Abstract
Using the rice (Oryza sativa) sp. japonica genome annotation, along with genomic sequence and clustered transcript assemblies from 184 species in the plant kingdom, we have identified a set of 861 rice genes that are evolutionarily conserved among six diverse species within the Poaceae yet lack significant sequence similarity with plant species outside the Poaceae. This set of evolutionarily conserved and lineage-specific rice genes is termed conserved Poaceae-specific genes (CPSGs) to reflect the presence of significant sequence similarity across three separate Poaceae subfamilies. The vast majority of rice CPSGs (86.6%) encode proteins with no putative function or functionally characterized protein domain. For the remaining CPSGs, 8.8% encode an F-box domain-containing protein and 4.5% encode a protein with a putative function. On average, the CPSGs have fewer exons, shorter total gene length, and elevated GC content when compared with genes annotated as either transposable elements (TEs) or those genes having significant sequence similarity in a species outside the Poaceae. Multiple sequence alignments of the CPSGs with sequences from other Poaceae species show conservation across a putative domain, a novel domain, or the entire coding length of the protein. At the genome level, syntenic alignments between sorghum (Sorghum bicolor) and 103 of the 861 rice CPSGs (12.0%) could be made, demonstrating an additional level of conservation for this set of genes within the Poaceae. The extensive sequence similarity in evolutionarily distinct species within the Poaceae family and an additional screen for TE-related structural characteristics and sequence discounts these CPSGs as being misannotated TEs. Collectively, these data confirm that we have identified a specific set of genes that are highly conserved within, as well as specific to, the Poaceae.
Comparative analysis of genomes is a robust strategy to identify evolutionarily conserved DNA sequences across a range of species (Eddy, 2005). Commonly, these methods entail comparative evaluation of either translated amino acid or nucleotide sequences to identify either structurally conserved genes or domains across broad expanses of evolutionary time (Thomas et al., 2003; Margulies et al., 2005). The core principle for conserved sequence identification is that selection has constrained variation of the nucleotides in functionally important sequences relative to those sequences that are presumed to be nonfunctional (Boffelli et al., 2004a; Hardison, 2004). Interspecies comparisons are oriented toward identifying genes that are germane to, as well as evolutionarily conserved within, a taxonomically related group of species (Kellis et al., 2003). One central component, which is inherent to interspecies comparative strategies, is that the closer these species are in their taxonomic rank generally the higher the degree of conservation; this fusion of genomics and evolution has been termed phylogenomics (Eisen and Fraser, 2003; Hardison, 2004; Eddy, 2005). Juxtaposed with the identification of broadly conserved genes is the identification of genes that are exclusive to a group of related species (e.g. lineage-specific genes) or even within a species (Boffelli et al., 2004a, 2004b). Identification and characterization of lineage-specific sets of genes has been successful across a range of eukaryotic species (Domazet-Loso and Tautz, 2003; Boffelli et al., 2004b; Graham et al., 2004; Mitreva et al., 2005), and this strategy is generally reliant upon having a completed genomic sequence and extensive collections of transcribed sequences (Allen, 2002; Margulies et al., 2005).
Within the plant kingdom, extensive analyses of genomic and cDNA sequences have revealed core sets of conserved genes within the Angiospermophyta (angiosperms; Rice Full-Length cDNA Consortium, 2003; Schoof and Karlowski, 2003; Choi et al., 2004; International Rice Genome Sequencing Project, 2005; Tuskan et al., 2006). Meanwhile, comprehensive identification of lineage-specific genes in plant families is ongoing. A preliminary comparative analysis using the finished Arabidopsis (Arabidopsis thaliana; belonging to the Brassicaceae family) genomic sequence and its annotation with assembled transcripts from species in the Fabaceae (legume family) and Solanaceae revealed a small number of family-specific genes (Allen, 2002). Subsequently, a more thorough interspecies sequence comparison was performed to identify family-specific sequences in the Fabaceae using the finished Arabidopsis genomic sequence, the partially completed rice (Oryza sativa sp. japonica) genome, and clustered Fabaceae ESTs from three species (Graham et al., 2004). Among these legume-specific sequences, three gene families were reported: F-box proteins, Cys-rich proteins, and Pro-rich proteins (Graham et al., 2004). A comparative strategy, using clustered ESTs from six solanaceous species, found that between 16% and 19% of the clustered ESTs are specific to the species sampled (Rensink et al., 2005). Systematic analysis using the Arabidopsis and rice genomic annotations in conjunction with clustered ESTs from 30 plant species identified 7,882 rice proteins that lacked significant homology (SH) to any other plant sequence, and these were defined as orphan or species-specific proteins (Vandepoele and Van de Peer, 2005). In a recent analysis of the annotated rice genome, a set of rice genes (7,669) for which no sequence similarity in 184 other species from the plant kingdom was identified, suggesting these genes may be species specific and evolved after speciation or that they are potential artifacts of the annotation process (Zhu and Buell, 2007).
Using these previous comparative analyses as a guide, our analysis has incorporated the finished rice genome sequence and its annotation in combination with the genomic sequence and EST resources present for 184 evolutionarily diverse species in the plant kingdom to define and characterize a set of genes conserved within, as well as specific to, the Poaceae. This set of 861 rice genes has been termed conserved Poaceae-specific genes (CPSGs). In addition to their presence in rice, which is in the Ehrhartoideae subfamily (BEP clade) of the Poaceae, similar sequences are present in at least four other species, which are classified into two Poaceae subfamilies, namely, Panicoideae (in the PACCAD clade) and Pooideae (BEP clade; Grass Phylogeny Working Group, 2000; Kellogg, 2001). The vast majority of CPSGs encode proteins that lack similarity to genes with known function or lack a characterized protein-encoded domain; these genes are functionally annotated as either hypothetical (based solely upon ab initio gene prediction) or expressed (i.e. hypothetical genes supported by expression; Yuan et al., 2005; Ouyang et al., 2007; http://rice.tigr.org). It is notable that these rice hypothetical and expressed genes do possess significant sequence similarity across a range of evolutionarily distinct Poaceae species. Further, this broad evolutionary conservation across the Poaceae indicates that the CPSGs are not artifacts of annotation or unclassified transposable elements (TEs; Bennetzen et al., 2004; Kellogg and Bennetzen, 2004); rather, they represent a bona fide set of lineage-specific genes and largely lack any known function.
RESULTS
Sequences Used in This Analysis
Pair-wise sequence comparisons with the 42,653 The Institute for Genomic Research (TIGR) Version 4 non-TE rice genes were performed with plant genomic sequences and TIGR Version 1 plant transcript assemblies (TAs) using TBLASTN (Altschul et al., 1997; Childs et al., 2007; Ouyang et al., 2007; http://plantta.tigr.org). An E value of 10−5 was defined as the minimal cutoff for significant sequence similarity at the translated protein level. TBLASTN was performed with the rice non-TE genes separately against five genomic sequences: (1) a finished genomic sequence for Arabidopsis (Arabidopsis Genome Initiative, 2000); (2) bacterial artificial chromosome (BAC)-based sequence assemblies and annotation for a model species in the Fabaceae, Medicago (Medicago truncatula; Cannon et al., 2005; Town, 2006); (3) hi-Cot and methylation filtration genomic assemblies for maize (Zea mays; AZMs; Palmer et al., 2003; Whitelaw et al., 2003; Yuan et al., 2003; Chan et al., 2006); (4) methylation filtration genomic assemblies for sorghum (Sorghum bicolor; ASBs; Bedell et al., 2005; ftp://ftp.tigr.org/pub/data/MAIZE/Sorghum_assembly/ASB.gz); and (5) whole-genome shotgun assemblies for the model species in the Salicaceae, poplar (Populus trichocarpa; Tuskan et al., 2006). The TAs were constructed from 185 plant species (excluding rice in this analysis, hence 184 plant species) using publicly available ESTs and full-length cDNA collections (Childs et al., 2007; http://plantta.tigr.org). These TAs exclude all virtual transcripts, which are derived from the annotation of genomic sequence.
Identification of CPSGs
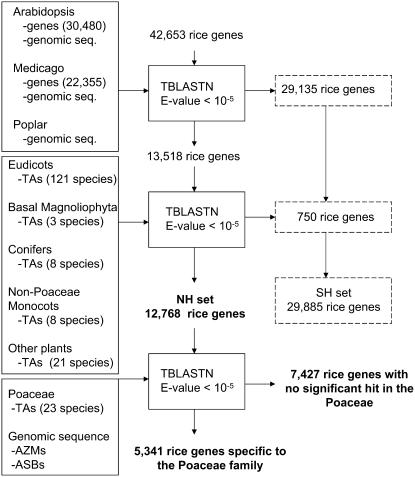
Using the TBLASTN results of the rice non-TE loci with the various genomic sequences and TAs, a filtering strategy was employed to identify a core set of rice genes with highly similar sequences within the Poaceae that lack similarity to sequences from plant families outside the Poaceae. Starting with a total of 42,653 non-TE rice genes, any significant TBLASTN hit with a sequence (either genomic or TA-based) from a species outside the Poaceae was flagged and removed from further analysis. Figure 1 depicts a schematic for the filtering strategy employed to identify a set of candidate rice genes that may be Poaceae specific. From this strategy, a total of 29,135 rice non-TE loci were identified as having a TBLASTN hit with an Arabidopsis, Medicago, or poplar genomic sequence and/or their annotated genes. Extending this search by using the phylogenetically clustered TAs (except those TAs from species in the Poaceae) identified an additional 750 genes with similarity to non-Poaceae species. After removing the 29,885 rice genes having similarity with species outside the Poaceae (SH), 12,768 rice loci remain and are defined as the nonhomologous (NH) set.
Figure 1.
Diagram illustrating the strategy to identify the CPSG, NH, and SH gene sets. Shown is the filtering strategy employed to identify the NH and SH sets of genes from the total set of non-TE rice genes using genomic sequence, genome annotations, and TAs. The dotted boxes reflect the identification of the SH set. The TAs are clustered into six phylogenetic groupings: (1) eudicots; (2) nasal Magnoliophyta; (3) conifers; (4) non-Poaceae monocots; (5) Poaceae; and (6) other plants. The rice TAs were excluded from this analysis so the total number of TAs represents 184 plant species.
Subsequently, a combinatorial strategy was employed using the NH set to define the set of rice genes that possess significant sequence similarity (E value <10−5) (1) with sequences from one (or more) of the non-rice Poaceae TAs (Supplemental Table S1); (2) in the sorghum genomic sequence assemblies; and/or (3) in the maize genomic sequence assemblies. From these analyses, a total of 5,341 genes were identified that may be specific to, and conserved within, the Poaceae (Fig. 1).
Using this set of 5,341 rice genes, we then screened for significant sequence similarity in at least four of the five Poaceae species (simultaneously) with extensive transcript and/or genomic sequences. These five species with extensive sequence resources represent three subfamilies among two clades within the Poaceae family. Within the PACCAD clade, there are three species that are within the Panicoideae subfamily: (1) maize; (2) sugarcane (Saccharum officinarum); and (3) sorghum. Within the BEP clade, there are three species represented among two subfamilies. Rice is grouped within the Ehrhartoideae subfamily, whereas barley (Hordeum vulgare) and wheat (Triticum aestivum) are present in the Pooideae subfamily (Grass Phylogeny Working Group, 2000; Kellogg, 2001). From this comparative analysis within the Poaceae, we found 1,119 rice genes that have significant similarity with at least four of the five non-rice species simultaneously. A more rigid requirement of all five simultaneously was too stringent because the barley and sugarcane TAs have relatively lower coverage of their respective transcriptomes when compared with the more extensive genomic and/or transcript assemblies from wheat, sorghum, and maize.
Annotated TE-related rice loci were not included in this analysis. The TE annotation relies upon sequence similarity to the TIGR Plant Repeat Database and the presence of repetitive element-related Pfam domains (Ouyang and Buell, 2004; Ouyang et al., 2007). To screen for misannotated TE-related genes that might be absent or underrepresented in this repeat library, we performed a secondary and more refined screen for TE-related sequences based upon structural features and sequence homology to class 1 and class 2 TEs, including (1) the presence of terminal inverted repeats present in most class 2 DNA-mediated TEs; (2) long terminal repeats in direct orientation common to class 1 long terminal repeat-containing retrotransposons; (3) target site duplications, which are present for most class 1 and class 2 TEs; (4) Helitron elements, which are not associated with terminal inverted repeats or target site duplications, but insert into an AT dinucleotide and start with the dinucleotide TC and end with the consensus sequence CTRR (where R is either an A or a G); and (5) sequence similarity to non-long terminal repeat retrotransposons (e.g. long interspersed nuclear elements and short interspersed nuclear elements; Kumar and Bennetzen, 1999; Feschotte et al., 2002; Choi et al., 2007). From the original set of 1,119 conserved genes, a total of 258 genes were flagged as possessing structural features and/or sequence homology with either class 1 or class 2 TEs. Within this set of 258 TE-related genes, a broad collection of class 1 and class 2 TE superfamilies was represented. Among class 1, both of the non-long terminal repeat retrotransposon subclasses (i.e. long and short interspersed nuclear elements) and a variety of long terminal repeat retrotransposons were present. For class 2 TEs, elements from six superfamilies were represented (e.g. hAT, CACTA, Mutator-like, PIF/Pong-like, Tc1/mariner, and Helitrons). Using this approach, we identified a total of 861 rice genes that possess similar sequences simultaneously within three subfamilies of the Poaceae and do not possess any structural or sequence homology to characterized plant TE superfamilies. Supplemental Table S2 shows the 861 CPSGs and their matches to the five Poaceae species. A multi-FASTA file of the CPSG and the top matches to the five Poaceae species (if detected) is available in Supplemental Data S1.
Characterizing CPSGs
Functional annotation of the CPSGs revealed an enrichment of genes without a known function: 46.5% (400) are annotated as hypothetical genes and 40.2% (346) are annotated as expressed genes. For the remaining genes, 8.8% (76) are annotated as containing an F-box motif and 4.5% (39) are annotated with a known function (Table I).
Table I.
The CPSGs grouped by common functional assignment and the top 10 common functional assignments are listed in descending order of occurrence
| Functional Assignment | No. |
|---|---|
| Hypothetical protein | 400 |
| Expressed protein | 346 |
| F-box domain-containing protein | 45 |
| F-box domain-containing protein, expressed | 31 |
| CsAtPR5, putative, expressed | 4 |
| Ribosome-inactivating protein, expressed | 4 |
| Protease inhibitor/seed storage/LTP family protein, expressed | 3 |
| Pectinesterase inhibitor domain-containing protein, expressed | 3 |
| VQ motif family protein, expressed | 2 |
| Zinc finger, C2H2-type family protein, expressed | 1 |
Given that approximately 87% of the CPSGs have no known functional assignment, we compared this set to the SH set (those having significant sequence similarity to species outside the Poaceae), as well as the 13,237 rice genes annotated as TEs, to discern whether there are significant differences in their genic features. For the SH and TE sets, the average exon number is 5.2 and 4.3, respectively, whereas the CPSGs have an average exon number of 2.5 (Table II). This relative increase in exon number leads to the far larger average gene length for the SH and TE sets when compared with the CPSGs (Table II). The reduced average length of the CPSGs is consistent with previously published data (i.e. rice genes lacking significant similarity within the Arabidopsis genome [i.e. NH set] are approximately one-half the length of genes with a homolog in Arabidopsis [Yu et al., 2002]). Given that nearly one-half of the CPSGs are functionally classified as hypothetical genes and, as such, do not have updated structural annotation from ESTs or full-length cDNA evidence, this most likely contributes to the significantly decreased average length due to the exclusion of the untranslated regions as well as incomplete coding sequence structures. Additionally, the CPSG set has a longer intron length relative to the exon length, similar to the SH set. For both the CPSG and SH sets, both exon and intron GC content are consistent with previously published results for chromosome-wide annotation (Feng et al., 2002; Rice Chromosome 10 Sequencing Consortium, 2003; Rice Chromosome 3 Sequencing Consortium, 2005).
Table II.
The mean and median values for the three classes of genes
The SH set comprises rice genes with significant SH with a species outside the Poaceae. The TE set comprises all annotated TEs in rice.
| Feature | CPSG
|
SH
|
TE
|
|||
|---|---|---|---|---|---|---|
| Mean (sd) | Median | Mean (sd) | Median | Mean (sd) | Median | |
| Exons per gene | 2.5 (2.0) | 2 | 5.2 (4.8) | 4 | 4.3 (3.1) | 3 |
| Exon length | 458 (490) | 266 | 317 (426) | 159 | 559 (702) | 301 |
| Intron length | 476 (644) | 252 | 402 (639) | 163 | 333 (443) | 158 |
| Gene length | 1,819 (1,485) | 1,414 | 3,309 (2,612) | 2,744 | 3,503 (2,318) | 3,141 |
| Exon GC content | 54 (12) | 53 | 50 (11) | 46 | 50 (9) | 49 |
| Intron GC content | 38 (10) | 36 | 36 (7) | 35 | 42 (10) | 41 |
| Gene GC content | 55 (10) | 54 | 49 (10) | 46 | 49 (8) | 47 |
| CDS/ORF GC content | 61 (9) | 62 | 57 (10) | 55 | 51 (8) | 50 |
| First position GC | 62 (9) | 62 | 59 (8) | 58 | 57 (8) | 55 |
| Second position GC | 50 (9) | 49 | 46 (8) | 45 | 44 (8) | 42 |
| Third position GC | 71 (16) | 72 | 66 (19) | 63 | 54 (12) | 53 |
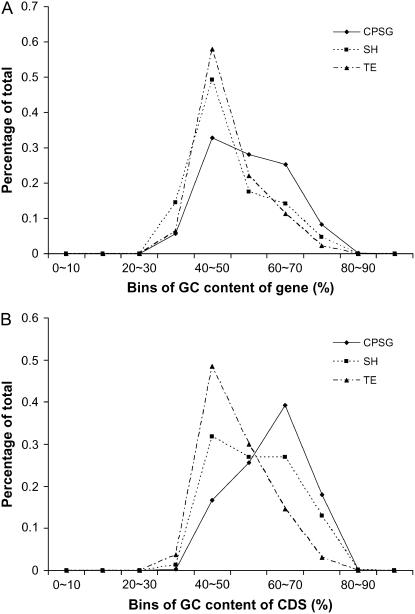
Both average whole-gene GC content (here, whole gene describes all exons and introns in the annotated gene) and GC content of the coding sequence for the CPSGs are elevated relative to the SH and TE sets. GC content of the coding sequence of rice genes has been used previously, with histograms to assess distribution of the data points that comprise the average (Carels et al., 1998; Carels and Bernardi, 2000). GC histograms for the whole gene as well as the coding sequence GC content were separately generated for the three sets of genes (CPSG, SH, and TE). These histograms use the percentage of total on the y axis and bin the genes into 10% bins on the x axis (Fig. 2). For whole-gene GC content, the CPSG sets display broader distribution with a maximum in the 40% to 50% bin relative to the TE and SH sets (Fig. 2A). This whole-gene GC distribution for the CPSGs is distinctly different from the more unimodal GC distribution for the TE and SH sets. By contrast, the histogram for GC content for the coding sequence shows that the CPSGs have a more unimodal distribution shifted toward a higher percentage of GC content in the 60% to 70% bin and the SH set has a broader distribution (Fig. 2B). This elevation of the coding sequence GC content for the CPSGs is reflected in the elevated GC mean values of the first, second, and third codon positions (Table II). These data support the hypothesis that the CPSGs, having higher GC content for the coding sequence as well as across the whole gene, are not TE related.
Figure 2.
Histograms showing the GC content for the CPSG, SH, and TE sets broken into bins of 10%. A, The histograms show the whole gene (including untranslated regions, exons, and introns) GC content. B, The histograms show the coding sequence GC content.
Distribution of CPSGs in the Rice Genome
Given that the CPSGs lack similar sequences outside the Poaceae family, we analyzed the spatial distribution of this class of genes. Supplemental Figure S1 has the CPSGs mapped across the 12 rice pseudomolecules and this distribution can be directly compared against the distribution of the SH set in this figure. The CPSGs lack an even distribution across the pseudomolecules. Further, there is clear clustering of CPSGs on particular pseudomolecules with the most apparent pseudomolecule 10. An analysis to screen for tandem duplications revealed that 214 (or 24.9%) of the CPSGs have an adjacent sequence that is highly similar. This compares to a tandem duplication rate of 17% for the SH set. A χ2 analysis indicated that this difference is significant (P < 0.00001). These data suggest that CPSGs have a higher rate of tandem duplication than those genes that have a significantly similar sequence outside the Poaceae family. A total of 21,998 proteins from rice release 4 were clustered into 3,865 paralogous protein families based on protein domain compositions that utilized both Pfam and BLASTP-based novel domains (http://rice.tigr.org/tdb/e2k1/osa1/para.family/para.method.shtml). A total of 44% (375/861) of CPSG genes and 63% (18,958/29,885) of SH genes were classified in paralogous protein families, respectively (χ2 test P < 0.00001).
CPSGs Are Overrepresented among Pack-Mutator-Like Elements
Gene and gene fragment amplification within the rice genome via TEs has been recently identified in rice and maize (Jiang et al., 2004; Lai et al., 2004; Juretic et al., 2005; Morgante et al., 2005). In rice, Mutator-like elements (MULEs) have been shown to capture whole genes or gene fragments within the boundary of this class of TEs as well as the ability to amplify these gene fragments within the rice genome via transposition (Jiang et al., 2004; Juretic et al., 2005; Diao et al., 2006). This particular class of elements has been termed alternatively Pack-MULEs or MULE-mediated transduplication (Jiang et al., 2004; Juretic et al., 2005). An analogous case of a TE capturing either genes or gene fragments has been demonstrated in maize with the Helitron TE (Lai et al., 2005; Morgante et al., 2005). Potentially, this transposon-mediated amplification of genic sequences could lead to amplification of the CPSGs in the rice genome via Pack-MULEs. To address whether CPSGs are being preferentially amplified by Pack-MULEs, the CPSG set was compared with a manually curated set of rice Pack-MULEs. In total, 76 (8.8%) CPSGs are contained within rice Pack-MULEs from the total of 861 CPSGs. When compared to the 1,324 rice genes that are contained in Pack-MULEs from the 42,653 non-TE rice genes, 3.1% of annotated genes are contained in Pack-MULEs. This result suggests that CPSGs are more likely to be contained within Pack-MULEs when compared to all non-TE annotated rice genes. Although Pack-MULEs are TEs, the genes inside Pack-MULEs are considered as non-TE genes in this study because they are derived from non-TE sequences.
To assess whether particular CPSGs have been amplified via Pack-MULEs, the CPSG-containing Pack-MULEs were further examined. Terminal inverted repeats are used to classify Pack-MULEs into subfamilies, and, from this classification scheme, the 76 CPSGs contained in Pack-MULEs were sorted by subfamily and tallied (Supplemental Table S3). Twenty of the CPSGs were captured by the Pack-MULE subfamily Os0037. This could potentially represent localized expansion of a CPSG via this Pack-MULE subfamily. A multiple sequence alignment (MSA) of the coding sequences for the 20 CPSGs within the 20 Os0037 Pack-MULEs is presented in Supplemental Figure S2. This MSA clearly shows that the genic sequences captured within the OS0037 subfamily have no significant sequence similarity to one another and that this Pack-MULE subfamily is not amplifying any particular CPSG sequence. This result is consistent with previous studies where Pack-MULEs have conserved terminal regions, but differ in their respective internal sequence. Further, the remaining 56 CPSGs are scattered in small numbers from a range of different Pack-MULE subfamilies. These data suggest that Pack-MULEs do capture CPSGs at an elevated rate relative to the genome as a whole, but these recognizable Pack-MULEs are not responsible for large-scale amplification of a particular subset of CPSGs in the rice genome.
Confirmation and Identification of Conserved Motifs in the CPSGs
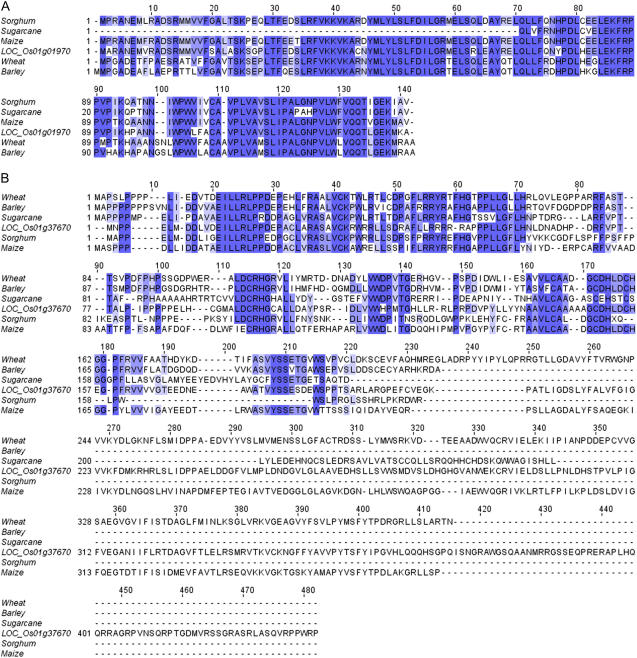
Cross-species comparisons within the Poaceae can be used to identify evolutionarily conserved domains and/or whole proteins. LOC_Os01g01970 is an expressed protein lacking significant similarity with proteins of known function or Pfam domains above the trusted cutoff. Three rice ESTs that map to this gene are present in either drought-stressed panicle or callus cDNA libraries. For these Poaceae TAs with significant similarity, the translated open reading frames (ORFs) are comparable both in length and BLASTP E values. A T-Coffee-generated MSA shows that all of these ORFs have sequence identity, which indicates that this protein structure is well conserved across the Poaceae (Fig. 3A; Supplemental Table S4). The ESTs, which constitute the TAs from maize and wheat, are derived from the developing seeds from either the ear (maize) or spike (wheat), whereas the barley ESTs are derived from a shoot cDNA library and the sorghum ESTs are derived from cDNA libraries representing multiple tissues. This striking evolutionary conservation, in conjunction with the tissue-specific expression patterns across the Poaceae, suggests that each species has altered the expression patterns while maintaining the coding sequence.
Figure 3.
MSAs of the CPSGs with translated ORFs from TAs or cDNAs from other Poaceae species. A, LOC_Os01g01970. B, LOC_Os01g37670.
LOC_Os01g37670, annotated as a protein containing an F-box motif (Pfam ID PF00646), has a total hidden Markov model (HMM) score of 30.2 (trusted cutoff is 13.2). The predicted F-box domain is positioned at the N terminus and is 50 amino acids in length, which is consistent with annotation for this domain (Pfam ID PF00646). LOC_Os01g37670 is supported by a full-length cDNA, which lacks information to indicate the precise conditions or tissues where this rice gene is expressed. The MSA for LOC_Os01g37670, along with five other Poaceae TAs/ESTs, has strong conservation across the first 60 amino acids, where the F-box domain is annotated, whereas all of the C termini are relatively divergent (Fig. 3B; Supplemental Table S4), contributing to the failure to detect similarity in other species.
In addition to LOC_Os01g37670, another 75 loci in the CPSG set (76 total) are annotated as F-box-containing proteins during whole-genome annotation (Ouyang et al., 2007) and these were rescreened with the PF00646 HMM (F-box). Of the 76 F-box-containing proteins, 72 were found to have an F-box motif above the trusted cutoff (data not shown), whereas the remaining four genes annotated as F-box domain-containing proteins were identified by significant homology to previously characterized F-box proteins. Lineage-specific enrichment of F-box domain-containing proteins has been reported previously in the Fabaceae (Graham et al., 2004). The F-box domain mediates interactions with the Skp1 and Cullin orthologs across a range of eukaryotic species (Cardozo and Pagano, 2004). Interestingly, the C-terminal substrate-binding motifs of F-box domain-containing proteins have been reported to be under positive selection (e.g. rapidly evolving) in both nematodes and Arabidopsis, whereas the N-terminal F-box domain, which mediates interaction with the Skp/Cullin complex, is evolutionarily conserved (Thomas, 2006). Evaluation of the C-terminal sequences of these 76 F-box proteins in the CPSG set was consistent with the data from Arabidopsis and nematodes; the C termini are highly divergent (data not shown). Construction of the F-box HMM PF00646 utilized a number of diverse eukaryotic species with a seed of 534 sequences and a full alignment of 3,442 sequences that, in part, may explain why this domain was not identified by the TBLASTN filtration strategy.
Synteny between Rice and Sorghum
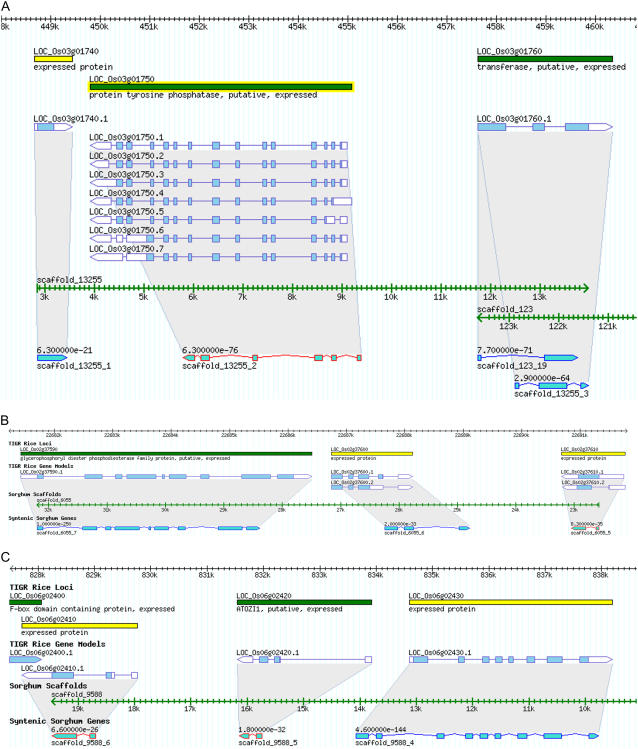
A recent draft sequence assembly for sorghum has been released (http://www.jgi.doe.gov). FGENESH trained for monocot gene structures was used for ab initio gene prediction in these sorghum genome assemblies (Salamov and Solovyev, 2000). These FGENESH gene predictions within the sorghum assemblies were then aligned with the annotated rice gene set to identify localized synteny between rice and sorghum. Syntenic regions were defined minimally as a CPSG and two flanking rice genes having significant similarity and collinear arrangement with adjacent FGENESH predictions derived from the sorghum genomic assemblies. For the CPSG set, 103 were found to possess a syntenic ortholog in sorghum.
The first example using LOC_Os03g01740, which is annotated as an expressed protein, has two additional orthologs when compared with the gene predictions in sorghum assembly 13255 (Fig. 4A). LOC_Os03g01740, which is the CPSG, has a BLASTP E value of 6.3e−21 with sorghum prediction 13255_1. The two adjacent rice genes (LOC_Os03g01750 annotated as a protein Tyr phosphatase and LOC_Os03g01760 annotated as a putative transferase) have significant similarity with sorghum FGENESH predictions (13255_2 [E value 6.3e−76] and 13255_3 [E value 2.9e−64]), respectively. Not only is the syntenic order conserved, but also the transcriptional orientation is likewise conserved (Fig. 4A). MSAs for LOC_Os03g01740 with translated ORFs from TAs in sorghum, sugarcane, maize, wheat, and barley show extensive conservation (Supplemental Fig. S3A).
Figure 4.
Synteny between regions encoding rice CPSGs and sorghum. A, LOC_Os03g01740. B, LOC_Os02g37610. C, LOC_Os06g02410.
A second example, LOC_Os02g37610, is annotated as an expressed gene and has two flanking orthologs when comparing the rice sequence with FGENESH predictions for the sorghum assembly 6055 (Fig. 4B). LOC_Os02g37610 has a BLASTP E value of 8.3e−35 with sorghum prediction 6055_5. The two rice orthologs of sorghum are annotated as a glycerophosphoryl diester phosphordiesterase family protein (LOC_Os02g37590) and an expressed protein (LOC_Os02g37600) with the respective sorghum genome assembly predictions (6055_7 and 6055_6). Not only are these orthologs highly similar (1.0e−250 and 2.0e−33, respectively) but their transcriptional orientation is also conserved. These rice and sorghum orthologs also possess strong evolutionary conservation with TAs from sorghum, maize, sugarcane, and wheat (Supplemental Fig. S3B).
A third example, LOC_Os06g02410, is annotated as an expressed gene and has two additional flanking orthologs on the sorghum assembly 9588. LOC_Os06g02410 has a BLASTP E value of 6.6e−26 when compared with its sorghum ortholog 9588_6. The two flanking orthologs for rice are annotated as ATOZI1 (LOC_OS06g02420) and an expressed protein (LOC_Os06g02430; Fig. 4C). A MSA with TAs from wheat, barley, and sugarcane demonstrates that this rice/sorghum sequence similarity is conserved broadly within the Poaceae (Supplemental Fig. S3C).
DISCUSSION
Using the finished rice genome and its annotation against the rich and extensive sequence resources that have recently become available for species in the plant kingdom, we have identified a set of genes conserved within, and specific to, the Poaceae. The use of TBLASTN searches to filter out rice genes using non-Poaceae TAs and genomic sequence presumptively act to purge domains that have broad evolutionary conservation (e.g. kinases, phosphatases, etc.). Therefore, the functional classification for the rice CPSG set is primarily either hypothetical or expressed.
Whereas prior research for genes lacking significant sequence similarity outside the Poaceae has shown them to have reduced average length and elevated GC content, it has been suggested that these genes may be artifacts of genome annotation (Cruveiller et al., 2003; Bennetzen et al., 2004; Jabbari et al., 2004; Yu et al., 2005). In contrast to these previous analyses, we have identified 861 CPSGs that lack significant similarity to sequences from species outside the Poaceae yet have similar sequences within the Poaceae. Furthermore, we have manually curated these CPSGs to remove any genes that have features of TEs. Certainly, we cannot rule out the possibility that some of the CPSGs are ancient TEs where the feature of TEs (e.g. the terminal inverted repeat of Pack-MULEs) is no longer recognizable due to mutations yet the coding region is conserved because of functional constraints. If that is the case, they should be considered as normal genes and it provides one mechanism of how those genes arose.
CPSGs have an overall reduction in total average gene length when compared to the SH set (primarily due to a reduction in total number of exons) and have a slight elevation in mean GC content. The histograms, particularly for the coding sequences for the CPSGs, are clearly skewed toward a higher total GC percentage relative to the distributions for the TE and SH sets (Fig. 2). To identify features that can be exploited for functional characterization, cross-species comparisons have been used to demonstrate extensive sequence conservation for species in differing Poaceae subfamilies (Fig. 3; Supplemental Fig. S3). Comparative analyses can be enriched by incorporating expression evidence from supporting ESTs (or their clustered TAs) to identify possible tissue-, organ-, or condition-specific expression.
Of those 115 genes that have a known functional classification among the CPSGs, more than one-half are predicted to possess an F-box domain, whereas the remaining genes comprise a sundry collection when grouped by functional assignment (Table I). The presence of F-box domain-containing proteins within this lineage-specific gene set is not surprising given that the C-terminal (substrate-binding) domains of these proteins are under strong positive selection in both plants and nematodes (Thomas, 2006). Conversely, strong N-terminal conservation is observed in the MSAs of cross-species comparisons and has been noted previously in a comparative genomics analysis involving Arabidopsis and Caenorhabditis elegans (Thomas, 2006).
The rice CPSG genes appeared and diversified during Poaceae evolution, or, alternatively, they have been lost subsequent from the divergence from the last common ancestor between the Poaceae and non-Poaceae families. Taxonomic and fossil records indicate that the grasses appeared between 55 and 70 million years ago (Jacobs et al., 1999). Within the Poaceae, a detailed phylogeny has been developed from plastid and nuclear genes as well as morphological variation of plant structures when compared to the current taxonomic assignments. The Poaceae family contains approximately 10,000 species that inhabit a wide range of environmental niches and possess unique developmental and physiological characteristics, which include: (1) spikelets that have evolved in several steps from other flowering structures in the angiosperms; (2) the adaptation of some clades for drought tolerance and dry habitats; (3) the multiple appearance of the C4 photosynthetic pathway and its attendant anatomical architecture; and (4) the unique developmental pattern of the fruit (Jacobs et al., 1999; Kellogg, 1999, 2000, 2001; Grass Phylogeny Working Group, 2000). The evolutionary changes that appeared within this family during the past 55 to 70 million years would presumptively require diversification of existing genes for novel functions to drive these alterations in morphology. Our identification of lineage-specific proteins within the Poaceae is a starting point to identify those genes that may have been involved in the highly successful adaptation and radiation of the Poaceae species across the planet.
MATERIALS AND METHODS
Identification of the CPSG Set
Datasets and methods used to identify the CPSG set in this study are similar, but not identical, to that described by Zhu and Buell (2007). For this study, the final pseudomolecule assembly for Arabidopsis (Arabidopsis thaliana) was obtained from TIGR (http://www.tigr.org/tdb/e2k1/ath1). The nucleotide coding sequences for all nuclear-encoded Arabidopsis genes were obtained from The Arabidopsis Information Resource Version 6 (http://www.arabidopsis.org/index.jsp). The Medicago (Medicago truncatula) BAC assemblies and their annotation were obtained from TIGR (http://www.tigr.org/tdb/e2k1/mta1). The poplar (Populus trichocarpa) genome assemblies were downloaded from the Joint Genome Institute (JGI; Tuskan et al., 2006; http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html). The maize (Zea mays) and sorghum (Sorghum bicolor) genome assemblies were obtained from TIGR (ftp://ftp.tigr.org/pub/data/MAIZE/Sorghum_assembly/ASB.gz; http://maize.tigr.org). All Version 1 plant TA assemblies were obtained from the plant TA resource (Childs et al., 2007; http://plantta.tigr.org). The Version 4 rice non-TE amino acid coding sequences (http://rice.tigr.org) were pair-wise matched with genomic and TA sequences using TBLASTN. All scores were filtered for an E-value cutoff of 10−5. Customized Perl scripts were used in conjunction with the parsed TBLASTN output during the filtration strategy outlined in Figure 1. Customized Perl scripts were also used with the TBLASTN output during the combinatorial steps to identify the CPSG set.
Functional Assignments of the Rice Genes
The functional assignment for each gene in the CPSG set was obtained from Version 4 of the TIGR Rice Annotation Project (http://rice.tigr.org). The hypothetical proteins are promoted to expressed proteins based upon supporting transcript, massively parallel signature sequencing, serial analysis of gene expression, or proteomic data (Ouyang et al., 2007). The PF00646 HMM was downloaded from Pfam and the F-box domain was identified using the program hmmsearch with the trusted cutoff option (- -cut_tc).
Genic Features
The mean and median values for the exon and intron lengths, exon, intron, gene, and CDS GC content, and exon counts were determined from Version 4 of the TIGR Rice Annotation Project (http://rice.tigr.org).
Tandem Duplication
All genes in either the CSPG set or the SH set were screened for tandem duplication using a method previously adapted for use in Arabidopsis (Arabidopsis Genome Initiative, 2000). Protein sequences were subjected to a BLASTP search against themselves. Two genes were assumed to be duplicated if they had a BLASTP E value <10−20. Genes were deemed to be tandemly duplicated if there was no more than one unrelated gene between the genes displaying similarity.
MSAs
The reading frame for the TBLASTN output for each of the plant TAs having significant matches to the CPSGs was identified. The translated amino acid sequence of the longest ORF in the correct translational frame (from the TBLASTN output) was generated using the ORF finder at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/projects/gorf). MSAs of the multi-FASTA amino acid sequence files were generated using the T-Coffee program (Notredame et al., 2000) with the default parameters. Jalview was used to visualize and customize the presentations of these MSAs (Clamp et al., 2004; http://www.jalview.org).
Synteny Identification
Sorghum scaffolds were downloaded from the JGI (https://www.jgi.doe.gov/downloads/Sorghum_bicolor/assembly20060630/scaffolds/sequences/Sorghum_bicolor.main_genome.scaffolds.fasta; dated on March 2, 2007). The sorghum genes predicted by FGENESH (Salamov and Solovyev, 2000; http://www.softberry.com) were searched against non-TE-related rice genes using BLASTP with E value <10−5. The FGENESH monocot matrix was used for ab initio gene prediction in the sorghum assemblies. The DAGchainer package (Haas et al., 2004) was then applied to the BLAST results to remove repetitive elements and identify syntenic blocks with at least three aligned pairs. DAGchainer settings were −g 20000 (defining the length of the gap between the syntenic genes), −D 100000 (the maximal distance allowed between two syntenic genes), −s, −I, and −A 3 (requiring a minimum of at least three collinear pairs).
Identification of TEs
TEs among the candidate CPSGs were identified through comparison to a known rice repeat library that has been described previously (Jiang et al., 2003). Specifically, repetitive sequences in rice were identified with RECON (Version 1.03; Bao and Eddy 2002). The resulting 3,300 repeat families (within each family over 90% of the sequence can be aligned between any two members based on the shorter sequence) were examined individually and those derived from TEs were analyzed further. If a sequence is similar to a known TE (BLASTX or BLASTN E < 10−10) at the nucleotide level or protein level, it is considered to be the relevant TE. If a sequence was not similar to any known TE, the following procedure was used to define the repetitive sequences. First, the relevant sequence was used to search the rice genome database and at least 20 hits (if there are 20 or more hits, BLASTX E < 10−10) and the 100-bp flanking sequence on each side of the hits were recovered. The recovered sequences were then aligned using pileup in GCG (Wisconsin GCG program suite, Version 10.1), with the resulting output examined for the presence of a possible border between putative elements and their flanking sequences. A border was defined if the sequence homology stops at the same position for more than one-half of the aligned sequences and the sequence at the most termini of the putative element was compared with known TEs. Furthermore, the sequence immediately flanking the border was examined for the possible presence of target site duplication. Finally, the putative terminal sequence was aligned (directly and inversely) using the gap program in GCG to detect possible inverted or direct repeats. All the above information was used to determine the identity of relevant sequences (see “Results” for the structural features about each superfamily of TEs).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Whole-genome distribution of the CPSG set and the SH set across the 12 pseudomolecules of rice.
Supplemental Figure S2. MSA for the coding sequence for the 20 CPSGs contained in the Pack-MULE subfamily Os0037.
Supplemental Figure S3. MSA for the coding sequence for the three rice/sorghum orthologs with translated ORFs from other Poaceae species presented in Figure 4.
Supplemental Table S1. Statistics of the Poaceae TAs used in this study.
Supplemental Table S2. CPSGs and significant matches with five Poaceae species.
Supplemental Table S3. Pack-MULEs identified within the CPSG set are tallied by common subfamily.
Supplemental Table S4. Summary statistics for the sequences used in the MSAs in Figure 3.
Supplemental Data S1. Multi-FASTA files (861) of the CPSGs and the top match from the five Poaceae species.
Supplementary Material
Acknowledgments
We wish to thank Michael Scanlon and Clifford Weil for contributions regarding the biological significance of the analysis during its formative stages. Daniel Haft's discussions regarding Pfam analysis and MSAs were both informative and insightful. We also acknowledge Francoise Thibaud-Nissen for her significant contributions to improve this work through its development.
This work was supported by the National Science Foundation (grant no. DBI–0321538 to C.R.B.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: C. Robin Buell (buell@msu.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Allen KD (2002) Assaying gene content in Arabidopsis. Proc Natl Acad Sci USA 99 9568–9572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–813 [DOI] [PubMed] [Google Scholar]
- Bao Z, Eddy SR (2002) Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res 12 1269–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell JA, Budiman MA, Nunberg A, Citek RW, Robbins D, Jones J, Flick E, Rholfing T, Fries J, Bradford K, et al (2005) Sorghum genome sequencing by methyl filtration. PLoS Biol 3 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Coleman C, Liu R, Ma J, Ramakrishna W (2004) Consistent over-estimation of gene number in complex plant genomes. Curr Opin Plant Biol 7 732–736 [DOI] [PubMed] [Google Scholar]
- Boffelli D, Nobrega MA, Rubin EM (2004. a) Comparative genomics at the vertebrate extremes. Nat Rev Genet 5 456–465 [DOI] [PubMed] [Google Scholar]
- Boffelli D, Weer CV, Weng L, Lewis KD, Shoukry MI, Pachter L, Keys DN, Rubin EM (2004. b) Intraspecies sequence comparison for annotating genomes. Genome Res 14 2406–2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, Crow JA, Heuer ML, Wang X, Cannon EK, Dwan C, Lamblin AF, Vasdewani J, Mudge J, Cook A, et al (2005) Databases and information integration for the Medicago truncatula genome. Plant Physiol 138 38–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo T, Pagano M (2004) The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol 5 739–751 [DOI] [PubMed] [Google Scholar]
- Carels N, Bernardi G (2000) Two classes of genes in plants. Genetics 154 1819–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carels N, Hatey P, Jabbari K, Bernardi G (1998) Compositional properties of homologous coding sequences from plants. J Mol Evol 46 45–53 [DOI] [PubMed] [Google Scholar]
- Chan AP, Pertea G, Cheung F, Lee D, Zheng L, Pontraroli AC, SanMiguel P, Yuan Y, Bennetzen J, Barbazuk WB, et al (2006) The TIGR maize database. Nucleic Acids Res 34 D771–D776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs KL, Hamilton JP, Zhu W, Ly E, Cheung F, Wu H, Rabinowicz PD, Town CD, Buell CR, Chan AP (2007) The TIGR plant transcript assemblies database. Nucleic Acids Res 35 D846–D851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HK, Mun JH, Kim DJ, Zhu H, Baek JM, Mudge J, Roe B, Ellis N, Doyle J, Kiss GB, et al (2004) Estimating genome conservation between crop and model legume species. Proc Natl Acad Sci USA 101 15289–15294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JD, Hoshino A, Park KI, Park IS, Iida S (2007) Spontaneous mutations caused by a Helitron transposon, Hel-It1, in morning glory, Ipomoea tricolor. Plant J 49 924–934 [DOI] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ (2004) The Jalview Java Alignment Editor. Bioinformatics 20 426–427 [DOI] [PubMed] [Google Scholar]
- Cruveiller S, Kabbari K, Clay O, Bernardi G (2003) Compositional features of eukaryotic genomes for checking predicted genes. Brief Bioinform 4 43–52 [DOI] [PubMed] [Google Scholar]
- Diao X, Freeling M, Lisch D (2006) Horizontal transfer for a plant transposon. PLoS Biol 4 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Loso T, Tautz D (2003) An evolutionary analysis of orphan genes in Drosophila. Genome Res 13 2213–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR (2005) A model of the statistical power of comparative genome sequence analysis. PLoS Biol 3 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA, Fraser CM (2003) Phylogenomics: intersection of evolution and genomics. Science 300 1706–1707 [DOI] [PubMed] [Google Scholar]
- Feng Q, Zhang Y, Hao P, Wang S, Fu G, Huang Y, Li Y, Zhu J, Liu Y, Hu X, et al (2002) Sequence and analysis of rice chromosome 4. Nature 420 316–320 [DOI] [PubMed] [Google Scholar]
- Feschotte C, Jiang N, Wessler SR (2002) Plant transposable elements: where genetics meets genomics. Nat Rev Genet 3 329–341 [DOI] [PubMed] [Google Scholar]
- Graham MA, Silverstein KAT, Cannon SB, VandenBosch KA (2004) Computational identification and characterization of novel genes from legumes. Plant Physiol 135 1179–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass Phylogeny Working Group (2000) A phylogeny of the grass family (Poaceae) as inferred from eight character sets. In SWL Jacobs, JE Everett, eds, Grasses: Systematics and Evolution. Commonwealth Scientific and Industrial Research Organization, Victoria, Australia, pp 3–7
- Haas BJ, Delcher AL, Wortman JR, Salzberg SL (2004) DAGchainer: a tool for mining segmental genome duplications and synteny. Bioinformatics 20 3643–3646 [DOI] [PubMed] [Google Scholar]
- Hardison RC (2004) Comparative genomics. PLoS Biol 1 156–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436 793–800 [DOI] [PubMed] [Google Scholar]
- Jabbari K, Cruveiller S, Clay O, Le Saux J, Bernardi G (2004) The new genes of rice: a closer look. Trends Plant Sci 9 281–285 [DOI] [PubMed] [Google Scholar]
- Jacobs BF, Kingston JD, Jacobs LL (1999) The origin of grass-dominated ecosystems. Ann Mo Bot Gard 86 590–643 [Google Scholar]
- Jiang N, Bao Z, Zhang X, Eddy SR, Wessler SR (2004) Pack-MULE transposable elements mediate gene evolution in plants. Nature 431 569–573 [DOI] [PubMed] [Google Scholar]
- Jiang N, Bao Z, Zhang X, Hirochika H, Eddy SR, McCouch SR, Wessler SR (2003) An active DNA transposon family in rice. Nature 421 163–167 [DOI] [PubMed] [Google Scholar]
- Juretic N, Hoen DR, Huynh ML, Harrison PM, Bureau TE (2005) The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genet Res 15 1292–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES (2003) Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423 241–254 [DOI] [PubMed] [Google Scholar]
- Kellogg EA (1999) Phylogenetic aspects of the evolution of C4 photosynthesis. In RF Sage, RK Monson, eds, C4 Plant Biology. Academic Press, San Diego, pp 411–444
- Kellogg EA (2000) The grasses: a case study in macroevolution. Annu Rev Ecol Syst 31 217–238 [Google Scholar]
- Kellogg EA (2001) Evolutionary history of the grasses. Plant Physiol 125 1198–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg EA, Bennetzen JL (2004) The evolution of nuclear genome structure in seed plants. Am J Bot 91 1709–1725 [DOI] [PubMed] [Google Scholar]
- Kumar A, Bennetzen JL (1999) Plant retrotransposons. Annu Rev Genet 33 479–532 [DOI] [PubMed] [Google Scholar]
- Lai J, Li Y, Messing J, Dooner HK (2005) Gene movement by Helitron transposons contributes to the haplotype variability of maize. Proc Natl Acad Sci USA 102 9068–9073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J, Ma J, Swigonova A, Ramakrishna W, Linton E, Llaca V, Tanyolac B, Park YJ, Jeong OY, Bennetzen JL, et al (2004) Gene loss and movement in the maize genome. Genome Res 14 1924–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies EH, Vinson JP, NISC Comparative Sequencing Program, Miller W, Jaffe DB, Lindblad-Toh K, Chang JL, Green ED, Lander ES, Mullikin JC, et al (2005) An initial strategy for the systematic identification of functional elements in the human genome by low-redundancy comparative sequencing. Proc Natl Acad Sci USA 102 4795–4800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitreva M, McCarter JP, Arasu P, Hawdon J, Martin J, Dante M, Wylie T, Xu J, Stajich JE, Kapulkin W, et al (2005) Investigating hookworm genomes by comparative analysis of two Ancylostoma species. BMC Genomics 6 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgante M, Brunner S, Pea G, Fengler K, Zuccolo A, Rafalski A (2005) Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat Genet 37 997–1002 [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302 205–217 [DOI] [PubMed] [Google Scholar]
- Ouyang S, Buell CR (2004) The TIGR Plant Repeat Databases: a collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res 32 D360–D363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Zhu W, Hamilton JH, Haining L, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, et al (2007) The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res 35 D883–D887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer LE, Rabinowicz PD, O'Shaughnessy AL, Balija VS, Nascimento LU, Dike S, de la Bastide M, Martinssen RA, McCombie WR (2003) Maize genome sequencing by methyl filtration. Science 302 2115–2117 [DOI] [PubMed] [Google Scholar]
- Rensink WA, Lee Y, Liu J, Iobst S, Ouyang S, Buell CR (2005) Comparative analyses of six solanaceous transcriptomes reveal a high degree of sequence conservation and sequence-specific transcripts. BMC Genomics 6 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice Chromosome 3 Sequencing Consortium (2005) Sequence, annotation, and analysis of synteny between rice chromosome 3 and diverged grass species. Genome Res 15 1284–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice Chromosome 10 Sequencing Consortium (2003) In-depth view of structure, activity, and evolution of rice chromosome 10. Science 300 1566–1569 [DOI] [PubMed] [Google Scholar]
- Rice Full-Length cDNA Consortium (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301 376–379 [DOI] [PubMed] [Google Scholar]
- Salamov AA, Solovyev VV (2000) Ab initio gene finding in Drosophila genomic DNA. Genome Res 10 515–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H, Karlowski WM (2003) Comparison of rice and Arabidopsis annotation. Curr Opin Plant Biol 6 106–112 [DOI] [PubMed] [Google Scholar]
- Thomas JH (2006) Adaptive evolution in two large families of ubiquitin-ligase adapters in nematodes and plants. Genome Res 16 1017–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JW, Touchman JW, Blakesley RW, Bouffard GG, Beckstrom-Sternber SM, Margulies EH, Blanchette M, Siepel AC, Thomas PJ, McDowell JC, et al (2003) Comparative analyses of multi-species sequences from targeted genomic regions. Nature 424 788–793 [DOI] [PubMed] [Google Scholar]
- Town CD (2006) Annotating the genome of Medicago truncatula. Curr Opin Plant Biol 9 122–127 [DOI] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr & Gray). Science 313 1596–1604 [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Van de Peer Y (2005) Exploring the plant transcriptome through phylogenetic profiling. Plant Physiol 137 31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw CA, Barbazuk WB, Pertea G, Chan AP, Cheung F, Lee Y, Zheng L, van Heeringen S, Karamycheva S, Bennetzen JL, et al (2003) Enrichment of gene-coding sequences in maize by genome filtration. Science 302 2118–2120 [DOI] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296 79–92 [DOI] [PubMed] [Google Scholar]
- Yu J, Wang J, Lin W, Li S, Li H, Zhou J, Ni P, Dong W, Hu S, Zeng C, et al (2005) The genomes of Oryza sativa: a history of duplications. PLoS Biol 3 e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan YN, SanMiguel PJ, Bennetzen JL (2003) High-Cot sequence analysis of the maize genome. Plant J 34 249–255 [DOI] [PubMed] [Google Scholar]
- Yuan Q, Ouyang S, Wang A, Zhu W, Maiti R, Lin H, Hamilton J, Haas B, Sultana R, Cheung F, et al (2005) The Institute for Genomic Research Osa1 rice genome annotation database. Plant Physiol 138 18–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Buell CR (2007) Improvement of whole-genome annotation of cereals through comparative analyses. Genome Res 17 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.