Abstract
The galactolipid digalactosyldiacylglycerol (DGDG) is present in the thylakoid membranes of oxygenic photosynthetic organisms such as higher plants and cyanobacteria. Recent x-ray crystallographic analysis of protein-cofactor supercomplexes in thylakoid membranes revealed that DGDG molecules are present in the photosystem II (PSII) complex (four molecules per monomer), suggesting that DGDG molecules play important roles in folding and assembly of subunits in the PSII complex. However, the specific role of DGDG in PSII has not been fully clarified. In this study, we identified the dgdA gene (slr1508, a ycf82 homolog) of Synechocystis sp. PCC6803 that presumably encodes a DGDG synthase involved in the biosynthesis of DGDG by comparison of genomic sequence data. Disruption of the dgdA gene resulted in a mutant defective in DGDG synthesis. Despite the lack of DGDG, the mutant cells grew as rapidly as the wild-type cells, indicating that DGDG is not essential for growth in Synechocystis. However, we found that oxygen-evolving activity of PSII was significantly decreased in the mutant. Analyses of the PSII complex purified from the mutant cells indicated that the extrinsic proteins PsbU, PsbV, and PsbO, which stabilize the oxygen-evolving complex, were substantially dissociated from the PSII complex. In addition, we found that heat susceptibility but not dark-induced inactivation of oxygen-evolving activity was notably increased in the mutant cells in comparison to the wild-type cells, suggesting that the PsbU subunit is dissociated from the PSII complex even in vivo. These results demonstrate that DGDG plays important roles in PSII through the binding of extrinsic proteins required for stabilization of the oxygen-evolving complex.
Thylakoid membranes are sites of the primary reactions of photosynthesis in plants, algae, and cyanobacteria (Malkin and Niyogi, 2000; Nelson and Ben-Shem, 2004). The lipid composition of thylakoid membranes is highly conserved among oxygenic photosynthetic organisms. They are composed of uncharged lipids, monogalactosyldiacylglycerol (MGDG), and digalactosyldiacylglycerol (DGDG), as well as anionic lipids, sulfoquinovosyldiacylglycerol (SQDG), and phosphatidylglycerol (PG; Somerville et al., 2000). Although phospholipids are the major components of cellular membranes in animal cells and bacteria, as well as nonchloroplastic cellular membranes in plants, the majority (80%–90%) of lipids in thylakoid membranes are glycolipids, namely MGDG, DGDG, and SQDG. PG is the only phospholipid that is present in the thylakoid membranes. Based on a structural point of view, lipids in thylakoid membranes can be divided into three different classes: (1) bulk lipids that form lipid bilayers; (2) annular lipids that surround protein-cofactor supercomplexes and directly interact with the outer surface of the complexes; and (3) integral lipids that are embedded in protein-cofactor supercomplexes (Loll et al., 2007). The integral lipids are found at the interfaces of subunits within the supercomplexes and possibly have important functions in the folding and assembly of protein subunits (Jones, 2007; Loll et al., 2007).
The physiological importance of thylakoid lipids in photosynthesis has been studied using mutants that are defective in the biosynthesis of individual lipid classes. We previously made a pgsA mutant of Synechocystis sp. PCC6803 that cannot synthesize PG because of the disruption of the pgsA gene encoding PG phosphate synthase (Hagio et al., 2000). The mutant requires exogenous PG for its survival, as it grows only in medium supplemented with PG. When PG is supplemented in the growth medium, the mutant cells can grow almost like wild-type cells. However, the PG content decreases after cells are transferred to PG-free medium because of dilution with other newly synthesized lipids. Concomitant with the decrease in PG content, photosynthetic activity of the mutant cells decreases and their growth is inhibited (Hagio et al., 2000; Sakurai et al., 2003). A cdsA mutant of Synechocystis sp. PCC6803 incapable of synthesizing PG was also made by disruption of the cdsA gene for CDP-diacylglycerol synthase (Sato et al., 2000). The cdsA mutant had phenotypes similar to the pgsA mutant. Through detailed analyses of the pgsA mutant, we demonstrated an important role of PG in the acceptor side of PSII at the QB site (Gombos et al., 2002; Sakurai et al., 2006) as well as in the donor side of PSII for the binding of extrinsic proteins required for sustaining the manganese (Mn) cluster (Sakurai et al., 2007).
SQDG-deficient mutants have been isolated from Synechococcus sp. PCC7942 (Güler et al., 1996), Synechocystis sp. PCC6803 (Aoki et al., 2004), Chlamydomonas reinhardtii (Sato et al., 1995), and Arabidopsis (Arabidopsis thaliana; Yu et al., 2002), and roles of SQDG in photosynthesis have been studied with these mutants. An analysis of the mutants indicated that the level of responsibility of SQDG for photosynthesis differed among the organisms. The mutants of Synechococcus sp. PCC7942 and Arabidopsis showed no detrimental phenotypes with respect to growth and photosynthetic activity (Güler et al., 1996; Yu et al., 2002). By contrast, in the mutants of C. reinhardtii and Synechocystis sp. PCC6803, PSII activity decreased, and the functional site of PSII damaged in the C. reinhardtii mutant was specified as the electron transfer site from water to Tyr Z in the donor site of PSII (Minoda et al., 2003).
DGDG-deficient mutants have been isolated from Arabidopsis (Dörmann et al., 1995; Härtel et al., 1997; Kelly et al., 2003). In Arabidopsis, two genes (DGD1 and DGD2) encoding DGDG synthase are involved in the biosynthesis of DGDG. A dgd1 mutant that has a point mutation in the DGD1 gene contained a strongly decreased amount of DGDG (1% of total lipids compared to 15% in the wild type), and its growth and photosynthetic activity were severely affected (Dörmann et al., 1995). Kelly et al. (2003) isolated a T-DNA insertional mutant (dgd2) of the DGD2 gene and generated the double mutant dgd1 dgd2. The double mutant containing only a trace amount of DGDG showed a dwarf phenotype and altered parameters of chlorophyll (Chl) fluorescence, suggesting that DGDG has a crucial function in normal plant development and photosynthesis.
In this study, we focused on the function of DGDG in photosynthesis. Recent x-ray crystallographic analysis of PSII complexes prepared from the thermophilic cyanobacterium Thermosynechococcus elongatus revealed that four DGDG molecules per monomer are present in the crystal structure (Loll et al., 2005). In accordance with the structural analysis, we detected six and three DGDG molecules per monomer in PSII complexes prepared from Thermosynechococcus vulcanus and Synechocystis sp. PCC6803, respectively, by biochemical analysis (Sakurai et al., 2006). These findings indicate that DGDG could have important roles in folding and assembly of protein subunits in PSII. However, the gene encoding DGDG synthase has not been identified in cyanobacteria, and a cyanobacterial mutant incapable of synthesizing DGDG has not been isolated; thus, no direct evidence for a requirement of DGDG in PSII has been provided in cyanobacteria. In higher plants, DGDG-deficient mutants have been isolated from Arabidopsis as mentioned above (Dörmann et al., 1995; Härtel et al., 1997; Kelly et al., 2003). Analysis of the double mutant (dgd1 dgd2) with a newly developed laser flash fluorometer indicated that the lack of DGDG affected the donor side of PSII, namely, the oxygen-evolving complex (Steffen et al., 2005). However, a direct interaction between DGDG molecules and the oxygen-evolving complex was not found in the crystal structure of the cyanobacterial PSII complex (Loll et al., 2005). Why the oxygen-evolving complex was affected by the lack of DGDG remains obscure. In this study, we identified the dgdA gene (slr1508) of Synechocystis sp. PCC6803 that is involved in the biosynthesis of DGDG. Disruption of the gene resulted in a mutant lacking DGDG. The data obtained by the analyses of the mutant demonstrated that DGDG is not essential for growth, but it plays important roles on the donor side of PSII through the binding of extrinsic proteins required for stabilization of the oxygen-evolving complex.
RESULTS
Identification of the dgdA Gene
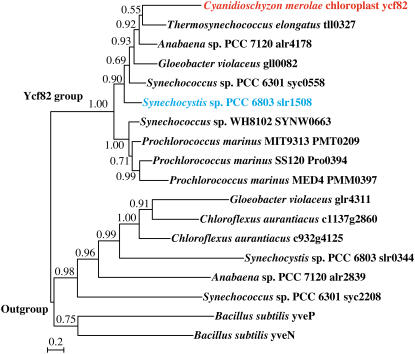
Because neither DGD1 nor DGD2 has homologs in cyanobacteria and the red alga Cyanidioschyzon merolae (Sato and Moriyama, 2007), we determined whether a new DGDG synthesis enzyme is present in cyanobacteria and red algae. We exploited the comparative genomics tools developed in the laboratory of one of the authors (Naoki Sato) to find putative glycosyltransferases that are shared by cyanobacteria and C. merolae but not by green plants. A supervised phylogenetic profiling of the proteins in various photosynthetic and nonphotosynthetic organisms was performed using Gclust software (Sato et al., 2005; N. Sato, M. Ishikawa, N. Sasaki, and M. Fujiwara, unpublished data). The data are now available from the Gclust server (Sato, 2006; http://gclust.c.u-tokyo.ac.jp/). We found two putative glycosyltransferases that meet the criterion. One of them (dataset CZ20x0, cluster 2825) was a putative glycosyltransferase (Ycf82) shared by cyanobacteria and the C. merolae plastid genome (Fig. 1). We therefore decided to analyze the involvement of the ycf82 homolog (slr1508) in the biosynthesis of DGDG in Synechocystis sp. PCC6803. We call this gene dgdA because disruption of the gene resulted in a mutant incapable of synthesizing DGDG, as described below.
Figure 1.
Bayesian Inference tree of the Ycf82 family. The alignment was produced from a cluster in an old version of the CZ16Y dataset using the Gclust software (CZ16Yf1; not available in the Gclust server now). Each value at a branch indicates a confidence level (posterior probability). [See online article for color version of this figure.]
Generation of a DGDG-Deficient Mutant by Disruption of the dgdA Gene
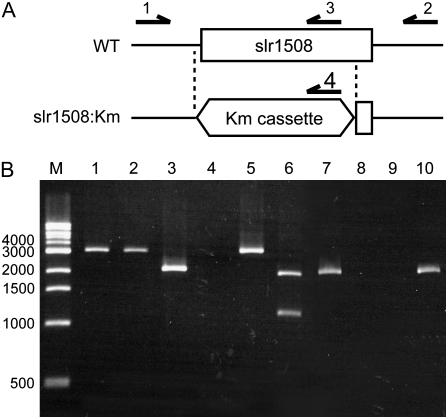
To confirm whether the dgdA gene (slr1508) is involved in the biosynthesis of DGDG, we disrupted the dgdA gene in a CP47-His strain of Synechocystis sp. PCC6803 (Sakurai et al., 2006), which was used as a wild-type (or parent) strain in this study. The open reading frame of the dgdA gene was replaced with a cassette of the kanamycin-resistance gene (Km) from pUC4K to disrupt the dgdA gene (Fig. 2A). Because cyanobacterial cells normally contain many copies of chromosomal DNA (Herdman et al., 1979), complete replacement of all copies was checked by PCR analysis (Fig. 2B). As expected, PCR using a primer set (primer 1 and primer 2) resulted in the amplification of DNA fragments of a similar size in both wild-type (lane 1) and transformed cells (lane 2); however, PCR with another primer set (primer 1 and primer 3) amplified DNA fragments only in wild-type cells (lane 3). To distinguish the amplified DNA fragments in wild-type and transformed cells (lane 1 and lane 2), we treated them with XhoI, which cleaves the Km cassette. The XhoI treatment digested the amplified DNA fragments of transformed cells (lane 6) but not those of wild-type cells (lane 5). A second PCR was performed with two primer sets (primer 1 and primer 3, primer 1 and primer 4) using the DNA fragments amplified by the first PCR as templates (lanes 7–10). The second PCR with a certain primer set (primer 1 and primer 3) amplified DNA fragments only with the DNA of wild-type cells (lane 7), whereas that with another primer set (primer 1 and primer 4) amplified DNA fragments only with the DNA of transformed cells (lane 10). These results clearly demonstrate that all copies of the dgdA gene were disrupted in the transformed cells.
Figure 2.
Disruption of slr1508. A, Design of replacement of slr1508 by a Km cassette and primers used for PCR analysis are shown. B, Replacement of slr1508 by a Km cassette confirmed by PCR analysis. In lanes 1 to 4, PCR was performed using genomic DNA of the wild type (lanes 1 and 3) or the slr1508∷Km (lanes 2 and 4) mutant with a primer set (primers 1 and 2; lanes 1 and 2) or another primer set (primers 1 and 3; lanes 3 and 4). In lanes 5 and 6, amplified DNA fragments in the wild type and the slr1508∷Km mutant were treated with XhoI, which cleaves only within the Km cassette. In lanes 7 to 10, a second PCR was performed with a primer set (primers 1 and 3; lanes 7 and 8) or another primer set (primers 1 and 4; lanes 9 and 10). As templates, DNA fragments amplified by the first PCR in the wild type (lanes 7 and 9) or the slr1508∷Km mutant (lanes 8 and 10) were used.
Table I shows the lipid composition of thylakoid membranes prepared from the wild-type and the dgdA mutant cells. In the dgdA mutant, DGDG was not detected, and the content of MGDG was elevated with respect to that in the wild-type cells, whereas the contents of SQDG and PG were not significantly affected. In both the wild-type and the dgdA mutant cells, trigalactosyldiacylglycerol, which is synthesized by a processive galactolipid:galactolipid galactosyltransferase in higher plants (Benning and Ohta, 2005), was not detected. It is therefore not likely that the alternative biosynthetic pathway to DGDG involving the processive enzyme is operative in Synechocystis sp. PCC6803. These results demonstrate that the dgdA gene is required for the biosynthesis of DGDG, presumably because it encodes a component of the DGDG synthase. The resultant dgdA mutant entirely lacked DGDG, which makes it useful in the investigation of the function of DGDG in photosynthesis.
Table I.
Lipid composition of thylakoid membranes prepared from wild-type and dgdA mutant cells
Values represent averages ± sd of independent samples (n > 3). nd, Not detected (<0.5 mol%).
| Strain | Total | MGDG | DGDG | SQDG | PG |
|---|---|---|---|---|---|
| nmol/μg Chl | mol% | ||||
| Wild type | 2.39 ± 0.08 | 37.4 ± 1.9 | 20.0 ± 1.4 | 28.9 ± 2.3 | 13.7 ± 0.9 |
| dgdA mutant | 2.92 ± 0.05 | 56.0 ± 2.1 | nd | 29.9 ± 1.8 | 14.1 ± 1.0 |
Growth and Photosynthetic Activity of the dgdA Mutant
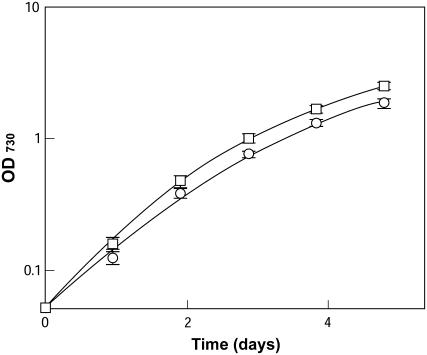
Figure 3 shows the growth profile of wild-type and dgdA mutant cells. Despite the lack of DGDG, the mutant cells grew well but at a slightly lower growth rate than the wild-type cells, suggesting that DGDG is not essential for the growth of Synechocystis under standard growth conditions.
Figure 3.
Growth of wild-type and dgdA mutant cells in BG-11 medium. Cells grown in BG-11 medium were inoculated to fresh BG-11 medium at a cell concentration where optical density at 730 nm = 0.05, and the growth of the wild-type (□) and dgdA mutant (○) cells was monitored.
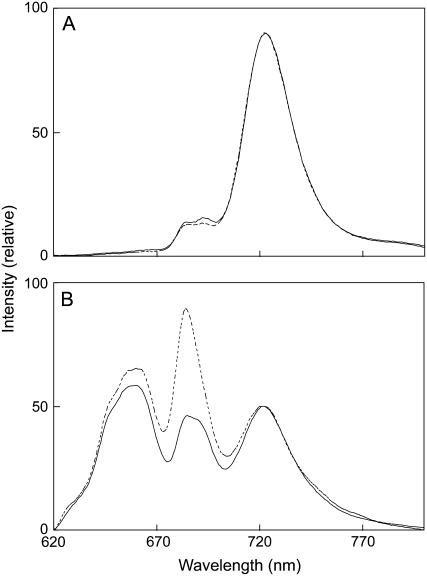
To investigate the effects of the lack of DGDG on photosynthesis, we measured parameters of Chl fluorescence at room temperature in the wild-type and mutant cells. As shown in Table II, the mutant cells displayed a higher level of fluorescence in the dark (F0) than did the wild-type cells. The level of Fv/Fm, which reflects the photochemical efficiency of PSII, was much lower than that of the wild-type cells. These results indicate that the function of PSII is partially impaired by the lack of DGDG. In the search for alterations in the antenna system of the mutant cells, low-temperature (77 K) Chl fluorescence emission spectra were measured. As shown in Figure 4A, excitation of Chl at 440 nm resulted in almost identical emission spectra in both wild-type and mutant cells. However, the emission peak at 695 nm arising from PSII core antenna protein CP47 (Vermaas et al., 1986) was slightly decreased in the mutant cells compared to the wild-type cells. By contrast, excitation of phycobilisomes at 590 nm resulted in a drastic increase in two emission peaks, one at 685 nm from the PSII core antenna protein CP43 (Nakatani et al., 1984) and the other at 660 nm from phycobilisomes, in the mutant cells (Fig. 4B). These results suggest that the efficiency of energy transfer from the antenna Chl to the reaction center and from phycobilisomes to the reaction center (or within phycobilisomes) was decreased in the mutant cells.
Table II.
Characteristics of room-temperature Chl fluorescence
Each sample was adjusted to a Chl concentration of 5 μg mL−1. Values represent averages ± sd of independent samples (n > 3).
| Parameters | Wild Type | dgdA Mutant |
|---|---|---|
| F0 | 857 ± 91 | 1,004 ± 113 |
| Fm | 1,680 ± 118 | 1,600 ± 49 |
| Fv | 823 ± 9 | 595 ± 48 |
| Fv/Fm | 0.504 ± 0.032 | 0.370 ± 0.200 |
Figure 4.
The 77-K fluorescence emission spectra from wild-type and dgdA mutant cells excited at 440 nm (A) or 590 nm (B). Solid lines and dotted lines represent emission spectra from wild-type and mutant cells, respectively. In each case, Chl concentrations were adjusted to 10 μg mL−1.
Table III shows photosynthetic oxygen-evolving activity of intact cells and thylakoid membranes of the wild type and the mutant. Photosynthetic oxygen-evolving activity of the mutant cells that is dependent on electron transport from water to CO2 was similar to that of wild-type cells. However, oxygen-evolving activity of the mutant cells with 2,6-dichloro-p-benzoquinone (DCBQ) as an electron acceptor of PSII was much lower than that of the wild-type cells. A similar difference in PSII activity was observed in thylakoid membranes. Compared to thylakoid membranes of the wild type, PSII activity of thylakoid membranes of the mutant cells was about 30%. Consistent with the findings obtained by the measurement of Chl fluorescence at room temperature, these results suggest that PSII was impaired in the dgdA mutant cells.
Table III.
Photosynthetic oxygen-evolving activity of intact cells and thylakoid membranes of wild type and dgdA mutant
For the measurement of PSII activity, 1 mm DCBQ was added. Values represent averages ± sd of independent samples (n > 3).
| Sample | Strain | Net | PSII |
|---|---|---|---|
| μmol O2 mg Chl−1 h−1 | |||
| Cell | Wild type | 351 ± 12 | 493 ± 9 |
| dgdA mutant | 328 ± 4 | 279 ± 27 | |
| Thylakoid | Wild type | – | 191 ± 5 |
| dgdA mutant | – | 56 ± 8 | |
To understand the critical site of PSII impairment in the mutant cells, we measured PSII activity of thylakoid membranes by means of photoreduction of 2,6-dichlorophenolindophenol (DCIP) using water or 1,5-diphenylcarbazide (DPC) as an electron donor (Table IV). The DCIP photoreduction activity of thylakoid membranes from the mutant cells without DPC was 22% of the activity of thylakoid membranes from the wild-type cells. The activity of thylakoid membranes of the wild type did not depend on the presence of DPC, whereas the activity of thylakoid membranes of the mutant increased by the presence of DPC, which directly donates electrons to Tyr Z. These results indicate that the lack of DGDG induced impairment of PSII at the donor side, namely, at the oxygen-evolving complex.
Table IV.
DCIP photoreduction activity of thylakoid membranes isolated from wild-type and dgdA mutant cells
Values represent averages ± sd of independent samples (n > 3).
| Strain | −DPC | +DPC |
|---|---|---|
| μmol DCIP mg Chl−1h−1 | ||
| Wild type | 102 ± 4 | 100 ± 5 |
| dgdA mutant | 22 ± 1 | 46 ± 4 |
Characterization of PSII Complex Prepared from the dgdA Mutant
As mentioned above, the lack of DGDG resulted in impairment of PSII. To clarify the function of DGDG in PSII, we purified monomer and dimer complexes of PSII from the wild-type and the dgdA mutant cells and characterized the complexes with respect to oxygen-evolving activity and subunit composition. Table V shows oxygen-evolving activity of PSII monomer and dimer complexes from the wild-type and the mutant cells. Potassium ferricyanide (Fecy) and DCBQ were used as electron acceptors. The activities of PSII dimers purified from the wild-type cells with Fecy and DCBQ were 1,975 and 725 μmol O2 mg Chl−1 h−1, respectively. These activities were much higher than those of the monomer. In the case of the mutant cells, the activities of PSII dimers with Fecy and DCBQ were much higher than those of the PSII monomer. However, the activities of the monomer and dimer complexes of the mutant cells were much lower overall than those of the monomer and dimer complexes of the wild-type cells. These findings also suggest the lack of DGDG-induced impairment of PSII.
Table V.
Photosynthetic oxygen-evolving activity of PSII complexes
Values represent averages ± sd of independent preparations (n > 3).
| Strain | Sample | Acceptor
|
|
|---|---|---|---|
| Fecy | DCBQ | ||
| μmol O2 mg Chl−1 h−1 | |||
| Wild type | Monomer | 470 ± 69 | 243 ± 60 |
| Dimer | 1,975 ± 235 | 725 ± 133 | |
| dgdA mutant | Monomer | 94 ± 18 | 131 ± 20 |
| Dimer | 604 ± 83 | 336 ± 30 | |
Table VI shows lipid composition of PSII monomer and dimer complexes of the wild-type and mutant cells. DGDG was not detected in both monomer and dimer complexes of the mutant cells as described in the thylakoid membranes (Table I). The contents of SQDG and PG in monomer and dimer complexes of the mutant were similar to those of monomer and dimer complexes of the wild type, whereas the content of MGDG in monomer and dimer complexes of the mutant was much higher than that of MGDG in monomer and dimer complexes of the wild-type cells. The content of MGDG in the mutant PSII was almost equal to the sum of the contents of MGDG and DGDG in the wild-type PSII.
Table VI.
Lipid composition of PSII complexes
Values represent averages ± sd of independent preparations (n > 3). Numbers in parentheses indicate the numbers of molecules bound to PSII per monomer complex. nd, Not detected (<0.5 mol%).
| Strain | Sample | Total | MGDG | DGDG | SQDG | PG |
|---|---|---|---|---|---|---|
| nmol/μg Chl | mol% (molecules/PSII) | |||||
| Wild type | Monomer | 0.77 ± 0.06 | 30.3 (7.5) | 14.3 (3.5) | 29.2 (7.2) | 26.2 (6.5) |
| Dimer | 0.60 ± 0.02 | 28.7 (5.6) | 16.4 (3.2) | 25.5 (5.0) | 29.4 (5.8) | |
| dgdA mutant | Monomer | 0.78 ± 0.02 | 45.8 (11.4) | nd | 25.5 (6.4) | 28.7 (7.2) |
| Dimer | 0.61 ± 0.02 | 47.1 (9.2) | nd | 23.6 (4.6) | 29.3 (5.7) | |
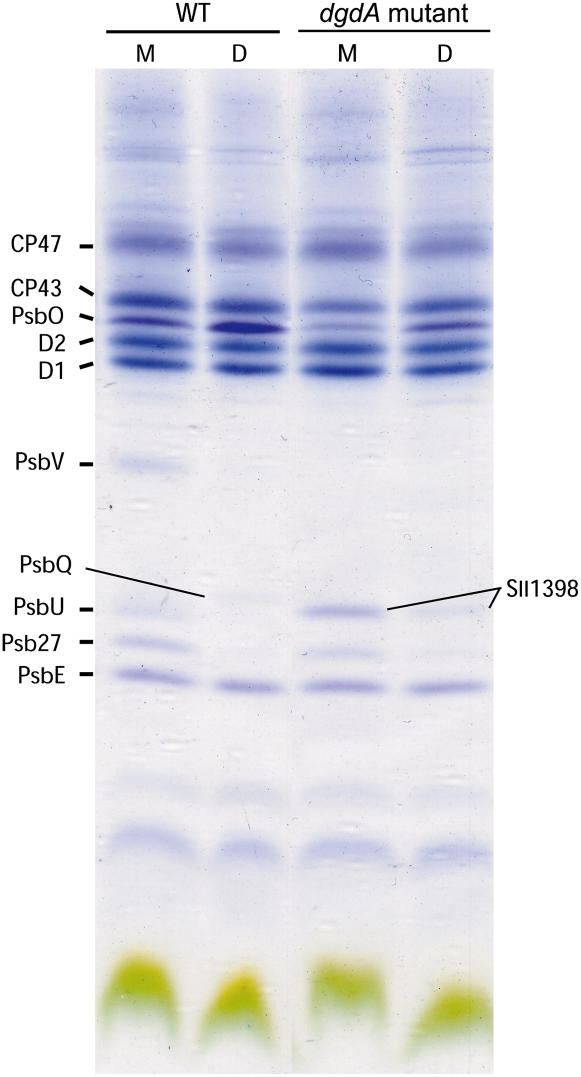
Figure 5 shows protein subunits of PSII from wild-type and mutant cells analyzed by SDS-PAGE. In protein subunits that are complexes of the PSII core (CP47, CP43, D1, D2, and PsbE), a notable difference between the wild type and the mutant was not observed both in monomer and dimer components. However, the amounts of extrinsic proteins were significantly changed in the mutant. PsbU and PsbV primarily present in the wild-type monomer were not detected in the mutant PSII. Psb27 and PsbQ preferentially found in the wild-type monomer and dimer, respectively, were also less abundant in the mutant, as was the PsbO subunit. These extrinsic proteins bind to the luminal side of the PSII complex and play important roles in stabilization of the Mn cluster (Roose et al., 2007). These findings suggest that the decreases in these subunits in the mutant PSII make the Mn cluster unstable and reduce the oxygen-evolving activity of mutant PSII. In addition, accumulation of Sll1398 (Psb28) was observed in the monomer of mutant PSII.
Figure 5.
SDS-PAGE analysis of protein subunits of PSII monomers (M) and dimers (D) prepared from wild-type and dgdA mutant cells. PSII corresponding to 5 μg Chl was loaded in each lane. Identification of peptide bands was performed by subsequent matrix-assisted laser desorption ionization time-of-flight mass spectrometry analysis.
Evidence for the Dissociation of Extrinsic Proteins in Vivo
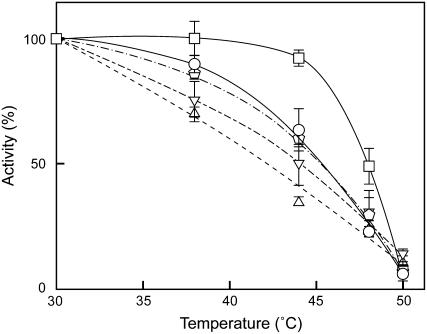
Although we found that the amount of the extrinsic proteins of PSII was decreased in the mutant, it was still obscure whether these proteins are dissociated from PSII in vivo. To address this question, heat susceptibility of the mutant cells was analyzed and compared to that of the wild-type cells. As shown in Figure 6, incubation of the wild-type cells for 20 min did not affect their photosynthetic activity up to 45°C, but the oxygen-evolving activity of dgdA mutant cells began decreasing at a lower temperature. The property of heat susceptibility found in the dgdA mutant cells was similar to that of the mutants deficient in the extrinsic proteins, such as ΔpsbO, ΔpsbV, and ΔpsbU, in which the Mn cluster is not protected properly and oxygen-evolving activity is easily inactivated by heat treatment (Nishiyama et al., 1994, 1997; Shen et al., 1995; Clarke and Eaton-Rye, 1999; Kimura et al., 2002).
Figure 6.
Comparison of heat susceptibility of photosynthetic oxygen-evolving activity. Wild-type (□), dgdA (○), ΔpsbO (▵), ΔpsbV (▿), and ΔpsbU (pentagons) cells were incubated for 20 min at the designated temperatures, and photosynthetic oxygen-evolving activity was measured at 30°C. One-hundred percent activities reflecting the activities after incubation at 30°C for 20 min were 994, 972, 840, 821, and 959 μmol O2 108 cells−1 h−1 for wild-type, dgdA, ΔpsbO, ΔpsbV, and ΔpsbU cells, respectively.
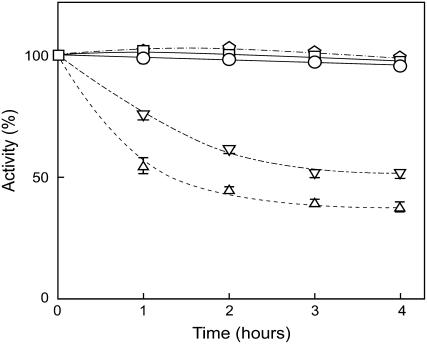
In mutant cells lacking PsbO or PsbV, Mn ions in the Mn cluster are reduced and released from PSII under dark conditions causing oxygen-evolving activity of the mutants to decrease under dark conditions (Burnap et al., 1996; Shen et al., 1998). Thus, a dark-induced decrease in oxygen-evolving activity is a useful marker to monitor dissociation of extrinsic proteins from PSII in vivo. Figure 7 shows changes in oxygen-evolving activity of wild-type, ΔpsbO, ΔpsbV, ΔpsbU, and dgdA mutant cells under dark conditions. Consistent with previous reports, the oxygen-evolving activity of ΔpsbO and ΔpsbV mutant cells decreased during dark incubation, whereas the dgdA mutant cells as well as the wild-type and ΔpsbU mutant cells sustained oxygen-evolving activity, suggesting that the binding of PsbO and PsbV is not affected in the dgdA mutant cells but PsbU is dissociated from PSII even in vivo.
Figure 7.
Changes in photosynthetic oxygen-evolving activity during dark incubation. Wild-type (□), dgdA (○), ΔpsbO (▵), ΔpsbV (▿), and ΔpsbU (pentagons) cells were incubated under dark conditions for the designated time, and photosynthetic oxygen-evolving activity was measured at 30°C. One-hundred percent activities reflecting the initial activities just prior to moving to the dark conditions were 1,130, 903, 810, 851, and 913 μmol O2 108 cells−1 h−1 for wild-type, dgdA, ΔpsbO, ΔpsbV, and ΔpsbU cells, respectively.
DISCUSSION
In this study, we identified the dgdA gene (ycf82 or slr1508) involved in the biosynthesis of DGDG and successfully constructed a DGDG-deficient mutant by disruption of the dgdA gene. Growth of the resultant mutant cells was almost the same as that of wild-type cells in normal BG-11 medium, indicating that DGDG is not an essential component for the growth of Synechocystis (Fig. 3). This result is consistent with the finding that the DGDG-deficient mutant of Arabidopsis can grow photoautotrophically under standard growth conditions (Kelly et al., 2003). The dgdA mutant cells presented slightly lower photosynthetic oxygen-evolving activity than wild-type cells; this activity is dependent on electron transport from water to CO2 (Table III). Low-temperature fluorescence spectra of intact cells excited at 590 nm indicated that fluorescence from PSII increased significantly in the dgdA mutant cells compared to the wild-type cells (Fig. 4B). Because the fluorescence from PSII excited at 440 nm was not significantly different between wild-type and dgdA mutant cells (Fig. 4A), the increased PSII fluorescence in the dgdA mutant cells excited at 590 nm could not be explained by the change in the ratio of PSI to PSII. It is likely that efficiency of energy transfer from phycobilisomes to the reaction center of PSII and from antenna Chl to the reaction center of PSII decreased in the dgdA mutant cells. Veerman et al. (2005) previously reported similar changes in fluorescence emission spectra in ΔpsbU mutant cells, and we found increased fluorescence from PSII in ΔpsbO and ΔpsbV mutant cells (data not shown). These findings suggest that the lack of DGDG affects energy transfer in PSII because of the dissociation of extrinsic proteins.
We demonstrated that the extrinsic proteins required for the stabilization of the Mn cluster are dissociated from PSII of dgdA mutant cells (Fig. 5). This could be a reason for the impairment and instability of PSII in the dgdA mutant cells. It was also found that photosynthetic oxygen-evolving activity of dgdA mutant cells was decreased by heat treatment (Fig. 6), which is similar to our previous finding that inactivation of oxygen evolution was induced in PG-depleted pgsA mutant cells (Sakurai et al., 2007). In the PG-depleted pgsA mutant cells, inactivation of oxygen evolution was also observed during dark incubation as in the ΔpsbO and ΔpsbV mutant cells (Sakurai et al., 2007). However, in the dgdA mutant cells, no decrease in activity was found under dark conditions as in the ΔpsbU mutant cells (Fig. 7). These results indicate that the effect of lack of DGDG on the binding of extrinsic proteins is different from that of PG observed in the pgsA mutant cells. In the dgdA mutant, changes in photosynthetic oxygen-evolving activity during heat treatment and dark incubation were similar to those found in ΔpsbU mutant cells. These findings suggest that DGDG is required for the binding of PsbU subunits. It is very likely that PsbO and PsbV subunits were dissociated during the preparation of PSII, but they are not dissociated in vivo. This is consistent with the finding that PsbO and PsbV were dissociated from PSII prepared from the ΔpsbU mutant (Inoue-Kashino et al., 2005). DGDG is required for the binding for PsbU, so it stabilizes the binding of PsbO and PsbV subunits to PSII. In contrast to the dissociation of extrinsic proteins, accumulation of Sll1398 was found in PSII monomer prepared from dgdA mutant cells (Fig. 5). Interestingly, accumulation of Sll1398 was also observed in PSII of a PG-depleted pgsA mutant (Sakurai et al., 2007). Although subsequent analyses are required to investigate the function of Sll1398, it is assumed that Sll1398 has an important function in the assembly of subunits in PSII.
Recently, Steffen et al. (2005) conducted spectroscopic analyses of a DGDG-deficient mutant of Arabidopsis using a flash laser fluorometer. They showed that the DGDG-deficient mutant exhibited Chl fluorescence that differed from the wild type and could be caused by the defect on the donor side of PSII. Their findings are consistent with this study. Loll et al. (2005) reported the crystal structure of PSII from T. elongatus and revealed that four DGDG molecules per monomer are present in the PSII. Polar head groups of all of DGDG molecules face the luminal side of the PSII complex. Three of the DGDG molecules (DGDG2, DGDG5, and DGDG6) are located between CP43 and D1 subunits, whereas the other DGDG molecule (DGDG8) is located between CP47 and D2 subunits. In the binding sites of DGDG molecules, the following subunits were identified as neighboring subunits: CP43, D1, and PsbJ subunits for DGDG5 and DGDG6; CP43 and D1 subunits for DGDG2; and CP47, D2, and PsbH subunits for DGDG8. The polar head groups and acyl groups in the DGDG molecules interact with the amino acid residues of the subunits. However, a direct interaction of the DGDG molecules with the extrinsic proteins was not found in the crystal structure. It is likely that the lack of DGDG molecules induces a conformational change in the luminal loops of CP43 and CP47 subunits that interact with the extrinsic proteins and cause the dissociation of extrinsic proteins.
In this study, we focused on the function of DGDG on the donor side of PSII. However, it should be noted that DGDG plays important roles not only on the donor side but also on the acceptor side of PSII. As presented in Table III, oxygen-evolving activity of the mutant cells was somewhat inhibited by addition of DCBQ, an artificial quinone used as an electron acceptor of PSII, whereas the activity of wild-type cells was increased by the addition of DCBQ. This result indicates that physical properties in the quinone-exchange cavity of PSII might be changed by the lack of DGDG. A similar phenotype was also found in the PG-depleted pgsA mutant, although greater inhibition was induced in the case of the pgsA mutant (Hagio et al., 2000). Several constituents, including hydrophobic amino acids, phytol chains of pigments, and acyl chains of lipids, contribute to form a hydrophobic environment in the quinone-exchange cavity (Loll et al., 2007). Because acyl groups of DGDG5 and DGDG6 molecules in PSII are located on the surface of the cavity, structural changes in the cavity can be induced by a lack of DGDG molecules.
As shown in Table I, the content of DGDG in the dgdA mutant cells was under the detection limit; however, total amounts of lipids in the thylakoid membranes and the PSII of dgdA mutant cells were similar to those of the wild-type cells (Table VI). The contents of SQDG and PG were not significantly changed, and only the content of MGDG increased in the mutant. These results suggest that DGDG molecules were substituted with MGDG in the PSII of the mutant cells. However, the defects found in the mutant PSII indicate that DGDG has a specific role, which could not be compensated with MGDG. It could be that hydroxyl groups in the second (outer) Gal moiety interact with amino acid residues of some subunits in PSII. Recently, Hölzl et al. (2006) presented interesting results that a growth defect in the dgd1 mutant of Arabidopsis could be complemented by the production of glucosylgalactosyldiacylglycerol; however, the function of PSII was not compensated with this lipid. Their findings indicate an important role of the second Gal moiety of DGDG molecules in the function of PSII.
In conclusion, we identified a ycf82 homolog, dgdA (slr1508), which is involved in the biosynthesis of DGDG in Synechocystis. Analyses of the dgdA mutant, in which the content of DGDG was below the detection limit, demonstrated that DGDG is not essential for the growth of Synechocystis. However, we found that efficiency of energy transfer in PSII is decreased in the mutant cells and the oxygen-evolving complex of the mutant PSII is unstable because of the dissociation of extrinsic proteins. These results demonstrate that DGDG plays an important role in stabilization of the oxygen-evolving complex through the binding of extrinsic proteins.
MATERIALS AND METHODS
Phylogenetic Profiling and Phylogenetic Analysis
Supervised phylogenetic profiling was performed using Gclust and associated software (Sato et al., 2005; Sato, 2006). The ycf82 family was detected as a cluster shared by nine cyanobacteria and a red alga using the CZ20x0 dataset in the Gclust server (http://gclust.c.u-tokyo.ac.jp/). In a different version of the Gclust database, clustering was done with different parameters, and a group of paralogs was added as in the dataset CZ16Y (Fig. 1). A phylogenetic tree was constructed with this cluster using the paralogs as outgroups. Construction of amino acid alignment, file conversion, and phylogenetic analysis by Bayesian Inference were performed as described by Terasawa et al. (2007). MrBayes version 3.1 software (Ronquist and Huelsenbeck, 2003) was used for the Bayesian Inference analysis, with ngen = 40,000; samplefreq = 100; and aamodelpr = mixed.
Organisms and Growth Conditions
CP47-His transformants expressing a CP47 subunit of the PSII complex with six His residues as a tag at the C terminus were constructed with the wild type and a dgdA mutant of Synechocystis sp. PCC6803 as described by Sakurai et al. (2006). Disruption of the dgdA gene (slr1508) in the Glc-tolerant strain (Kazusa strain) was performed as described below, and then the mutant allele was brought into the CP47-His strain. CP47-His transformants of the wild type and the dgdA mutant are simply referred to as the wild type and the dgdA mutant in this study. The transformants were grown photoautotrophically at 30°C in BG-11 medium with 20 μg mL−1 spectinomycin under the continuous white light of fluorescent lamps. In the case of the dgdA mutant, 20 μg mL−1 kanamycin was added to the growth medium. Growth of the cells was monitored by determination of the optical density at 730 nm.
Construction of the dgdA Mutant
The entire coding region of the slr1508 gene was removed by homologous recombination. The upstream (49up) and the downstream (49dn) regions of the slr1508 were amplified by PCR using the following two sets of primers: TTGCTTTCGGGCAGTGGAGGAATG (49upF; primer 1 in Fig. 2) and ACATCAGAGATTTTGAGACACAACGTGGCTCATGCAGATAACCAGACCGTGAACGAA (49upR-4KLf), and CACCAACTGGTCCACCTACAACAAAGCTCTCCAAGCCCATTCTACGAACCTAAGT (49dnF-4KRf) and ATCAATGGTGTTAAAGCCCGCTGTCCG (49dnR; primer 2 in Fig. 2). The underlined parts are complementary to the ends (4KLf and 4KRr, see below) of the Km cassette from pUC4K, which was also amplified by PCR in two overlapping parts, 4KL and 4KR, using the following two sets of primers: AGCCACGTTGTGTCTCAAAATCTCTGATGT (4KLf) and GAGAAATCACCATGAGTGACGACTGAATCC (4KLr), and AAGCTTTTGCCATTCTCACCGGATTCAGTC (4KRf) and AGAGCTTTGTTGTAGGTGGACCAGTTGGTG (4KRr; primer 4 in Fig. 2). We thus used four fragments, 49up, 4KL, 4KR, and 49dn, which were then linked by successive PCR to finally obtain a disruption cassette. This is a rapid and high-throughput method that was used to generate 40 mutants of Synechocystis in a previous study (Sato et al., 2005). The DNA was mixed with the wild-type cells, and mutants were then selected by screening with 30 μg mL−1 kanamycin. Several resistant colonies were isolated and analyzed by PCR using the primers 49upF and 49dnR. The internal primer TTGCACCATATTGGCCGCAAAGGTG (49iR; primer 3 in Fig. 2) was also used to check the remaining wild-type copy of the slr1508 gene. Complete segregation was confirmed by PCR.
Analyses of Oxygen-Evolving Activity and Chl Fluorescence Parameters
Photosynthetic activity was monitored by a Clark-type oxygen electrode following Gombos et al. (1991). Oxygen-evolving activity of PSII was measured as described previously (Sakurai et al., 2006). The DCIP photoreduction activity of thylakoid membranes was measured in a reaction buffer (50 mm MES-NaOH, pH 6.0, 10 mm NaCl, 20 mm CaCl2, 1 m Suc) containing 50 μm DCIP. Where indicated, 1 mm DPC was added to the reaction medium. The samples were illuminated with white light filtered through thermocutting and red filters, and photoreduction of DCIP was measured by the change in absorption at 600 nm (UV-3000, Shimadzu) at room temperature. Parameters of Chl fluorescence were measured at room temperature with a fluorescence monitoring system (FMS1, Hansatech Instruments) using a cell suspension adjusted at 5 μg mL−1 Chl in BG-11 medium (Sakurai et al., 2007). Using the cells suspended in the BG-11 medium at 10 μg mL−1 Chl, 77-K fluorescence emission spectra were recorded with a spectrofluorophotometer (RF-5300PC, Shimadzu). Chl concentrations were determined by the method of Arnon et al. (1974).
Purification of Thylakoid Membranes and PSII Complexes
Thylakoid membranes were prepared from 10 L of cell culture with 5 mg L−1 Chl concentration according to Kashino et al. (2002). PSII containing CP47-His was purified using Ni-affinity column chromatography (Ni-NTA column; Qiagen). Monomers and dimers of PSII were separated by ultracentrifugation with a 5% to 30% linear glycerol density gradient as described previously (Sakurai et al., 2007).
Lipid Analysis
Lipids were extracted from thylakoid membranes and PSII by the method of Bligh and Dyer (1959). Lipid classes were separated on thin-layer chromatography and were quantified by gas chromatography according to Wada and Murata (1989). The numbers of lipid molecules in PSII were calculated based on the numbers of Chl molecules obtained in our previous study (Sakurai et al., 2006).
Protein Analysis
Polypeptide compositions of PSII were analyzed by SDS-PAGE according to the method described by Kashino et al. (2001), with a gradient gel of 18% to 24% polyacrylamide containing 6 m urea. Polypeptides separated on the gels were visualized with Coomassie Brilliant Blue R250 (Fulka). Identification of protein subunits was performed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry as described previously (Sakurai et al., 2006).
This work was supported by Grants-in-Aid for Scientific Research (no. 18770029 to N.M., nos. 18017005 and 16GS0304 to N.S.) and a Research Fellowship for Young Scientists (no. 11578 to I.S.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Naoki Sato (naokisat@bio.c.u-tokyo.ac.jp).
Some figures in this article are displayed in color online but in black and white in the print edition.
Open Access articles can be viewed online without a subscription.
References
- Aoki M, Sato N, Meguro A, Tsuzuki M (2004) Differing involvement of sulfoquinovosyl diacylglycerol in photosystem II in two species of unicellular cyanobacteria. Eur J Biochem 271 685–693 [DOI] [PubMed] [Google Scholar]
- Arnon DI, McSwain BD, Tsujimoto HY, Wada K (1974) Photochemical activity and components of membrane preparations from blue-green algae. I. Coexistence of two photosystems in relation to chlorophyll a and removal of phycocyanin. Biochim Biophys Acta 357 231–245 [DOI] [PubMed] [Google Scholar]
- Benning C, Ohta H (2005) Three enzyme systems for galactoglycerolipid biosynthesis are coordinately regulated in plants. J Biol Chem 280 2397–2400 [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37 911–917 [DOI] [PubMed] [Google Scholar]
- Burnap RL, Qian M, Pierce C (1996) The manganese-stabilizing protein of photosystem II modifies the in vivo deactivation and photoactivation kinetics of the H2O oxidation complex in Synechocystis sp. PCC6803. Biochemistry 35 874–882 [DOI] [PubMed] [Google Scholar]
- Clarke SM, Eaton-Rye JJ (1999) Mutation of Phe-363 in the photosystem II protein CP47 impairs photoautotrophic growth, alters chloride requirement, and prevents photosynthesis in the absence of either PSII-O or PSII-V in Synechocystis sp. PCC 6803. Biochemistry 38 2707–2715 [DOI] [PubMed] [Google Scholar]
- Dörmann P, Hoffmann-Benning S, Balbo I, Benning C (1995) Isolation and characterization of an Arabidopsis mutant deficient in the thylakoid lipid digalactosyl diacylglycerol. Plant Cell 7 1801–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gombos Z, Várkonyi Z, Hagio M, Iwaki M, Kovács L, Masamoto K, Itoh S, Wada H (2002) Phosphatidylglycerol requirement for the function of electron acceptor plastoquinone QB in the photosystem II reaction center. Biochemistry 41 3796–3802 [DOI] [PubMed] [Google Scholar]
- Gombos Z, Wada H, Murata N (1991) Direct evaluation of effects of fatty-acid unsaturation on the thermal properties of photosynthetic activities, as studied by mutation and transformation of Synechocystis PCC6803. Plant Cell Physiol 32 205–211 [Google Scholar]
- Güler S, Seeliger A, Härtel H, Renger G, Benning C (1996) A null mutant of Synechococcus sp. PCC7942 deficient in the sulfolipid sulfoquinovosyl diacylglycerol. J Biol Chem 271 7501–7507 [DOI] [PubMed] [Google Scholar]
- Hagio M, Gombos Z, Várkonyi Z, Masamoto K, Sato N, Tsuzuki M, Wada H (2000) Direct evidence for requirement of phosphatidylglycerol in photosystem II of photosynthesis. Plant Physiol 124 795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härtel H, Lokstein H, Dörmann P, Grimm B, Benning C (1997) Changes in the composition of the photosynthetic apparatus in the galactolipid-deficient dgd1 mutant of Arabidopsis thaliana. Plant Physiol 115 1175–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdman M, Janvier M, Ripkka R, Stanier RY (1979) Genome size of cyanobacteria. J Gen Microbiol 111 73–85 [Google Scholar]
- Hölzl G, Witt S, Kelly AA, Zähringer U, Warnecke D, Dörmann P, Heinz E (2006) Functional differences between galactolipids and glucolipids revealed in photosynthesis of higher plants. Proc Natl Acad Sci USA 103 7512–7517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue-Kashino N, Kashino Y, Satoh K, Terashima I, Pakrasi HB (2005) PsbU provides a stable architecture for the oxygen-evolving system in cyanobacterial photosystem II. Biochemistry 44 12214–12228 [DOI] [PubMed] [Google Scholar]
- Jones MR (2007) Lipids in photosynthetic reaction centers: structural roles and functional holes. Prog Lipid Res 46 56–87 [DOI] [PubMed] [Google Scholar]
- Kashino Y, Koike H, Satoh K (2001) An improved sodium dodecyl sulfate-polyacrylamide gel electrophoresis system for the analysis of membrane protein complexes. Electrophoresis 22 1004–1007 [DOI] [PubMed] [Google Scholar]
- Kashino Y, Lauber WM, Carroll JA, Wang Q, Whitmarsh WJ, Satoh K, Pakrasi HB (2002) Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium Synechocystis sp. PCC 6803 reveals the presence of novel polypeptides. Biochemistry 41 8004–8012 [DOI] [PubMed] [Google Scholar]
- Kelly AA, Froehlich JE, Dörmann P (2003) Disruption of the two digalactosyldiacylglycerol synthase genes DGD1 and DGD2 in Arabidopsis reveals the existence of an additional enzyme of galactolipid synthesis. Plant Cell 15 2694–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A, Eaton-Rye JJ, Morita EH, Nishiyama Y, Hayashi H (2002) Protection of the oxygen-evolving machinery by the extrinsic proteins of photosystem II is essential for development of cellular thermotolerance in Synechocystis sp. PCC 6803. Plant Cell Physiol 43 932–938 [DOI] [PubMed] [Google Scholar]
- Loll B, Kern J, Saenger W, Zouni A, Biesiadka J (2005) Towards complete cofactor arrangement in the 3.0 Å resolution structure of photosystem II. Nature 438 1040–1044 [DOI] [PubMed] [Google Scholar]
- Loll B, Kern J, Saenger W, Zouni A, Biesiadka J (2007) Lipids in photosystem II: interactions with protein and cofactors. Biochim Biophys Acta 1767 509–519 [DOI] [PubMed] [Google Scholar]
- Malkin R, Niyogi K (2000) Photosynthesis. In BB Buchanan, W Gruissem, RL Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 568–628
- Minoda A, Sonoike K, Okada K, Sato N, Tsuzuki M (2003) Decrease in the efficiency in the electron donation to tyrosine Z of photosystem II in an SQDG-deficient mutant of Chlamydomonas. FEBS Lett 553 109–112 [DOI] [PubMed] [Google Scholar]
- Nakatani HY, Ke B, Dolan E, Arntzen CJ (1984) Identity of the photosystem II reaction center polypeptide. Biochim Biophys Acta 765 347–352 [Google Scholar]
- Nelson N, Ben-Shem A (2004) The complex architecture of oxygenic photosynthesis. Nat Rev Mol Cell Biol 5 971–982 [DOI] [PubMed] [Google Scholar]
- Nishiyama Y, Hayashi H, Watanabe T, Murata N (1994) Photosynthetic oxygen evolution is stabilized by cytochrome c550 against heat inactivation in Synechococcus sp. PCC 7002. Plant Physiol 105 1313–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama Y, Los DA, Hayashi H, Murata N (1997) Thermal protection of the oxygen-evolving machinery by PsbU, an extrinsic protein of photosystem II, in Synechococcus species PCC 7002. Plant Physiol 115 1473–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19 1572–1574 [DOI] [PubMed] [Google Scholar]
- Roose JL, Wegener KM, Pakrasi HB (2007) The extrinsic proteins of photosystem II. Photosynth Res 92 369–387 [DOI] [PubMed] [Google Scholar]
- Sakurai I, Hagio M, Gombos Z, Tyystjärvi T, Paakkarinen V, Aro EM, Wada H (2003) Requirement of phosphatidylglycerol for maintenance of photosynthetic machinery. Plant Physiol 133 1376–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai I, Mizusawa N, Ohashi S, Kobayashi M, Wada H (2007) Effects of the lack of phosphatidylglycerol on the donor side of photosystem II. Plant Physiol 144 1336–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai I, Shen JR, Leng J, Ohashi S, Kobayashi M, Wada H (2006) Lipids in oxygen-evolving photosystem II complexes of cyanobacteria and higher plants. J Biochem (Tokyo) 140 201–209 [DOI] [PubMed] [Google Scholar]
- Sato N (2006) Gclust Server: phylogenetic profiling with pre-defined organism sets. Genome Inform 17 P156 [Google Scholar]
- Sato N, Hagio M, Wada H, Tsuzuki M (2000) Requirement of phosphatidylglycerol for photosynthetic function in thylakoid membranes. Proc Natl Acad Sci USA 97 10655–10660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Ishikawa M, Fujiwara M, Sonoike K (2005) Mass identification of chloroplast proteins of endosymbiont origin by phylogenetic profiling based on organism-optimized homologous protein groups. Genome Inform 16 56–68 [PubMed] [Google Scholar]
- Sato N, Moriyama T (2007) Genomic and biochemical analysis of lipid biosynthesis in the unicellular rhodophyte Cyanidioschyzon merolae: lack of plastidic desaturation pathway results in mixed pathway of galactolipid synthesis. Eukaryot Cell 6 1006–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Tsuzuki M, Matsuda Y, Ehara T, Osafune T, Kawaguchi A (1995) Isolation and characterization of mutants affected in lipid metabolism of Chlamydomonas reinhardtii. Eur J Biochem 230 987–993 [DOI] [PubMed] [Google Scholar]
- Shen JR, Qian M, Inoue Y, Burnap RL (1998) Functional characterization of Synechocystis sp. PCC 6803 ΔpsbU and ΔpsbV mutants reveals important roles of cytochrome c-550 in cyanobacterial oxygen evolution. Biochemistry 37 1551–1558 [DOI] [PubMed] [Google Scholar]
- Shen JR, Vermaas W, Inoue Y (1995) The role of cytochrome c-550 as studied through reverse genetics and mutant characterization in Synechocystis sp. PCC 6803. J Biol Chem 270 6901–6907 [DOI] [PubMed] [Google Scholar]
- Somerville C, Browse J, Jaworski JG, Ohlrogge JB (2000) Lipids. In BB Buchanan, W Gruissem, RL Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 456–527
- Steffen R, Kelly AA, Huyer J, Dörmann P, Renger G (2005) Investigations on the reaction pattern of photosystem II in leaves from Arabidopsis thaliana wild type plants and mutants with genetically modified lipid content. Biochemistry 44 3134–3142 [DOI] [PubMed] [Google Scholar]
- Terasawa K, Odahara M, Kabeya Y, Kikugawa T, Sekine Y, Fujiwara M, Sato N (2007) The mitochondrial genome of the moss Physcomitrella patens sheds new light on mitochondrial evolution in land plants. Mol Biol Evol 24 699–709 [DOI] [PubMed] [Google Scholar]
- Veerman J, Bentley FK, Eaton-Rye JJ, Mullineaux CW, Vasil'ev S, Bruce D (2005) The PsbU subunit of photosystem II stabilizes energy transfer and primary photochemistry in the phycobilisome-photosystem II assembly of Synechocystis sp. PCC 6803. Biochemistry 44 16939–16948 [DOI] [PubMed] [Google Scholar]
- Vermaas WFJ, Williams JGK, Rutherford AW, Mathis P, Arntzen CJ (1986) Genetically engineered mutant of the cyanobacterium Synechocystis 6803 lacks the photosystem II chlorophyll-binding protein CP-47. Proc Natl Acad Sci USA 83 9474–9477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H, Murata N (1989) Synechocystis PCC6803 mutants defective in desaturation of fatty acids. Plant Cell Physiol 30 971–978 [Google Scholar]
- Yu B, Xu C, Benning C (2002) Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limiting growth. Proc Natl Acad Sci USA 99 5732–5737 [DOI] [PMC free article] [PubMed] [Google Scholar]