μm CoCl2·6H2O, 100 μm FeSO4·7H2O, 100 μm Na2EDTA·2H2O, and 1% Suc. Plants were maintained at 65 μm m−2 s−1 photosynthetically active radiation and placed in vertical orientation in controlled-environment chambers (22°C, 16-h light).
Root Growth Experiments
Arabidopsis root length, lateral roots, and root hairs were photographed with a Leica MZ16FA stereomicroscope with Leica IM50 software. Root tissues were scanned and length was measured using SigmaScan Pro 5 software (Systat Software Inc.). For each set of experiments, at least 60 seedlings were measured.
Detection of GFP Fluorescence and GUS Staining
For GUS staining, Arabidopsis seedlings were incubated for 4 h at 37°C (in 0.5 mg/mL of 5-bromo-4-chloro-3-indolyl-β-d-glucuronide in 100 mm sodium phosphate, pH 7.0) and stained seedlings were cleared and photographed. Fluorescence due to GFP-RGA in root cell nuclei was determined by Olympus laser confocal microscopy as described previously (Fu and Harberd, 2003).
Detection of Pi and Anthocyanin
Seedlings were grown on one-half-strength Murashige and Skoog medium for 3 d. Subsequently, seedlings were transferred to HP or LP medium with or without GA treatment for 12 d. Fifty milligrams (fresh weight) of seedlings were collected and anthocyanin content was measured as described before (Kim et al., 2003). For measurement of Pi content, seedlings were transferred to LP or HP medium for 7 d, then collected and dried at 80°C for 48 h. Pi content of 50 mg of seedlings (dry weight) was evaluated by the vanadate-molybdate colorimetric method (Hesse, 1971).
Transcript Analysis
Total RNA was extracted using TRIzol reagent (Invitrogen). Semiquantitative RT-PCR was performed as previously described (Fu et al., 2004). Real-time PCR was performed using SYBR green PCR master mix (Applied Biosystems) in optical 96-well reaction plates (Applied Biosystems) on an Eppendorf mastercycler system. All reactions were repeated at least three times. The relative quantity was based on the comparative Ct (threshold cycle) method and α-tubulin as control.
Primer pairs used for PCR amplification were as follows: GA20ox1, 5′-aaatccggtgagagtgttgg-3′ and 5′-cggacacaagaagaatgcaa-3′; GA20ox2, 5′-cggcagattctccactaagc-3′ and 5′-tcgcgctctctctattcaca-3′; GA20ox3, 5′-cttaatcagcactcgcacca-3′ and 5′-ccggaatattgaatcgctgt-3′; GA3ox1, 5′-ccgaaggtttcaccatcact-3′ and 5′-gaccccaaaggaatgctaca-3′; GA3ox2, 5′-tagatcgcatcccattcaca-3′ and 5′-tgaacctaatgcgaaccaca-3′; GA2ox1, 5′-ccgaggaacacacttagcaag-3′ and 5′-ggcttcaacaattcgaaagg-3′; GA2ox2, 5′-gagtgactcgtgcctgagac-3′ and 5′-ccttgtatgagagtagtcat-3′; GA2ox3, 5′-tggtagaggaagagctaaag-3′ and 5′-ctaagcttggtgactatagg-3′; RGA, 5′-agaagcaatccagcaga-3′ and 5′-gtgtactctcttcttaccttc-3′; SLY1, 5′-gcgcagtactaccgactctg-3′ and 5′-cgagaagatgagtttcactaag-3′; AtGID1a (At3g05120), 5′-actctttgtcgcaggcttgt-3′ and 5′-tcgggctaaacggattacac-3′; AtGID1b (At3g63010), 5′-taccaacctgcgtctctcct-3′ and 5′-ttccccaacactttgactcc-3′; AtGID1c (At5g27320), 5′-accgtcatctcgcagagttt-3′ and 5′-tccttgactcaaccgctctt-3′; F3′H, 5′-ggacaccgatggagactgtt-3′ and 5′-cagtccaccgtacttgctga-3′; LDOX, 5′-ctaacaacgcgagtggacaa-3′ and 5′-cggagactcaacactcacca-3′; UF3GT, 5′-gtgttctgcgctttcggtag-3′ and 5′-aaaaccagagtcagtcaaaacaca-3′; AtPAP1, 5′-tgtcccccttttctgttgtc-3′ and 5′-attcctacaacaccggcact-3′; 18S RNA, 5′-atacgtgcaacaaaccc-3′ and 5′-ctacctccccgtgtca-3′; TUB, 5′-tttggagcctgggactatggat-3′ and 5′-acgggggaatgggatgagat-3′.
GA Analysis
Wild-type seeds were surface sterilized and placed on GM medium plates at 4°C for 4 d to synchronize germination. Plates were placed in vertical orientation in controlled-environment chambers (22°C, 16-h light). Four-day-old seedlings were transferred to LP medium for 5 d as described above. Then, seedlings were collected and homogenized in 80% methanol. GA was extracted and separated using HPLC and GA was analyzed using gas chromatography-mass spectrometry methods as described (Eriksson et al., 2006).
Acknowledgments
We thank Tai-ping Sun for the pRGA:GFP-RGA line, and Daowen Wang and Long Mao for helpful discussion of the manuscript.
This work was supported by the National Natural Science Foundation of China (grant nos. 30525003, 30630009, and 30521001) and the Biotechnology and Biological Sciences Research Council (Response modes Core Strategic Grant to the John Innes Centre).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Xiangdong Fu (xdfu@genetics.ac.cn).
Open Access articles can be viewed online without a subscription.
References
- Achard P, Baghour M, Chapple A, Hedden P, Van Der Straeten D, Genschik P, Moritz T, Harberd NP (2007. a) The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc Natl Acad Sci USA 104 6484–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P, Cheng H, Grauwe LD, Decat J, Schoutteten H, Moritz T, Dominique VDS, Peng J, Harberd NP (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311 91–94 [DOI] [PubMed] [Google Scholar]
- Achard P, Liao L, Jiang C, Desnos T, Bartlett J, Fu X, Harberd NP (2007. b) DELLAs contribute to plant photomorphogenesis. Plant Physiol 143 1163–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Ghazi Y, Muller B, Pinloche S, Tranbarger TJ, Nacry P, Rossignol M, Tardieu F, Doumas P (2003) Temporal responses of Arabidopsis root architecture to phosphate starvation: evidence for the involvement of auxin signaling. Plant Cell Environ 26 1053–1066 [Google Scholar]
- Alvey L, Harberd NP (2005) DELLA proteins: integrators of multiple plant growth regulatory inputs? Physiol Plant 123 153–160 [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12 2383–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Hussain A, Cheng H, Peng J (2005) Loss of function of four DELLA genes leads to light- and gibberellin-independent seed germination in Arabidopsis. Planta 223 105–113 [DOI] [PubMed] [Google Scholar]
- Casimiro I, Marchant A, Bhalerao RP, Beeckman T, Dhooge S, Swarup R, Graham N, Inzé D, Sandberg G, Casero PJ, et al (2001) Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Qin L, Lee S, Fu X, Richards DE, Cao D, Luo D, Harberd NP, Peng J (2004) Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131 1055–1064 [DOI] [PubMed] [Google Scholar]
- Chiang HH, Hwang I, Goodman HM (1995) Isolation of the Arabidopsis GA4 locus. Plant Cell 7 195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Allona I, Rubio V, Leyva A, de la Pena A, Aragoncillo C, Paz-Ares J (1999) A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilizing/oxidative stress conditions. Plant J 19 579–589 [DOI] [PubMed] [Google Scholar]
- Dill A, Sun T (2001) Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun TP (2004) The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S, Bohlenius H, Moritz T, Nilsson O (2006) GA4 is active gibberellin in the regulation of leafy transcription and Arabidopsis floral initiation. Plant Cell 18 2172–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Harberd NP (2003) Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421 740–743 [DOI] [PubMed] [Google Scholar]
- Fu X, Richards DE, Ait-ali T, Hynes LW, Ougham H, Peng J, Harberd NP (2002) Gibberellin-mediated proteasome dependent degradation of the barley DELLA protein SLN1 repressor. Plant Cell 14 3191–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Richards DE, Fleck B, Xie D, Burton N, Harberd NP (2004) The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 16 1406–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert GA, Knight JD, Vance CP, Allan DL (2000) Proteoid root development of phosphorus deficient lupin is mimicked by auxin and phosphonate. Ann Bot (Lond) 85 921–928 [Google Scholar]
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Phillips AL, Hedden P, Sun TP, et al (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Ma Z, Brownk M, Lynch JP (2005) Assessment of inequality of root hair density in Arabidopsis thaliana using the Gini coefficient: a close look at the effect of phosphorus and its interaction with ethylene. Ann Bot (Lond) 95 287–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse PR (1971) Soil phosphorus: its measurements and its uptake by plants. Aust J Soil Res 35 228–239 [Google Scholar]
- Holford ICR (1997) Soil phosphorus: its measurements and its uptake by plants. Aust J Soil Res 35 227–239 [Google Scholar]
- Hooley R (1994) Gibberellins: perception, transduction and responses. Plant Mol Biol 26 1529–1555 [DOI] [PubMed] [Google Scholar]
- Jaffe FW, Jacob SR, Bogs J, Robinson SP, Walker AR, Takos AM (2006) Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol 142 1216–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Yi H, Choi G, Shin B, Song PS, Choi G (2003) Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King KE, Moritz T, Harberd NP (2001) Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159 767–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ (2004) Secondary metabolites and plant/environment interactions: a view through Arabidopsis thaliana tinged glasses. Plant Cell Environ 27 675–684 [Google Scholar]
- Koornneef M, van der Veen JH (1980) Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Heyhn. Theor Appl Genet 58 257–263 [DOI] [PubMed] [Google Scholar]
- Lai F, Thacker J, Li Y, Doerner P (2007) Cell division activity determines the magnitude of phosphate starvation responses in Arabidopsis. Plant J 50 545–556 [DOI] [PubMed] [Google Scholar]
- Lee S, Cheng H, King KE, Wang W, He Y, Hussain A, Lo J, Harberd NP, Peng J (2002) Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev 16 646–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenburg ME, O'Shea EK (1996) Signaling phosphate starvation. Trends Biochem Sci 21 383–387 [PubMed] [Google Scholar]
- López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L (2003) The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol 6 280–287 [DOI] [PubMed] [Google Scholar]
- López-Bucio J, de la Vega OM, Guevara-García A, Herrera-Estrella L (2000) Enhanced phosphorus uptake in transgenic tobacco plants that over-produce citrate. Nat Biotechnol 18 450–453 [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L (2002) Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol 129 244–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP, Brown KM (2001) Topsoil foraging: an architectural adaptation of plants to low phosphorus availability. Plant Soil 237 225–237 [Google Scholar]
- Ma Z, Baskin TI, Brown KM, Lynch JP (2003) Regulation of root elongation under phosphorus stress involves changes in ethylene responsiveness. Plant Physiol 131 1381–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AC, del Pozo JC, Iglesias J, Rubio V, Solano R, de la Pena Leyva A, Paz-Ares J (2000) Influence of cytokinins on the expression of phosphate starvation-responsive genes in Arabidopsis. Plant J 24 559–567 [DOI] [PubMed] [Google Scholar]
- McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun TP, Steber CM (2003) The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15 1120–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchhal US, Pardo JM, Raghothama KG (1996) Phosphate transporters from the higher plant Arabidopsis thaliana. Proc Natl Acad Sci USA 93 10519–10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muday GK, Haworth P (1994) Tomato root growth, gravitropism, and lateral development: correlation with auxin transport. Plant Physiol Biochem 32 193–203 [PubMed] [Google Scholar]
- Nakajima M, Shimada A, Takashi Y, Kim YC, Park SH, Ueguchi-Tanaka M, Suzuki H, Katoh E, Iuchi S, Kobayashi M, et al (2006) Identification and characterization of Arabidopsis gibberellin receptors. Plant J 46 880–889 [DOI] [PubMed] [Google Scholar]
- Penfield S, Gilday AD, Halliday KJ, Graham IA (2006) DELLA-mediated cotyledon expansion breaks coat-imposed seed dormancy. Curr Biol 16 2366–2370 [DOI] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AL, Ward DA, Uknes S, Appleford NE, Lange T, Huttly AK, Gaskin P, Graebe JE, Hedden P (1995) Isolation and expression of three gibberellin 20-oxidase cDNA clone from Arabidopsis. Plant Physiol 108 1049–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50 665–693 [DOI] [PubMed] [Google Scholar]
- Rausch C, Bucher M (2002) Molecular mechanisms of phosphate transport in plants. Planta 216 23–37 [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J (1993) Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed RC, Brady SR, Muday GK (1998) Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiol 118 1369–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards DE, King KE, Ait-ali T, Harberd NP (2001) How gibberellin regulates plant growth and development: a molecular genetic analysis of gibberellin signaling. Annu Rev Plant Physiol Plant Mol Biol 52 67–88 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Jeong DH, An G, Kitano H, Ashikari M, et al (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299 1896–1898 [DOI] [PubMed] [Google Scholar]
- Schikora A, Schmidt W (2001) Acclimative changes in root epidermal cell fate in response to Fe and P deficiency: a specific role for auxins? Protoplasma 218 67–75 [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Ciampaglio CN, Sun TP. (1998) The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Jung HS, Dill A, Kawaide H, Kamiya Y, Sun TP (2001) Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene KR, James WM (2000) A comparison of the effects of auxin on cluster root initiation and development in Grecil lea robusta Cunn. ex R. Br. (Proteaceae) and in the genus Lupinus (Leguminosae). Plant Soil 219 221–229 [Google Scholar]
- Stewart AJ, Chapman W, Jenkins GI, Graham I, Martin T, Crozier A (2001) The effect of nitrogen and phosphorus deficiency on Xavonol accumulation in plant tissues. Plant Cell Environ 24 1189–1197 [Google Scholar]
- Svistoonoff S, Creff A, Reymond M, Sigoillot-Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T (2007) Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet 39 792–796 [DOI] [PubMed] [Google Scholar]
- Thomas SG, Phillips AL, Hedden P (1999) Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci USA 96 4698–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torriani A (1990) From cell membrane to nucleotides: the phosphate regulation in Escherichia coli. Bioessays 12 371–376 [DOI] [PubMed] [Google Scholar]
- Wen CK, Chang C (2002) Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14 87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP (1993) Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5 757–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Ito T, Zhao Y, Peng J, Kumar P, Meyerowitz EM (2004) Floral homeotic genes are targets of gibberellin signaling in flower development. Proc Natl Acad Sci USA 101 7827–7832 [DOI] [PMC free article] [PubMed] [Google Scholar]