Abstract
RNA interference (RNAi) is a powerful tool for functional gene analysis, which has been successfully used to down-regulate the levels of specific target genes, enabling loss-of-function studies in living cells. Hairpin (hp) RNA expression cassettes are typically constructed on binary plasmids and delivered into plant cells by Agrobacterium-mediated genetic transformation. Realizing the importance of RNAi for basic plant research, various vectors have been developed for RNAi-mediated gene silencing, allowing the silencing of single target genes in plant cells. To further expand the collection of available tools for functional genomics in plant species, we constructed a set of modular vectors suitable for hpRNA expression under various constitutive promoters. Our system allows simple cloning of the target gene sequences into two distinct multicloning sites and its modular design provides a straightforward route for replacement of the expression cassette's regulatory elements. More importantly, our system was designed to facilitate the assembly of several hpRNA expression cassettes on a single plasmid, thereby enabling the simultaneous suppression of several target genes from a single vector. We tested the functionality of our new vector system by silencing overexpressed marker genes (green fluorescent protein, DsRed2, and nptII) in transgenic plants. Various combinations of hpRNA expression cassettes were assembled in binary plasmids; all showed strong down-regulation of the reporter genes in transgenic plants. Furthermore, assembly of all three hpRNA expression cassettes, combined with a fourth cassette for the expression of a selectable marker, resulted in down-regulation of all three different marker genes in transgenic plants. This vector system provides an important addition to the plant molecular biologist's toolbox, which will significantly facilitate the use of RNAi technology for analyses of multiple gene function in plant cells.
Posttranscriptional gene silencing (PTGS) is a powerful tool for functional gene analysis. PTGS reduces the level of target transcripts in a sequence-specific manner (Agrawal et al., 2003), enabling loss-of-function studies in living cells. PTGS was first discovered in transgenic petunia (Petunia hybrida) plants, which, upon overexpression of the chalcone synthase-encoding sequence, resulted in variegated plants instead of the predicted purple flowers (Napoli et al., 1990). This phenomenon was originally termed cosuppression because it suppressed the expression of both the transgene and the native genes (Napoli et al., 1990), and it was thus suggested that interaction between the transgenes and the native transcripts triggers a mechanism that leads to destruction of both transcripts or obstruction of the translation process and to gene silencing. Today we know that overexpression-mediated gene silencing results from in vivo hybridization between native and transgene RNA molecules and the production of double-stranded (ds)RNA molecules, which are then targeted for degradation by a complex cellular mechanism (for review, see Watson et al., 2005). Whereas the silencing effect can be directly attributed to the degradation of these dsRNA molecules, gene silencing can also occur via a range of epigenetic phenomena, including paramutation, epimutation, DNA methylation, and imprinting (McGinnis et al., 2007).
The mechanism of dsRNA-mediated gene silencing is probably similar in all eukaryotes (for review, see Horiguchi, 2004). Briefly, dsRNA molecules are processed by an RNase III-like enzyme (dicer) into small interfering RNA (siRNA) molecules. These 21- to 28-base-long siRNA molecules (Hamilton and Baulcombe, 1999; Elbashir et al., 2001) mediate the degradation of target RNAs and possibly also the methylation of genomic DNA by RNA-induced silencing-like effector complexes in a process generally known as RNA interference (RNAi; Mette et al., 1999; Hamilton et al., 2002; Zilberman et al., 2003). Interestingly, siRNAs can also move from cell to cell and systematically spread and deliver the silencing signal to the entire organism (Voinnet et al., 2000; Winston et al., 2002).
Whereas both sense- and antisense-mediated gene silencing have proven fruitful for PTGS in plant cells (Bruening, 1998; Waterhouse et al., 1998), RNAi induction can be more efficiently achieved by specialized expression cassettes that produce self-complementary hairpin (hp)-like RNA molecules (Horiguchi, 2004; Watson et al., 2005; Hirai et al., 2007). Such cassettes typically include plant promoter and terminator sequences that control the expression of two inversely repeated sequences of the target genes that are separated by a specific spacer (Fig. 1A). Upon delivery to plant cells, expression of an RNAi cassette will result in a dsRNA molecule composed of two distinct regions: a single-stranded loop, encoded by the spacer region, and a ds stem, encoded by the inverted repeats. It is the stem region that is used as a substrate by the dicer, but, because the spacer itself can potentially be recognized as a substrate as well, intron sequences are often used in the construction of such RNAi vectors (e.g. Meyer et al., 2004; Miki and Shimamoto, 2004; Hirai et al., 2007). RNAi (or hpRNA) plant expression can potentially be delivered to plant cells by various means of transformation but are typically used by mounting on binary plasmids to be delivered to plant cells by Agrobacterium-mediated transformation.
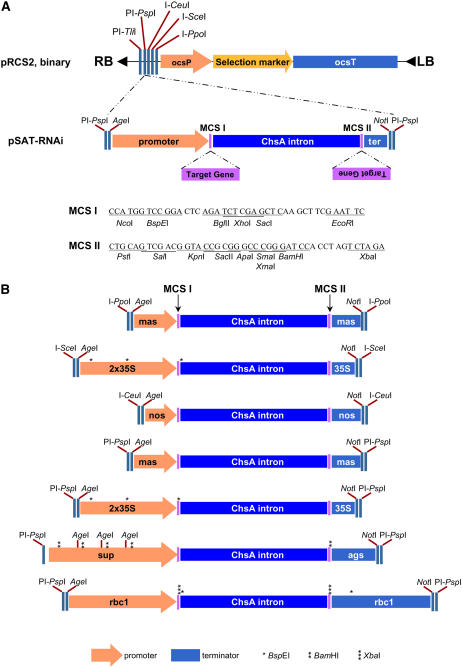
Figure 1.
Structural features of the pSAT-RNAi vector system. A, The general structure of a pSAT-RNAi plasmid and its cloning compatibility with pRCS2-based binary plasmid. A typical pSAT-RNAi plasmid is composed of a promoter and terminator region, the ChsA intron, and two unique multicloning sites, MCS-I and MCS-II, which should be used for cloning the two target gene sequences in reverse orientations. The entire hpRNA cassette is flanked by AgeI and NotI, allowing its transfer between different pSAT plasmid backbones. The hpRNA cassette is also flanked with rare cutters (e.g. PI-PspI), which can be used for its cloning into the pRCS2-based binary plasmid. B, Outline of the different pSAT-RNAi plasmids produced in this study. Expression of hpRNA is controlled by various promoters and terminators and each of the various RNAi cassettes can be transferred to the pRCS2-based binary plasmid using different combinations of rare-cutting endonucleases, as indicated. Both MCS-I and MCS-II are present in all plasmids and restriction recognition sites that are not unique in all plasmids are indicated by asterisks. Plasmid names from top to bottom are: pSAT3.masP.RNAi, pSAT4.35SP.RNAi, pSAT5.nosP.RNAi, pSAT6.masP.RNAi, pSAT6.35SP.RNAi, pSAT6.supP.RNAi, and pSAT6.rbcP.RNAi. 35S, CaMV 35S promoter and terminator; sup, superpromoter; mas, manopine synthase promoter and terminator; rbc1, Rubisco small-subunit promoter and terminator; nos, nopaline synthase promoter and terminator; ags, agropine synthase terminator. One, two, and three asterisks represent BspEI, BamHI, and XbaI restriction sites, respectively.
RNAi-mediated gene suppression has been highly instrumental in basic plant cell research and biotechnology (for review, see Tenllado et al., 2004; Small, 2007). RNAi vectors have been used, for example, in determining the importance of flavone synthases for nodulation in Medicago truncatula (Zhang et al., 2007), in revealing the role of a homeobox gene in rice (Oryza sativa) leaf development (Dai et al., 2007), and in engineering transgenic resistance to nematodes in Arabidopsis (Arabidopsis thaliana) plants (Huang et al., 2006). Biotechnologically, RNAi has been used, for example, to suppress the caffeine synthase gene, reducing caffeine content in coffee (Coffea arabica) plants (Ogita et al., 2003), to knock down a transcription factor that regulates the expression of a set of storage proteins in maize (Zea mays) plants while increasing the Lys content in maize (Segal et al., 2003), and to manipulate the expression of a cinnamyl alcohol dehydrogenase to produce tragenic flax (Linum usitatissimum) with reduced lignin and improved mechanical properties (Wrobel-Kwiatkowska et al., 2007).
Realizing the importance of RNAi for plant research, different reports have described the development of various vectors for the construction and expression hpRNA in plant cells. These include, for example, the pHANNNIBAL, pKANNIBAL, and pHELLSGATE vectors, which were designed for high-throughput gene silencing in plant cells and allow the assembly of hpRNA constructs by conventional cloning or Gateway recombination (Wesley et al., 2001; Helliwell and Waterhouse, 2003), or the pX7 vectors, which were specifically designed as either chemically inducible or constitutive RNAi vector systems (Guo et al., 2003). Other, more specialized vectors have also been developed and include, for example, the pANDA and pANDA-mini vectors, which were developed for stable and transient RNAi-mediated suppression of gene function in rice (Miki and Shimamoto, 2004), and a set of small, high-replicating pUC-based shuttle and binary RNAi vectors suitable for direct gene transfer and Agrobacterium-mediated transformation of poplar (Populus spp.) tissues (Meyer et al., 2004). These vectors, as well as many others that have been constructed for more specific tasks or as part of large families of plant vectors (e.g. Earley et al., 2006), have been successfully used for down-regulation and functional studies of various genes in various plant species (e.g. Stoutjesdijk et al., 2002; McGinnis et al., 2005, 2007; Travella et al., 2006).
Biotechnological applications and analysis of various cellular processes sometimes require suppression of more than just a single gene. Suppression of a multigene family or any other collection of related sequences can potentially be achieved using a single transgene construct and a specific design of the hpRNA expression cassette. Efficient simultaneous targeting of four different tospoviruses, for example, was reported by expression of a single hpRNA transgene designed to target the conserved N gene in four major tomato (Lycopersicon esculentum)-infecting tospoviruses (Bucher et al., 2006) and several members of the psbP gene family from tobacco (Nicotiana tabacum) were simultaneously suppressed using a single hpRNA that targeted the 3′-untranslated sequences of the four psbP tobacco isogenes (Ifuku et al., 2003). Nevertheless, whereas the suppression of several gene family members can be achieved by a single construct (e.g. Ifuku et al., 2003; Miki et al., 2005), cosuppression of unrelated genes requires using similar strategies for the delivery of multiple transgenes into plants cells. These include, for example, crossing two transgenic lines that are homozygous for each transgene (e.g. Chabannes et al., 2001; Pincon et al., 2001), retransformation (e.g. Zhong et al., 1998), and cotransformation (e.g. Ye et al., 2000). More recently, simultaneous suppression of three genes using a single chimeric construct was achieved by incorporating partial sequences from three different lignin biosynthetic genes under the control of a single promoter (Abbott et al., 2002). Whereas such chimeric constructs can certainly provide a possible technical solution for multiple gene suppression in plant cells, the assembly of several expression cassettes onto a single plasmid can provide a simpler and more efficient route for the delivery of multiple transgenes into plant cells. Such a strategy was used, for example, by Zhong et al. (2007), who assembled two RNAi constructs to simultaneously inhibit SND1 and NST1 genes in Arabidopsis plants. Nevertheless, more flexible and modular vectors than those used in many RNAi studies are needed to facilitate the construction of multiple RNAi expression cassette vectors and for the assembly of any complex gene expression system for that matter.
We previously described construction of the versatile pSAT plant expression vector system (Tzfira et al., 2005), which allows the assembly of multiple expression cassettes into specially designed Agrobacterium binary plasmids (Goderis et al., 2002). The pSAT plasmid, originally constructed as a set of modular satellite vectors in a system that supports fusions to five different autofluorescent tags under the control of the cauliflower mosaic virus (CaMV) 35S promoter (35SP; Tzfira et al., 2005), was later expanded to support the expression of foreign genes under the control of various promoters and terminators (Chung et al., 2005) and with the support of GATEWAY-mediated cloning (Chakrabarty et al., 2007). Relying on the system's flexible design, we sought to expand its cloning possibilities to include the expression of multiple hpRNAs in plant cells. Here, we describe the assembly of our pSAT-RNAi vectors and demonstrate their use for the simultaneous silencing of multiple transgenes in transgenic plants. Our pSAT-RNAi plasmids feature a ChsA intron sequence that is flanked by two unique multiple cloning sites (MCSs) suitable for insertion of target gene sequences in reverse orientations. Importantly, expression of hpRNA in different pSAT-RNAi plasmids is controlled by various promoter and terminator sequences and several hpRNA expression cassettes can be assembled into a single Agrobacterium binary vector. We believe that our vector system not only provides a unique and important addition to the growing collection of plant RNAi plasmids, but will also significantly facilitate the use of RNAi technology for the analysis of multiple gene function in plant cells.
RESULTS AND DISCUSSION
Design of a Modular Set of Plasmids for Expression of hpRNA
We decided to use the previously described basic design of our pSAT family of plasmids to address the difficulties involved in assembling multiple hpRNAi expression cassettes on a single plasmid (Tzfira et al., 2005). One crucial feature of the cloning system is that each expression cassette, whether it directs the expression of an untagged or autofluorescently tagged protein (Tzfira et al., 2005) under the control of various promoters and terminators (Chung et al., 2005) or whether it directs the expression of a hpRNA sequence (as described below), is flanked by different rare cutter sites that allow its transfer from pSAT plasmids into the corresponding sites of the pPZP-RCS1- or pPZP-RCS2-based (Goderis et al., 2002) Agrobacterium binary plasmids. This feature is exemplified in Figure 1A, where the hpRNA expression cassette from the generic pSAT-RNAi vector is flanked by PI-PspI and thus can be cloned directly into the MCS of pPZP-RCS1, pPZP-RCS2 (Goderis et al., 2002), or their descending plasmids pRCS2-ocs-bar, pRCS2-ocs-hpt, and pRCS2-ocs-nptII (Chung et al., 2005). The latter plasmids, exemplified by the generic pRCS2 binary plasmid (Fig. 1A), carry three different plant selection marker expression cassettes cloned near the left border of the pPZP-RCS2, which facilitates their direct use for plant transformation experiments (Chung et al., 2005).
Other key features of individual pSAT-RNAi plasmids are exemplified by the generic pSAT-RNAi illustrated in Figure 1. A complete hpRNA expression cassette is composed of four distinct regions: a promoter and terminator sequence, the ChsA intron sequence, and a dual MCS. The dual MCS results from cloning of the ChsA intron sequence into pSAT6-MCS (Tzfira et al., 2005) and dividing the original MCS into two new, distinct regions, designated MCS-I and MCS-II, which contain the following unique restriction endonuclease recognition sites: NcoI, BspEI, BglII, XhoI, SacI, and EcoRI in MCS-I and PstI, SalI, KpnI, SacII, ApaI, XmaI, SmaI, BamHI, and XbaI in MCS-II. The two MCS regions allow the successive cloning of the target gene sequence in reverse orientation and assembly of a hpRNA sequence (Fig. 1A). In pSAT6.35S.RNAi, for example (Fig. 1B), expression of hpRNA is controlled by tandem CaMV 35SP and CaMV 35S terminator (35ST), conferring a complete expression cassette that is flanked by PI-PspI.
Another important aspect of our extended family of pSAT plasmids is their flexible design, which allows the simple, one-step replacement of their regulatory elements and mobilization of the hpRNA expression cassettes between different pSAT plasmids. Exemplified by pSAT6.35S.RNAi, the promoter region in this plasmid (i.e. the tandem CaMV 35S) is flanked by AgeI and NcoI and the terminator (i.e. CaMV 35ST) is flanked by XbaI and NotI, allowing their replacement with different regulatory elements by successive cloning. Furthermore, the entire hpRNA expression cassette can be mobilized between various pSAT plasmid backbones as AgeI-NotI fragments (excluding pSAT6.sup.RNAi.ags, which lost its original AgeI site during cloning). Indeed, we employed these various cloning strategies while assembling additional pSAT-RNAi plasmids (Fig. 1B). Overall, we produced seven different pSAT.RNAi plasmids (i.e. pSAT3.masP.RNAi, pSAT4.35SP.RNAi, pSAT5.nosP.RNAi, pSAT6.masP.RNAi, pSAT6.35SP.RNAi, pSAT6.supP.RNAi, and pSAT6.rbcP.RNAi; Fig. 1B), in which the expression of hpRNA is under the control of the tandem CaMV 35SP, the superpromoter (supP; Ni et al., 1995), the manopine synthase promoter (masP), the strong Rubisco small-subunit promoter (rbcP) from Asteraceous chrysanthemum (http://www.pri.wur.nl/UK/products/ImpactVector), or the nopaline synthase promoter (nosP). In the resulting series of pSAT-RNAi constructs, the hpRNA expression cassettes are flanked by I-PpoI (pSAT3), I-SceI (pSAT4), I-CeuI (pSAT5), and PI-PspI (pSAT6; Fig. 1B), which allow their successive cloning into the MCS of pPZP-RCS1- or pPZP-RCS2-based plasmids (Goderis et al., 2002; Chung et al., 2005) and assembly of multiple hpRNA expression cassettes.
Production of Transgenic Plants Overexpressing Three Marker Genes
To functionally test our pSAT-RNAi plasmids in plant cells, we decided to first produce transgenic plants that overexpress three marker genes: the enhanced green autofluorescent protein (EGFP; CLONTECH), the red autofluorescent protein (DsRed2) from coral (Matz et al., 1999), and the nptII gene, which codes for kanamycin (KAN) resistance in transgenic plants. We chose to simultaneously express these three proteins in transgenic plants under the control of strong constitutive promoters, rationalizing that suppressing high levels of protein expression is the best method for evaluating the function of our new RNAi plasmids. We thus constructed the pRCS2-[EGFP][KAN][DsRed2] binary plasmid (Fig. 2), which was composed of three expression cassettes. Expression of the EGFP, KAN, and DsRed2 coding sequences was controlled by nopaline synthase, the tandem CaMV 35S, and the rbcP, respectively. Several KAN-resistant transgenic lines were produced and lines with a putative single T-DNA insertion (as determined by segregation of their offspring on KAN-containing medium) were analyzed for expression of EGFP and DsRed2. Two transgenic lines were selected in which the expressions of both EGFP and DsRed2 were at their highest levels, as determined by the relative expression of the transgene's fluorescence as compared with the chloroplast's autofluorescence using confocal laser-scanning microscopy. A sample of the confocal imaging of pRCS2-[EGFP][KAN][DsRed2]-transgenic plants, designated KGD (KAN-EGFP-DsRed2), is given in Figure 3, A to D.
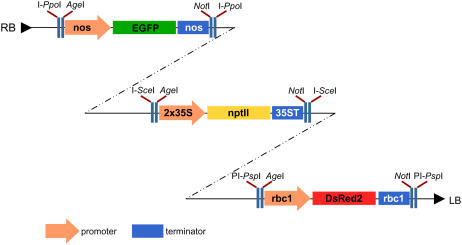
Figure 2.
Outline of the pRCS2-[EGFP][KAN][DsRed2] binary plasmid. The plasmids constructed to carry three cassettes for the stable overexpression of the EGFP, NPTII, and DsRed2 coding sequences. Expression cassettes were cloned as an AgeI-NotI fragment into the different pSAT plasmids and were successively transferred as I-PpoI, I-SceI, and PI-PspI fragments into the pRCS2 binary plasmid. 2x35S, Tandem CaMV 35S promoter; 35ST, CaMV 35S terminator; nos, nopaline synthase promoter and terminator; rbc1, Rubisco small-subunit promoter and terminator.
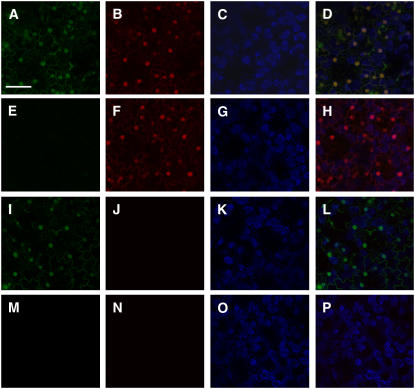
Figure 3.
Confocal microscopy analysis of EGFP and DsRed2 expression in single- and double-transformed plants. A to D, Confocal imaging of pRCS2-[EGFP][KAN][DsRed2] (designated KGD plants). E to H, Confocal imaging of KGD transformed by GFPi construct. I to L, Confocal imaging of KGD transformed by DsRed2i construct. M to P, Confocal imaging of KGD transformed by GFPi and DsRed2i constructs. A, E, I, and M, EGFP signal (green, when present). B, F, J, and N, DsRed2 signal (red, when present). C, G, K, and O, Plastid autofluorescence in blue. D, H, L, and P, Merge signals of the corresponding EGFP, DsRed2, and plastid autofluorescence. Images are single confocal sections (bar = 50 μm).
Suppression of a Single Marker Transgene
We began analyzing our RNAi system by constructing satellite plasmids for the suppression of EGFP, DsRed2, or KAN in KGD transgenic plants. We first tested the expression of hpRNA under the control of the supP and tandem 35SP by the assembly of functional GFP-RNAi (GFPi), KAN-RNAi (KANi), and DsRed2-RNAi (DsRed2i) expression cassettes. pSAT6.sup.GFPi, pSAT4.35SP.KANi, and pSAT4.35SP.DsRed2i were constructed by successive cloning of approximately 500-, 300-, and 310-bp-long sequences from the GFP, nptII, and DsRed2 coding sequences, respectively, in forward and reverse orientations. The simplicity of assembly of each cassette can be illustrated, for example, by the construction of pSAT6.sup.GFPi, which required only one pair of primers and a single PCR. More specifically, a pair of forward and reverse primers, carrying the NcoI and XhoI recognition sites, respectively, was designed and used for amplification of the 500-bp partial GFP sequence. The product was then digested with XhoI alone or double digested with NcoI and XhoI and the fragments were successively cloned into the SmaI and SalI sites in MCS-II (XhoI-digested fragment) and the XhoI and NcoI sites in MCS-I (XhoI-NcoI fragment). The location of the XhoI upstream of NcoI and the location of SalI (and its compatibility with XhoI) downstream of SmaI resulted in the inverted alignment of both GFP fragments. Similarly, two pairs of primers were used for successive cloning of the KAN or DsRed2 inverted repeats into the MCS-II and MCS-I for the production of KANi or DsRed2i expression cassettes in pSAT4.35SP.RNAi.
We next assembled the GFPi, DsRed2i, and KANi hpRNA expression cassettes into an Agrobacterium binary plasmid. To this end, we removed the GFPi, KANi, or DsRed2i expression cassettes from pSAT6.sup.GFPi, pSAT4.35SP.KANi, or pSAT4.35SP.DsRed2i as PI-PspI or I-SceI fragments and cloned them, individually, into the corresponding sites of pRCS2-ocs-bar (Chung et al., 2005), producing pRCS2-[bar][GFPi], pRCS2-[bar][KANi], and pRCS2-[bar][DsRed2i] (Table I). We mobilized these vectors into Agrobacterium cells and used them, as well as the empty vector pRCS2-[bar], to transform our KGD transgenic plants. Double-transformed plants were selected on basta-containing medium and were examined for their suppression phenotype. As expected, confocal microscopy analysis of KGD lines, transformed by pRCS2-[bar][GFPi], revealed that these doubly transformed plants (designated KGD-Gi) no longer produced GFP fluorescence (Fig. 3E) but were still capable of emitting red fluorescence from the stable expression of DsRed2 (Fig. 3F). Transformation of KGD plants by pRCS2-[bar][DsRed2i], on the other hand, affected the expression of DsRed2 and not of GFP in these doubly transformed KGD-Di plants (Fig. 3, I and J). No suppression of either GFP or DsRed2 was observed when KGD plants were retransformed by either pRCS2-[bar][KANi] (KGD-Ki plants) or the empty vector pRCS2-[bar] (data not shown). Collectively, these results indicate that the suppression effect of GFPi and DsRed2i was indeed specific and that expression of KANi or the bar selection marker did not have a negative effect on the expression of either GFP or DsRed2 in the KGD transgenic plants.
Table I.
Constructs used for analysis of RNAi cassettes in transgenic plants
| Plasmid Name | Target Genesab
|
||
|---|---|---|---|
| gfp | dsred2 | nptII | |
| pRCS2-[bar] | – | – | – |
| pRCS2-[bar][GFPi] | 6, supP, agsT | – | – |
| pRCS2-[bar][DsRed2i] | 4, 35SP, 35ST | ||
| pRCS2-[bar][KANi] | – | – | 4, 35SP, 35ST |
| pRCS2-[bar][GFPi][DsRed2i] | 5, nosP, nosT | 6, rbcP, rbcT | – |
| pRCS2-[bar][GFPi][DsRed2i][KANi] | 5, nosP, nosT | 3, masP, masT | 4, 35SP, 35ST |
Numbers indicate the pSAT vector serial numbers.
The pairs of regulatory elements used are: supP, super promoter; agsT, agropine synthase terminator; nosP, nopaline synthase promoter; nosT, nopaline synthase terminator; rbcP, Rubisco small-subunit promoter; rbcT, Rubisco small-subunit terminator; 35SP, CaMV 35S dual promoter; 35ST, CaMV 35S terminator; masP, manopine synthase promoter; and masT, manopine synthase terminator.
We next tested the effect of our single-gene RNAi constructs on silencing of the KAN resistance gene. To this end, we set up a regeneration assay in which we placed leaf discs on regeneration medium supplemented with KAN. As expected, whereas leaf discs from wild-type plants failed to regenerate new shoots on this selectable medium, KGD plants, which were originally selected by their resistance to KAN, maintained their resistance capacity and were capable of regeneration in the presence of KAN (compare wild type with KGD in Fig. 4). More importantly, doubly transformed plants, carrying a KANi (KGD-Ki; Fig. 4), but not DsRed2i (KGD-Di; Fig. 4) or GFPi (KGD-Gi; Fig. 4) expression cassette, failed to regenerate in the presence of KAN, thus indicating that the expression of hpRNA for nptII, but not for gfp or dsred2, suppresses the constitutive expression of the nptII expression cassettes in the KGD transgenic lines. Transformation of KGD plants with the empty vector pRCS2-[bar] did not affect their regeneration capacity (KGD-bar; Fig. 4), further indicating that expression of bar has no specific effect on the expression or suppression of our target transgenes.
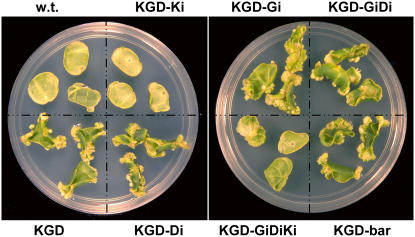
Figure 4.
KAN resistance regeneration assay of single- and double-transformed plants. Leaf discs of diverse transgenic tobacco plants were placed on shoot regeneration medium supplemented with KAN and documented following several weeks in culture. w.t., Wild type, nontransformed plants; KGD, single-transgenic plants produced by overexpressing the KAN, EGFP, and DsRed2 coding genes; KGD-Ki, KGD plants transformed with an nptII silencing cassette (KANi); KGD-Di, KGD plants transformed with a DsRed2-silencing cassette (DsRed2i); KGD-Gi, KGD plants transformed with a gfp silencing cassette (GFPi); KGD-GiDi, KGD plants transformed with GFPi and DsRed2i; KGD-GiDiKi, KGD plants transformed with GFPi, DsRed2i, and KANi; KGD-bar, KGD plants transformed with empty binary vector.
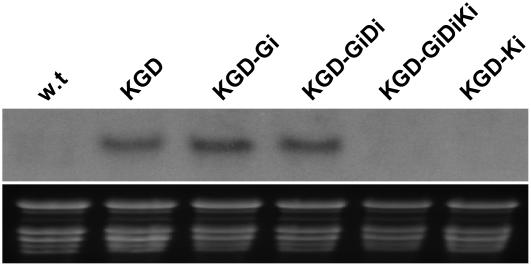
Northern-blot analysis of various transgenic plants revealed the molecular connection between the KANi expression cassette and the resistance to KAN in our transgenic plants. As expected, and in accordance with the KGD plant's resistance to KAN, the nptII-specific band could be observed in KGD but not in wild-type plants (Fig. 5). Furthermore, the nptII transcript was also visible in KGD-Gi, but not KGD-Ki transgenic plants. Here, again, nptII expression was directly linked with KGD-Ki's inability to regenerate on KAN-containing medium (Fig. 4) due to silencing of the nptII transcript by the KANi expression cassette.
Figure 5.
Northern-blot analysis of nptII expression in transgenic tobacco plants. Top, Detection of the nptII transcript by digoxigenin-labeled nptII probe; bottom, total RNA loading. w.t., Wild-type, nontransformed plants; KGD, single-transgenic plants produced by overexpressing the KAN, EGFP, and DsRed2 genes; KGD-Gi, KGD plants transformed with a gfp silencing cassette (GFPi); KGD-Ki, KGD plants transformed with an nptII silencing cassette (KANi); KGD-GiDi, KGD plants transformed with GFPi and dsRed2 silencing cassette (DsRed2i); KGD-GiDiKi, KGD plants transformed with GFPi, DsRed2i, and KANi.
Multiple Marker Gene Suppression
To demonstrate the suitability of our pSAT-RNAi vector system for the suppression of multiple genes from a single construct, we assembled pair and triplet hpRNA expression cassette onto binary plasmids. We first constructed the pSAT5.nosP.GFPi and pSAT6.rbcP.DsRed2i, in which the GFPi and DsRed2i hpRNA expression cassettes were driven under the control of the nosP and rbcP, respectively (Table I). We also constructed pSAT3.masP.DsRed2i in which the expression of DsRed2i hpRNA was driven under the control of the masP (Table I). Next, we mounted both GFPi and DsRed2i hpRNA expression cassettes as I-CeuI and PI-PspI fragments from pSAT5.nosP.GFPi and pSAT6.rbcP.DsRed2i, respectively, onto pRCS2-ocs-bar (Chung et al., 2005), producing pRCS2-[bar][GFPi][DsRed2i] (Table I). We also mounted all three (GFPi, DsRed2i, and KANi) hpRNA expression cassettes as I-CeuI, I-PopI, and I-SceI fragments from pSAT5.nosP.GFPi, pSAT3.masP.DsRed2i, and pSAT4.35SP.KANi, respectively, onto pRCS2-ocs-bar, producing pRCS2-[bar][GFPi][DsRed2i][KANi] (Table I). We then used these constructs in transformation experiments of our KGD transgenic plants. As expected, confocal microscopy analysis of KGD lines transformed by pRCS2-[bar][GFPi][DsRed2i] revealed that these double-transformed plants (designated KGD-GiDi) fail to produce either green (Fig. 3m) or red (Fig. 3N) fluorescence, but are still capable of regenerating on KAN-containing medium (compare wild type with KGD-GiDi in Fig. 4). Similarly, KGD lines transformed with pRCS2-[bar][GFPi][DsRed2i][KANi] failed to produce green or red fluorescence (data not shown). Furthermore, the latter plants (designated KGD-GiDiKi) failed to regenerate in the presence of KAN (compare wild type and KGD-Ki with KGD-GiDiKi in Fig. 4). Northern-blot analysis of KGD-GiDiKi and KGD-GiDi transgenic plants revealed the correlation between the absence (Fig. 5, lane KGD-GiDiKi) or presence (Fig. 5, lane KGD-GiDi) of a stable nptII transcript and their ability to regenerate in the presence of KAN. These results illustrate the suitability of our pSAT-RNAi system for assembly of multiple hpRNA expression cassettes and for suppression of multiple, unrelated genes in plant cells.
Inheritance of RNAi Silencing Cassettes
To further explore the relationship between our suppression cassettes and the phenotypes observed in our doubly transformed plants, we let some of our transgenic plants self-fertilize and set seed. We then analyzed the segregation of their progeny by their ability to germinate in the presence of KAN and/or basta. As shown in Table II, while our wild-type plants failed to germinate in the presence of either of these selection agents, 72% of the offspring of self-fertilized T1 KGD plants were able to grow on KAN-, but not basta-containing medium, indicating a ratio of 3:1 with respect to the KGD transgene. Segregation analyses of T1 KGD-Di plants revealed that while 73% or 74% of the seedlings germinated in the presence of KAN or basta, respectively, only 52% of the offspring germinated when both antibiotics were present in the medium (Table II). These data indicate that these two traits were inherited separately by their offspring at a ratio of 3:1 for each transgene and at a ratio of 9:7 when both selection agents were present in the medium. These segregations clearly fit dihybrid segregation with a complementary interaction between both selection genes and thus with both transgenes.
Table II.
Progeny segregation in single- and double-transformed transgenic plants
| Planta | Resistant Plantsbc
|
|||
|---|---|---|---|---|
| B | K | BK | No Selection | |
| %, survived plants/total seeds | ||||
| Wild type | 0 (0/134) | 0 (0/167) | 0 (0/145) | 98 (155/158) |
| KGD | 0 (0/156) | 72 (125/174) | 0 (0/178) | 96 (159/166) |
| KGD-Di | 73 (156/214) | 74 (115/155) | 52 (70/135) | 99 (183/185) |
| KGD-Ki | 73 (123/168) | 17 (36/213) | 0 (0/172) | 95 (157/165) |
| KGD-DiGiKi | 72 (133/186) | 16 (32/199) | 0 (0/144) | 96 (148/154) |
KGD, GFP, DsRed2, and KAN; Di, DsRed2-RNAi; Ki, KAN-RNAi, Gi, GFP-RNAi.
B, Basta-resistant plants; K, KAN-resistant plants; BK, Basta- and KAN-resistant plants.
Numbers in parentheses represent surviving plants out of the total planted seeds.
Next, we analyzed the segregation pattern of T1 KGD-Ki plants and found a 3:1 segregation ratio (with a germination rate of 73%) of the basta resistance gene (Table II). The rate of germination on KAN-containing medium, however, was much lower than that with KGD-Di plants and resulted in a 3:13 ratio (17%), fitting dominant epistasis-like segregation. This altered segregation ratio of the KAN resistance trait was due to the presence of the basta resistance gene (which is physically linked to the KANi expression cassette), which masks the expression of the KAN resistance gene in double-transformed plants. Similar ratios were also observed when the segregation of KGD-DiGiKi plants was analyzed (Table II), indicating the independent inheritance of both transgenes and the stability of the KANi expression cassette in the second generation. Confocal microscopic analyses of several T2 selected plants revealed a direct link between the presence of the basta resistance gene and the silencing of GFP- and/or DsRed2-encoding genes by their corresponding hpRNA expression cassettes (data not shown), indicating the integrity of the transgene inheritance by the offspring.
SUMMARY
Suppression of multiple unrelated genes in transgenic plants poses a technical challenge that we addressed here by constructing a new set of plant RNAi plasmids. The design of our vectors offers an easy cloning route for the assembly of hpRNA expression cassettes and the flexibility of being able to replace the plasmids' regulatory elements and to simply shuffle hpRNA expression cassettes among various pSAT vector backbones. Furthermore, our system provides the user with the ability to mount several hpRNAi expression cassettes onto a single Agrobacterium binary plasmid and use them in a single transformation event to silence several target genes. The compatibility of our system with previously described plasmids (Chung et al., 2005; Tzfira et al., 2005; Chakrabarty et al., 2007) further expands the user's flexibility because it potentially allows the assembly of complex constructs carrying gene-expressing, fluorescence-tagging, and gene- silencing cassettes. We thus believe that these vectors will further facilitate the discovery of gene functions in plant cells.
MATERIALS AND METHODS
Construction of pSAT-RNAi Vectors
The approximately 1.4-kb-long ChsA intron was PCR amplified from pFGC5941 (Kerschen et al., 2004) using the primers 5′-GGAATTCGTGTAAGAATTTCTTATGTTAC and 5′-TCCCCGCGGTACCGTCGACTGCAGACCTGCAAATTGACCAAAAAAGATG and cloned into the EcoRI-SacII sites of pSAT6-MCS (Tzfira et al., 2005), which resulted in construction of the pSAT6.35S.RNAi plasmid. Insertion of the ChsA intron resulted in splitting of the original single MCS of pSAT6-MCS and assembly of two new MCSs, designated MCS-I and MCS-II, which contained the following unique restriction endonuclease recognition sites: NcoI, BglII, XhoI, SacI, and EcoRI in MCS-I and PstI, SalI, KpnI, SacII, ApaI, XmaI, SmaI, BamHI, and XbaI in MCS-II. In this plasmid, the intron plant gene expression cassette was controlled by the tandem CaMV 35SP and the tobacco etch virus leader sequence from pRTL2-GUS (Restrepo et al., 1990), which was located between the AgeI and NcoI recognition sites and the CaMV 35ST sequence from pRTL2-GUS (Restrepo et al., 1990), which was located between the XbaI and NotI recognition sites. The entire expression cassette was flanked by the PI-PspI recognition sites.
To produce pSAT6.supP.RNAi, we first PCR amplified the approximately 1.2-kb supP sequence from pMSP-1 (Ni et al., 1995) using the primers 5′-TCCCCCCGGGGCGTTGGCCGATTCATTAATGC and 5′-CATGCCATGGCTAGAGTCGATTTGGTGTATC and cloned it as an XmaI-NcoI fragment into the AgeI-NcoI sites of pSAT6-MCS, replacing the tandem CaMV 35SP with the supP and producing pSAT6.supP.MCS.35ST. We next PCR amplified the 406-bp agropine synthase gene poly(A) terminator (agsT) signal from pMSP-1 (Ni et al., 1995) using the primers 5′-GCTCTAGAGAATTAACAGAGGTGGATGGACAGACCC and 5′-ATAAGAATGCGGCCGCATCATCAATATTATAATAAAAATATCC and cloned it as an XbaI-NotI fragment into the same sites of pSAT6.supP.MCS.35ST, replacing the CaMV 35ST with agsT and producing pSAT6.supP.MCS.agsT. We finally cloned the ChsA intron as an EcoRI-KpnI fragment from pSAT6.35SP.RNAi into the same sites of pSAT6.supP.MCS.agsT, producing the final plasmid pSAT6.supP.RNAi.
We transferred the masP and manopine synthase terminator (masT)-controlled expression cassette from pSAT3.masP.MCS.masT (Chung et al., 2005) as an AgeI-NotI fragment into the same sites of pSAT6-MCS (Tzfira et al., 2005; replacing the CaMV 35S-controlled expression cassette) to produce pSAT6.masP.MCS.masT. We also transferred the nosP- and nopaline synthase terminator (nosT)-controlled expression cassette from pSAT2.nosP.MCS.nosT (Chung et al., 2005) as an AgeI-NotI fragment into the same sites of pSAT5-MCS (Tzfira et al., 2005; replacing the CaMV 35S-controlled expression cassette) to produce pSAT5.nosP.MCS.nosT.
To make pSAT3.masP.RNAi, pSAT5.nosP.RNAi, pSAT4.35SP.RNAi, pSAT6.rbcP.RNAi, and pSAT6.masP.RNAi, we cloned the ChsA intron as an EcoRI-KpnI fragment from pSAT6.35SP.RNAi into the same sites of pSAT3.masP.MCS.masT, pSAT5.nosP.MCS.nosT, pSAT4.35SP.MCS.35ST (Tzfira et al., 2005), pSAT6.rbcP.MCS.rbcT (Chung et al., 2005), and pSAT6.masP.MCS.masT (Chung et al., 2005), respectively.
All PCRs were performed using a high-fidelity Pfu DNA polymerase (Stratagene) and their products were verified by DNA sequencing. Full DNA sequences of pSAT6.35SP.RNAi, pSAT6.supP.RNAi, pSAT3.masP.RNAi, pSAT5.nosP.RNAi, pSAT4.35SP.RNAi, pSAT6.rbcP.RNAi, and pSAT6.masP.RNAi vectors were deposited in GenBank and assigned the following accession numbers: EU049862, EU049861, EU049860, EU049865, EU049859, EU049864, and EU049863, respectively.
Construction of Binary Plasmid for Transgenic Overexpression
We cloned the EGFP-encoding sequence as an NcoI-XbaI fragment from pSAT6-EGFP-C1 into the same sites of pSAT2.nosP.MCS.nosT (Chung et al., 2005), producing pSAT2.nosP.EGFP.nosT. We cloned the DsRed2-encoding sequence as XhoI-XbaI from pSAT6-DsRed2-N1 into the same sites of pSAT6.rbcP.MCS.rbcT (Chung et al., 2005), producing pSAT6.rbcP.DsRed2.rbcT. We next transferred the EGFP and DsRed2 expression cassettes as AscI-I-PopI and PI-PspI-PI-PspI fragments from pSAT2.nosP.EGFP.nosT and pSAT6.rbcP.DsRed2.rbcT into the corresponding sites of pRCS2-nptII (Tzfira et al., 2005), producing pRCS2-[EGFP][KAN][DsRed2].
Cloning of Tested Genes into pSAT-RNAi Vectors
We constructed several vectors capable of expressing long hp-dsRNAs suitable for suppressing the expression of GFP, DsRed2, and KAN in plant cells. More specifically, about 500 bp of the GFP open reading frame (ORF) were cloned as a PCR-amplified fragment from pSAT6-EGFP-C1 using the primers 5′-CATGCCATGGGGATCCAAGTTCAGCGTGTCCGGCG and 5′-CCGCTCGAGGGGCCGTCGCCGATGGGGG. This fragment was digested by either NcoI-XhoI or XhoI alone, and the fragments were successively cloned, first into the NcoI-XhoI sites of MCS-I and then into the SmaI-SalI sites of MCS-II of pSAT6.sup.RNAi, producing pSAT6.sup.GFPi. A similar strategy was employed using the plasmid pSAT5.nosP.RNAi to produce pSAT5.nosP.GFPi. In addition, about 310 bp of the DsRed2 ORF were cloned as a PCR-amplified fragment from pSAT6-DsRed2-C1 (Tzfira et al., 2005) using the primers 5′-CATGCCATGGGGATCCCCTGGGTCACGGTCGC and 5′-CCGCTCGAGAACGTCATCACCGAGTTCATGC. This fragment was digested by either BamHI-XhoI or NcoI-XhoI, and the fragments were successively cloned, first into the BamHI-SalI sites of MCS-II and then into the NcoI-XhoI sites of MCS-I of pSAT4.35SP.RNAi, producing pSAT4.35S.DsRed2i. A similar strategy was employed using the plasmids pSAT3.masP.RNAi and pSAT6.rbcP.RNAi to produce pSAT3.masP.DsRed2i and pSAT6.rbcP.DsRed2i, respectively. Finally, about 310 bp of the nptII ORF were cloned as a PCR-amplified fragment from pSAT4-nptII (Tzfira et al., 2005) using the primers 5′-CATGCCATGGGGATCCAAGCGGGAAGGGACTGGC and 5′-CCGCTCGAGCGACGAGATCATCGCCGTC. This fragment was digested by either BamHI-XhoI or NcoI-XhoI alone, and the fragments were successively cloned, first into the BamHI-SalI sites of MCS-II and then into the NcoI-XhoI sites of MCS-I of pSAT4.35SP.RNAi, producing pSAT4.35S.KANi.
Construction of Binary RNAi Plasmids
For down-regulation of GFP alone, we transferred the GFPi expression cassette from pSAT6.sup.GFPi as a PI-PspI-PI-PspI fragment and cloned it into the corresponding site of pRCS2-ocs-bar (Chung et al., 2005), producing pRCS2-[bar][GFPi].
For down-regulation of DsRed2 alone, we transferred the DsRed2i cassette from pSAT4.35S.DsRed2i as a I-SceI-I-SceI fragment and cloned it into the corresponding site of pRCS2-ocs-bar, producing pRCS2-[bar][DsRed2i]. For down-regulation of nptII alone, we transferred the KANi cassette from pSAT4.35S.KANi as a I-SceI-I-SceI fragment and cloned it into the corresponding site of pRCS2-ocs-bar, producing pRCS2-[bar][KANi]. For the combined down-regulation of GFP and DsRed2, we transferred the GFPi and DsRed2 cassettes from pSAT5.nosP.GFPi and pSAT6.rbcP.DsRed2i as I-CeuI-I-CeuI and PI-PspI-PI-PspI fragments, respectively, and cloned them into the corresponding sites of pRCS2-ocs-bar, producing pRCS2-[bar][GFPi][DsRed2i]. For the combined down-regulation of GFP, DsRed2, and nptII, we transferred the GFPi, DsRed2, and KANi cassettes from pSAT5.nosP.GFPi, pSAT3.masP.DsRed2i, and pSAT4.35S.KANi as I-CeuI-I-CeuI, I-PopI-I-PopI, and PI-PspI-PI-PspI fragments, respectively, and cloned them into the corresponding sites of pRCS2-ocs-bar, producing pRCS2-[bar][GFPi][DsRed2i][KANi].
Production and Analysis of Transgenic Plants
Binary plasmids were introduced into Agrobacterium tumefaciens strain EHA105 using the calcium chloride transformation protocol (Tzfira et al., 1997). The resulting bacterial cultures were used to genetically transform tobacco (Nicotiana tabacum) turk leaf discs according to Horsch et al. (1985). Transgenic plants were first produced using the pRCS2-[EGFP][KAN][DsRed2] plasmid and selected in the presence of 50 μg/mL KAN. Selected lines were retransformed using various RNAi binary plasmids and double-transformed plants were selected in the presence of 20 μg/mL basta. Plants were maintained in tissue culture on basal Murashige and Skoog medium (Murashige and Skoog, 1962) with no exogenous growth regulators. An in vitro regeneration assay was performed by placing aseptic leaf discs from in vitro-grown plants on regeneration medium (Horsch et al., 1985) supplemented with KAN. Selected lines were cultivated in commercial compost in a regular growth chamber. Transformants were allowed to self-pollinate and seeds from individual plants were pooled, surface sterilized, and allowed to germinate on one-half-strength Murashige and Skoog medium (0.8% [w/v] agar and 3% [w/v] Suc) supplemented with the appropriate combinations of KAN and basta. Resistant plants were counted and the transgene segregation ratio was calculated.
Confocal Microscopy
Plant tissues were viewed directly under a Leica confocal microscope. Two to five independent clones were analyzed for expression of EGFP and DsRed2 and four to five leaves were analyzed from each individual plant. In addition, 15 to 20 seedlings were randomly analyzed for expression of EGFP and DsRed2 for each self-fertilized clone.
RNA Gel-Blot Analysis
Total RNA was isolated from approximately 200 mg of leaf tissue as previously described. RNA samples (10 μg/lane) were electrophoresed on a 1.2% formaldehyde/agarose gel, blotted onto Hybond N+ membranes, and probed with digoxigenin-labeled nptII probe followed by autoradiography using standard hybridization and detection protocols. rRNA within the analyzed RNA preparation was detected by ethidium bromide staining of agarose gels and served as an internal control for equal loading of the lanes.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EU049859 to EU049865.
Acknowledgments
The plasmid pFGC5941 was kindly provided by the Arabidopsis Biological Resource Center.
This work was supported by grants from the Human Frontiers Science Program, the Biotechnology Research and Development Cooperation, and University of Michigan startup funds.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Tzvi Tzfira (ttzfira@umich.edu).
Open Access articles can be viewed online without a subscription.
References
- Abbott JC, Barakate A, Pincon G, Legrand M, Lapierre C, Mila I, Schuch W, Halpin C (2002) Simultaneous suppression of multiple genes by single transgenes: down-regulation of three unrelated lignin biosynthetic genes in tobacco. Plant Physiol 128 844–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal N, Dasaradhi PV, Mohmmed A, Malhotra P, Bhatnagar RK, Mukherjee SK (2003) RNA interference: biology, mechanism, and applications. Microbiol Mol Biol Rev 67 657–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruening G (1998) Plant gene silencing regularized. Proc Natl Acad Sci USA 95 13349–13351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher E, Lohuis D, van Poppel PM, Geerts-Dimitriadou C, Goldbach R, Prins M (2006) Multiple virus resistance at a high frequency using a single transgene construct. J Gen Virol 87 3697–3701 [DOI] [PubMed] [Google Scholar]
- Chabannes M, Barakate A, Lapierre C, Marita JM, Ralph J, Pean M, Danoun S, Halpin C, Grima-Pettenati J, Boudet AM (2001) Strong decrease in lignin content without significant alteration of plant development is induced by simultaneous down-regulation of cinnamoyl CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD) in tobacco plants. Plant J 28 257–270 [DOI] [PubMed] [Google Scholar]
- Chakrabarty R, Banerjee R, Chung S-M, Farman M, Citovsky V, Hogenhout SA, Tzfira T, Goodin M (2007) pSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: probing Nicotiana benthamiana-virus interactions. Mol Plant Microbe Interact 20 740–750 [DOI] [PubMed] [Google Scholar]
- Chung SM, Frankman EL, Tzfira T (2005) A versatile vector system for multiple gene expression in plants. Trends Plant Sci 10 357–361 [DOI] [PubMed] [Google Scholar]
- Dai M, Hu Y, Zhao Y, Liu H, Zhou DX (2007) A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiol 144 380–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45 616–629 [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411 494–498 [DOI] [PubMed] [Google Scholar]
- Goderis IJ, De Bolle MF, Francois IE, Wouters PF, Broekaert WF, Cammue BP (2002) A set of modular plant transformation vectors allowing flexible insertion of up to six expression units. Plant Mol Biol 50 17–27 [DOI] [PubMed] [Google Scholar]
- Guo HS, Fei JF, Xie Q, Chua NH (2003) A chemical-regulated inducible RNAi system in plants. Plant J 34 383–392 [DOI] [PubMed] [Google Scholar]
- Hamilton A, Voinnet O, Chappell L, Baulcombe D (2002) Two classes of short interfering RNA in RNA silencing. EMBO J 21 4671–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286 950–952 [DOI] [PubMed] [Google Scholar]
- Helliwell C, Waterhouse P (2003) Constructs and methods for high-throughput gene silencing in plants. Methods 30 289–295 [DOI] [PubMed] [Google Scholar]
- Hirai S, Oka S, Adachi E, Kodama H (2007) The effects of spacer sequences on silencing efficiency of plant RNAi vectors. Plant Cell Rep 26 651–659 [DOI] [PubMed] [Google Scholar]
- Horiguchi G (2004) RNA silencing in plants: a shortcut to functional analysis. Differentiation 72 65–73 [DOI] [PubMed] [Google Scholar]
- Horsch RB, Fry JE, Hoffman NL, Eichholtz D, Rogers SG, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227 1229–1231 [DOI] [PubMed] [Google Scholar]
- Huang G, Allen R, Davis EL, Baum TJ, Hussey RS (2006) Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc Natl Acad Sci USA 103 14302–14306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ifuku K, Yamamoto Y, Sato F (2003) Specific RNA interference in psbP genes encoded by a multigene family in Nicotiana tabacum with a short 3′-untranslated sequence. Biosci Biotechnol Biochem 67 107–113 [DOI] [PubMed] [Google Scholar]
- Kerschen A, Napoli CA, Jorgensen RA, Muller AE (2004) Effectiveness of RNA interference in transgenic plants. FEBS Lett 566 223–228 [DOI] [PubMed] [Google Scholar]
- Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA (1999) Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol 17 969–973 [DOI] [PubMed] [Google Scholar]
- McGinnis K, Chandler V, Cone K, Kaeppler H, Kaeppler S, Kerschen A, Pikaard C, Richards E, Sidorenko L, Smith T, et al (2005) Transgene-induced RNA interference as a tool for plant functional genomics. Methods Enzymol 392 1–24 [DOI] [PubMed] [Google Scholar]
- McGinnis K, Murphy N, Carlson AR, Akula A, Akula C, Basinger H, Carlson M, Hermanson P, Kovacevic N, McGill MA, et al (2007) Assessing the efficiency of RNA interference for maize functional genomics. Plant Physiol 143 1441–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mette MF, van der Winden J, Matzke MA, Matzke AJ (1999) Production of aberrant promoter transcripts contributes to methylation and silencing of unlinked homologous promoters in trans. EMBO J 18 241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Nowak K, Sharma VK, Schulze J, Mendel RR, Hansch R (2004) Vectors for RNAi technology in poplar. Plant Biol (Stuttg) 6 100–103 [DOI] [PubMed] [Google Scholar]
- Miki D, Itoh R, Shimamoto K (2005) RNA silencing of single and multiple members in a gene family of rice. Plant Physiol 138 1903–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki D, Shimamoto K (2004) Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol 45 490–495 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Napoli C, Lemieux C, Jorgensen RA (1990) Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Cui D, Einstein J, Narasimhulu S, Vergara C, Gelvin SB (1995) Strength and tissue specificity of chimaeric promoters derived from the octopine and mannopine synthase genes. Plant J 7 661–676 [Google Scholar]
- Ogita S, Uefuji H, Yamaguchi Y, Koizumi N, Sano H (2003) Producing decaffeinated coffee plants. Nature 423 823. [DOI] [PubMed] [Google Scholar]
- Pincon G, Chabannes M, Lapierre C, Pollet B, Ruel K, Joseleau JP, Boudet AM, Legrand M (2001) Simultaneous down-regulation of caffeic/5-hydroxy ferulic acid-O-methyltransferase I and cinnamoyl-coenzyme A reductase in the progeny from a cross between tobacco lines homozygous for each transgene: consequences for plant development and lignin synthesis. Plant Physiol 126 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo MA, Freed DD, Carrington JC (1990) Nuclear transport of plant potyviral proteins. Plant Cell 2 987–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal G, Song R, Messing J (2003) A new opaque variant of maize by a single dominant RNA-interference-inducing transgene. Genetics 165 387–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I (2007) RNAi for revealing and engineering plant gene functions. Curr Opin Biotechnol 18 148–153 [DOI] [PubMed] [Google Scholar]
- Stoutjesdijk PA, Singh SP, Liu Q, Hurlstone CJ, Waterhouse PA, Green AG (2002) hpRNA-mediated targeting of the Arabidopsis FAD2 gene gives highly efficient and stable silencing. Plant Physiol 129 1723–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenllado F, Llave C, Diaz-Ruiz JR (2004) RNA interference as a new biotechnological tool for the control of virus diseases in plants. Virus Res 102 85–96 [DOI] [PubMed] [Google Scholar]
- Travella S, Klimm TE, Keller B (2006) RNA interference-based gene silencing as an efficient tool for functional genomics in hexaploid bread wheat. Plant Physiol 142 6–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Jensen CS, Wangxia W, Zuker A, Altman A, Vainstein A (1997) Transgenic Populus: a step-by-step protocol for its Agrobacterium-mediated transformation. Plant Mol Biol Rep 15 219–235 [Google Scholar]
- Tzfira T, Tian G-W, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V (2005) pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol 57 503–516 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Lederer C, Baulcombe DC (2000) A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103 157–167 [DOI] [PubMed] [Google Scholar]
- Waterhouse PM, Graham MW, Wang MB (1998) Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Natl Acad Sci USA 95 13959–13964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JM, Fusaro AF, Wang M, Waterhouse PM (2005) RNA silencing platforms in plants. FEBS Lett 579 5982–5987 [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27 581–590 [DOI] [PubMed] [Google Scholar]
- Winston WM, Molodowitch C, Hunter CP (2002) Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295 2456–2459 [DOI] [PubMed] [Google Scholar]
- Wrobel-Kwiatkowska M, Starzycki M, Zebrowski J, Oszmianski J, Szopa J (2007) Lignin deficiency in transgenic flax resulted in plants with improved mechanical properties. J Biotechnol 128 919–934 [DOI] [PubMed] [Google Scholar]
- Ye X, Al-Babili S, Kloti A, Zhang J, Lucca P, Beyer P, Potrykus I (2000) Engineering the provitamin A (beta-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287 303–305 [DOI] [PubMed] [Google Scholar]
- Zhang J, Subramanian S, Zhang Y, Yu O (2007) Flavone synthases from Medicago truncatula are flavanone-2-hydroxylases and are important for nodulation. Plant Physiol 144 741–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Iii WH, Negrel J, Ye ZH (1998) Dual methylation pathways in lignin biosynthesis. Plant Cell 10 2033–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Richardson EA, Ye ZH (2007) Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 225 1603–1611 [DOI] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen SE (2003) ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299 716–719 [DOI] [PubMed] [Google Scholar]