Abstract
Transient expression is a rapid, useful approach for producing proteins of interest in plants. Tobacco mosaic virus (TMV)-based transient expression vectors can express very high levels of foreign proteins in plants. However, TMV vectors are, in general, not efficiently delivered to plant cells by agroinfection. It was determined that agroinfection was very efficient with a 35S promoter-driven TMV replicon that lacked the TMV coat protein gene sequence. This coat protein deletion vector had several useful features as a transient expression system, including improved ease of use, higher protein expression rates, and improved biocontainment. Using this TMV expression vector, some foreign proteins were expressed at levels of 3 to 5 mg/g fresh weight of plant tissue. It is proposed that this new transient expression vector will be a useful tool for expressing recombinant proteins in plants for either research or production purposes.
As interest in proteomics, biochemistry, and protein structure increases, there is an increasing need for efficient, easy-to-use recombinant protein expression systems. For researchers interested in expressing recombinant proteins in plants, there are multiple transient expression systems available. However, every protein expression system has inherent advantages and disadvantages. Improving transient expression vectors so they are easier to use, more cost effective, and produce higher levels of recombinant proteins will be of use to the wide variety of researchers who use recombinant proteins in research or development. A recent study has shown that producing recombinant plant proteins in plant cells, instead of yeast (Saccharomyces cerevisiae) or Escherichia coli cells, is more likely to result in the production of properly folded, active plant proteins (Popescu et al., 2007).
A variety of viral- and nonviral-based plant transient protein expression systems have been described in the literature (Voinnet et al., 2003; for review, see Scholthof et al., 1996; Pogue et al., 2002; Gleba et al., 2007). The technically simplest plant transient expression systems take advantage of the ability of Agrobacterium tumefaciens to transfer DNA into plant cells (Grimsley et al., 1986; Grimsley, 1995). A. tumefaciens cell suspensions infiltrated (or injected) into leaves can efficiently transfer sequences from the T-DNA region of a modified A. tumefaciens Ti (binary) plasmid into plant cells where the T-DNA becomes integrated into the cell DNA. If the T-DNA transferred into the plant cell contains a DNA sequence of interest joined to a plant-functional promoter, the transferred DNA will be transcribed in the plant nucleus. Because this system is so efficient, easy, and inexpensive to use, it has become a very commonly used strategy for producing proteins in plants (Popescu et al., 2007). One disadvantage of this approach is that expression from T-DNA is generally quite low and transient and expression drops off after 5 d or so. It was demonstrated that posttranscriptional gene silencing directed toward the transcribed T-DNA was being induced in the plant after agroinfiltration (Voinnet et al., 2003). It was determined that this could be at least partially overcome by using two different A. tumefaciens cultures to simultaneously cointroduce T-DNA for both a cauliflower mosaic virus (CaMV) 35S promoter-driven gene of interest and a 35S-driven RNA-silencing suppressor gene into cells. Ectopic transient expression of an RNA-silencing suppressor protein (such as the P19 protein from Tomato bushy stunt virus) suppressed the posttranscriptional gene silencing of the cointroduced T-DNA (Voinnet et al., 2003). This resulted in an increase in the amount of recombinant protein expressed. For some proteins, ectopic coexpression of P19 resulted in a nearly 50-fold increase in recombinant protein expression levels (Voinnet et al., 2003). Using this strategy, hundreds of plant proteins have been expressed in relatively high-throughput fashion (Popescu et al., 2007). One limitation of this approach is that relatively high concentrations of A. tumefaciens cell suspensions must be infiltrated into leaves to get the highest expression levels possible. For some plant species, infiltration of such high concentrations of A. tumefaciens can elicit negative (hypersensitive) responses from the plant (Wroblewski et al., 2005).
Other transient expression systems are based on plant viruses, such as Tobacco mosaic virus (TMV), for example. Detailed descriptions of TMV and TMV-based transient expression vectors have been described elsewhere (Pogue et al., 1998; Creager et al., 1999; Scholthof, 2004) and will therefore be only briefly described here. TMV is a rod-shaped virus that has a single-stranded plus-sense RNA genome. TMV expresses four proteins from three open reading frames (ORFs). Two viral genes (the viral movement protein and the capsid protein) are expressed from separate subgenomic promoters. TMV has typically been modified to express foreign genes by either replacing a viral gene (such as the coat protein [CP] gene, for example) with a gene of interest (for review, see Scholthof et al., 1996) or by inserting an additional subgenomic promoter (Dawson et al., 1989; Donson et al., 1991; Pogue et al., 1998) into the viral genome to drive the expression of an inserted foreign gene.
Plants can be inoculated with TMV vectors through a process called agroinfection. In agroinfection, A. tumefaciens was used to deliver T-DNA composed of 35S promoter-driven TMV cDNA to plant cells. Transcription of T-DNA in the plant nucleus generated RNA that was capable of initiating self-replication in the cytoplasm. Multiple reports have documented the low agroinfection efficiency of the typical 35S-driven TMV vector (Turpen et al., 1993; Marillonnet et al., 2005; Man and Epel, 2006).
Here, we report on the construction of an improved agroinfection-compatible TMV vector that lacks the TMV CP gene coding sequence. This modification resulted in a vector with several significant improvements, such as (1) much higher agroinfection efficiency; (2) higher recombinant protein expression levels; and (3) inability to form virus particles during its infection/replication cycle. This new expression vector is called the TMV RNA-based overexpression (TRBO) vector. Here, we demonstrate that the TRBO vector can produce up to 100 times more recombinant protein than the P19-enhanced agroinfiltration transient expression system described above.
It is proposed that, because of its efficacy and ease of use, the TRBO vector will be a useful transient expression vector for production of recombinant proteins in plants for either research or production purposes.
RESULTS
Plasmids Constructed for This Experiment
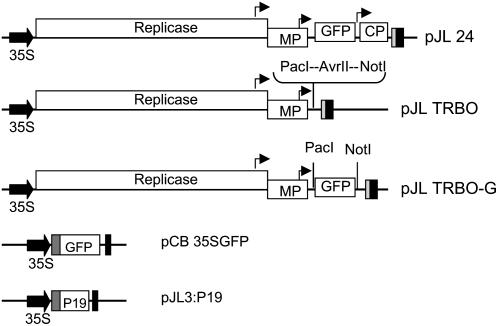
We previously constructed an agroinfection-compatible full-length TMV expression vector called pJL24 (Fig. 1) that expressed all of the TMV genes in addition to a foreign, inserted gene (Lindbo, 2007). It had been demonstrated by others that moving the foreign gene insertion site closer to the 3′ end of TMV RNA led to an increase in expression of the foreign insert (Culver et al., 1993). Because deletion of the virus CP gene sequence from the plasmid pJL24 in essence moves the foreign insert closer to the 3′ end of the viral RNA, we hypothesized that deletion of the CP gene sequence from pJL24 would increase the level of foreign gene expression from a TMV replicon. Initially, the GFP reporter gene (Chalfie et al., 1994; Chalfie, 1995; Crameri et al., 1996) was used to demonstrate the utility of the agroinfection-compatible TRBO replicon and to compare it to alternate nonviral or full-length TMV transient expression systems. Maps of the T-DNA regions of various modified Ti (binary) plasmids used in this study are shown in Figure 1.
Figure 1.
Maps of plasmids used in this project. The T-DNA regions of binary plasmids used in this project are represented. Block arrow, CaMV duplicated 35S promoter. Black box, CaMV polyA signal sequence/terminator. Dark gray box, Tobacco etch virus 5′-nontranslated leader sequence. Light gray box, Ribozyme. Bent arrows, Subgenomic promoters. ORFs are represented by white boxes. Identities of ORFs are labeled in white boxes. Replicase, TMV 126K/183K ORF; MP, movement protein; P19, 19-kD RNA-silencing suppressor gene from Tomato bushy stunt virus.
Agroinfection with the TRBO Replicon Is Very Efficient
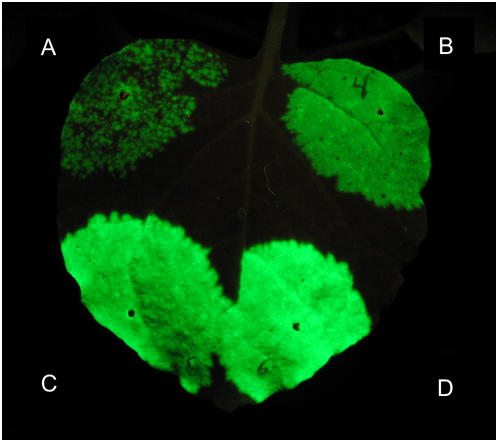
Previous work with the TMV vector contained in pJL24 determined that ectopic coexpression of an RNA-silencing suppressor gene (such as the P19 gene from Tomato bushy stunt virus) was needed to obtain the highest agroinfection rates for this vector (Lindbo, 2007). An example of this can be seen in Figure 2. The area of the leaf in Figure 2A was infiltrated with a suspension of A. tumefaciens cells carrying pJL24 (A.t./pJL24). About 4 d postinfiltration (dpi), when the leaf was viewed under UV illumination (to visualize expression of GFP from the TMV vector launched from the T-DNA), many discreet GFP-positive foci could be seen with the unaided eye. However, a significant portion of the infiltrated area did not express detectable levels of GFP. In contrast, when A.t./pJL24 cells were mixed with a suspension of A.t./pJL3:P19 cells and the mixture infiltrated into leaf tissue, GFP expression in the infiltrated zone appeared confluent (Fig. 2B). Very few, if any, non-GFP-expressing cells were observed even when examined under a fluorescent microscope (data not shown). Because the most efficient agroinfection rate with pJL24 required cointroduction of a 35S-driven P19 gene, we sought to determine whether coexpression of the RNA-silencing suppressor protein P19 was also needed to obtain a high agroinfection rate with pJL-TRBO-G (a GFP-expressing TRBO vector). To test this, A.t./pJL-TRBO-G cells alone (Fig. 2C) or mixed with A.t./pJL3:P19 cells (Fig. 2D) were infiltrated into separate areas of a leaf. Surprisingly, the agroinfection rate of the TRBO-G vector in the two treatments appeared identical. When infiltrated leaves were viewed under a hand-held long-wave UV lamp at 4 dpi, all cells in the area of the leaf infiltrated with A.t./pJL-TRBO-G or A.t./pJL-TRBO-G + A.t./pJL3:P19 cells appeared to be expressing GFP (Fig. 2, C and D). In addition, the GFP signal from the TRBO-G replicon was noticeably brighter than the GFP signal from JL24 (Fig. 2, A and B). These results were observed in dozens of repetitions of this experiment.
Figure 2.
Comparison of agroinfection efficiency of pJL24 and pJL-TRBO vectors. T-DNAs of TMV-based expression vectors were introduced into N. benthamiana by agroinfection. Sections of an N. benthamiana leaf were infiltrated with A. tumefaciens (A.t.) cell suspensions transformed with plasmids as follows. A, A.t./pJL24 (OD600 1.0). B, Mixture of A.t./pJL24 + A.t./pJL3:P19 (each at final OD600 of 0.5). C, A.t./pJL-TRBO-G (OD600 1.0). D, Mixture of A.t./pJL-TRBO-G + A.t./pJL3:P19 (each at final OD600 of 0.5). Photo taken under UV illumination 4 dpi. In grayscale, GFP fluorescence appears as a light color. [See online article for color version of this figure.]
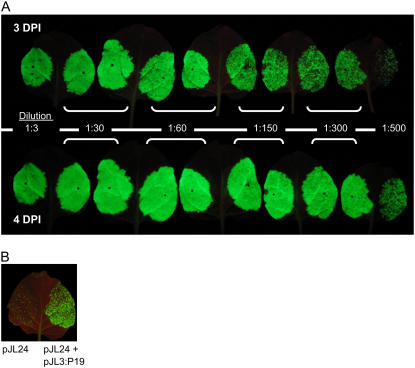
Because the pJL-TRBO-G vector had a higher rate of agroinfection than the vector pJL24, we determined whether dilute suspensions of A.t./pJL-TRBO-G alone could be used to efficiently inoculate leaves. Half-leaves of Nicotiana benthamiana plants were infiltrated with various dilutions of A.t./pJL-TRBO-G cells. Leaves were observed daily with a hand-held UV lamp to monitor the progress and extent of agroinfection, as demonstrated by GFP expression from the TRBO-G replicon. Results are shown in Figure 3. Even in leaves infiltrated with A. tumefaciens cells diluted 1:300 from a starting optical density (OD600) of 1.0, nearly all cells of the infiltrated zone expressed GFP by 4 dpi, as determined by visual inspection of infiltrated leaves under a hand-held UV lamp. Thus, plants can be efficiently agroinoculated with the TRBO-G replicon over a wide range of A.t./pJL-TRBO-G cell densities.
Figure 3.
Effect of A. tumefaciens (A.t.) cell density on agroinfection of plants with pJL-TRBO-G expression vector. Leaves of N. benthamiana plants were infiltrated with A.t. cell suspensions transformed with various binary (modified Ti) plasmids. A.t. cell suspensions were diluted, as noted in the figure, from an initial OD600 of 1.0. A, Individual leaves infiltrated with A.t./pJL-TRBO-G cell suspensions were photographed under UV illumination at 3 and 4 dpi as noted. B, Left half of leaf infiltrated with 1:100 dilution of A.t./pJL24. Right half of leaf infiltrated with a mixture of 1:100 dilution of A.t./pJL24 and 1:10 dilution of A.t./pJL3:P19. Photo taken under UV illumination 3 dpi. [See online article for color version of this figure.]
To further demonstrate that the agroinfection rate of A.t./pJL-TRBO-G is higher than that of A.t./pJL24, a 1:100 dilution (from a starting OD of 1.0) of A.t./pJL24 cells was infiltrated into N. benthamiana leaves with or without A.t./pJL3:P19 cells. The image in Figure 3B is a photograph (3 dpi) of such an infiltrated leaf under UV illumination. Again, coinfiltration of a mixture of A.t./pJL24 and A.t./pJL3:P19 cell suspensions dramatically increased the agroinfection rate of the pJL24 vector. The amount of GFP-expressing tissue in this treatment appeared similar visually to the amount of GFP-expressing tissue (at 3 dpi) in leaves infiltrated with 1:150 or 1:300 dilutions of A.t./pJL-TRBO-G cells (Fig. 3A). This further demonstrates that the agroinfection rate of pJL-TRBO-G is significantly higher than that of pJL24 and that even diluted A.t./pJL-TRBO-G cell suspensions can be used to efficiently inoculate leaves in the absence of an ectopically expressed RNA-silencing suppressor, such as P19.
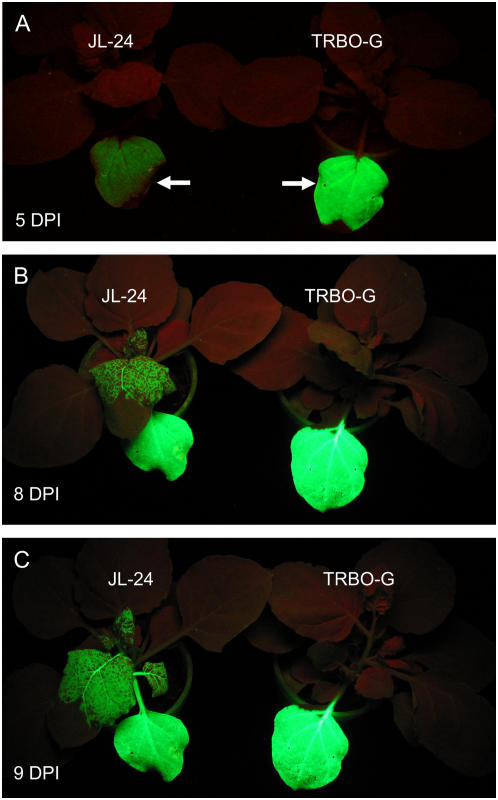
The TRBO Expression Vector Did Not Move Systemically in Plants
Because the TMV CP is required for systemic movement (Donson et al., 1991), the TRBO-G replicon was not expected to move systemically in plants. To test this, N. benthamiana plants were inoculated with the GFP-expressing vectors JL24 or TRBO-G by agroinfection. Plants were observed under UV illumination to visualize GFP expression from the vectors. Results are shown in Figure 4. The vector JL24 expressed GFP plus all of the genes of TMV, including the CP, and moved systemically (to noninoculated leaves) at about 6 dpi. By about 9 to 10 dpi, the majority of the tissue in systemically infected leaves (as viewed with the unaided eye under UV illumination) appeared to be expressing GFP. In contrast, the TRBO-G replicon did not move systemically in plants, even up to 14 dpi (data not shown). The TRBO-G replicon was never observed to move systemically in any of the dozens of agroinoculated plants in our experiments.
Figure 4.
TRBO-G replicon does not move systemically in plants. One leaf of an N. benthamiana plant was infiltrated with A. tumefaciens carrying pJL24 or pJL-TRBO-G plasmids. Plants were photographed under UV light to visualize the GFP expressed by either expression vector. [See online article for color version of this figure.]
TRBO Vector Expressed Very High Levels of Recombinant Protein
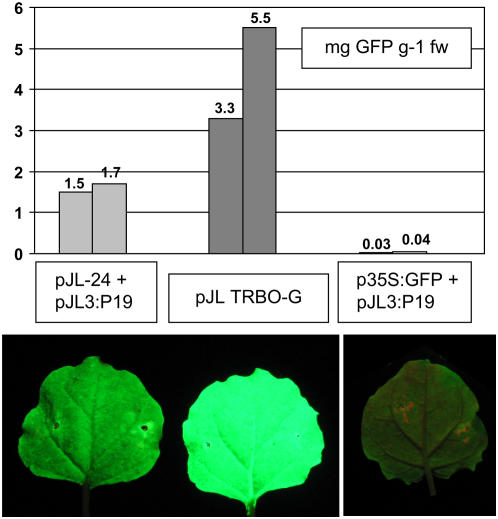
To compare the amounts of GFP produced from the TMV vectors JL24 and TRBO-G, or from the transient coexpression of 35S:GFP and the RNA-silencing suppressor protein P19, a plate-based GFP fluorescence assay was used. Purified His-6-tagged GFP, purified from TRBO-G-infected plants by metal affinity chromatography, was used as a standard. Leaves of N. benthamiana were infiltrated with one of the following A. tumefaciens cell suspensions: A.t./p35S:GFP + A.t./pJL3:P19 (each at an OD600 of 0.5); A.t./pJL24 + A.t./pJL3:P19 (each at an OD600 of 0.5); or A.t./pJL-TRBO-G (OD600 0.02). Protein samples from infiltrated tissues were prepared at 5 or 6 dpi. Dilutions of protein extracts and purified His-6-tagged GFP were transferred into wells of a 96-well plate (in triplicate). GFP fluorescence levels were recorded on a Perkin-Elmer HTS 7000 BioAssay plate reader. The results are shown in Figure 5. The TRBO-G replicon expressed up to 100 times more GFP than was obtained from cointroducing T-DNAs for 35S:GFP and 35S:P19 into plants, and 2 to 3 times more GFP than the TMV vector JL24. Similar results were also obtained from an ELISA assay, using anti-GFP-specific antibodies (data not shown). In multiple repetitions of this experiment, the relative expression levels from the different expression systems were always consistent. The TRBO-G replicon always expressed significantly more GFP than the other transient expression systems examined.
Figure 5.
Quantitative analysis of GFP expression levels from TMV vectors JL24 and TRBO-G. Leaves of N. benthamiana were infiltrated with A. tumefaciens cells transformed with plasmids identified in the figure. Bottom, Images of individual infiltrated leaves photographed under UV illumination at 4 dpi. Top, Quantitation of GFP fluorescence activity levels in extracts prepared from infiltrated leaves 6 dpi. Extracts were analyzed by a plate-based GFP fluorescence assay. Purified recombinant His-6-tagged GFP was used to generate a standard curve. Results are presented in micrograms GFP produced per gram of infiltrated tissue. [See online article for color version of this figure.]
Temporal Analysis of Protein Expression from TRBO
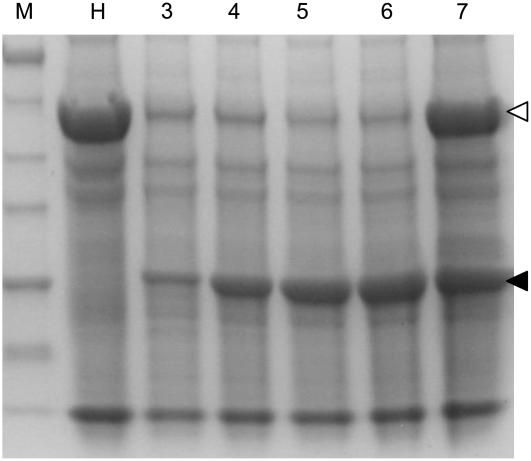
After pJL-TRBO-G T-DNA is transcribed, RNA initiates self-replication and gene expression in the cytoplasm. Because the TRBO-G replicon expressed the TMV movement protein, it moved cell to cell in the inoculated (infiltrated) leaf. The result of this movement is that individual GFP-expressing foci on a leaf enlarge as the virus moves cell to cell over time. This can be observed by comparing the sizes of individual GFP-expressing cell foci in 3- and 4-dpi images (Fig. 3A) of leaves infiltrated with 1:500 dilutions of A.t./pJL-TRBO-G. When leaves were infiltrated with higher concentrations of A.t./pJL-TRBO-G cells, the large number of GFP-expressing cells in the infiltrated zone made it difficult to identify an individual focus. Regardless, as replication and cell-to-cell movement of the replicon progressed, the amount of GFP expressed in the infiltrated leaf increased. After a certain point, the steady-state level of GFP in infiltrated tissue appeared to reach a plateau. To demonstrate the temporal nature of protein expression from the TRBO-G replicon, extracts were prepared from inoculated tissue at various days postinfiltration. Total soluble protein extracts were analyzed by SDS-PAGE and Coomassie Blue staining (Fig. 6). TRBO-G-expressed GFP accumulation appeared to plateau by 4 to 6 dpi, consistent with the increase in GFP activity that is observed by viewing infiltrated leaves under UV illumination. Samples in lanes 3 to 6 went through a freeze-thaw cycle that results in the precipitation of a significant portion of the Rubisco large subunit protein, with little or no effect on the solubility of the GFP (J. Lindbo, unpublished data).
Figure 6.
GFP expression from TRBO-G vector. N. benthamiana leaves were infiltrated with A. tumefaciens cultures transformed with pJL-TRBO-G. Total protein extracts were prepared from infiltrated leaf tissue from 3 to 7 dpi. Equal volumes of extracts were analyzed by SDS-PAGE, followed by staining with Coomassie Blue. Location of TRBO-expressed 27-kD GFP is noted by black arrowhead. Amount of Rubisco large subunit protein (white arrowhead) is greatly reduced in 3- to 6-dpi samples because they were subjected to a freeze-thaw cycle. Molecular mass of protein standards (kD) is noted at left of image. Lanes: M, molecular mass marker; H, healthy plant extract; 3 to 7, extracts from pJL-TRBO-G-agroinfiltrated leaves, 3 to 7 dpi, respectively.
Expression of Various Proteins from the TRBO Replicon
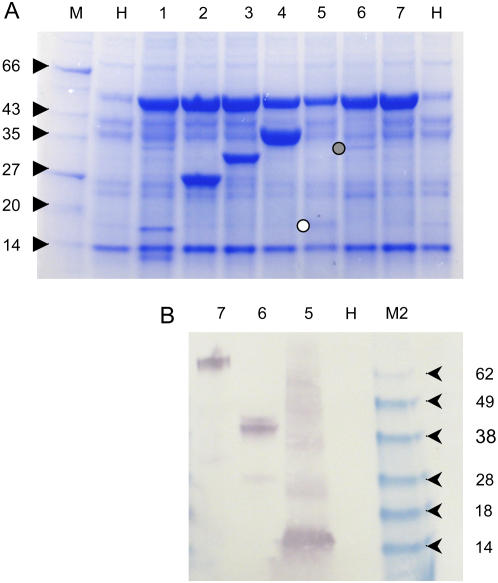
To further demonstrate the utility of the TRBO expression replicon, genes of various sizes were cloned into the pJL-TRBO plasmid. N. benthamiana plants were infiltrated with suspensions of A. tumefaciens cells transformed with the various plasmids. Several (4–6) days postinfiltration, total soluble protein extracts were prepared from agroinfiltrated tissue. Extracts were separated on SDS-PAGE gels and stained with Coomassie Blue (Fig. 7A). Because some of the recombinant proteins expressed from TRBO had C-terminal amino acid tags of His-6(HA)2 (where HA is the influenza hemagglutinin peptide YPYDVPDYA), some extracts (Fig. 7B) were also subjected to immunoblot analysis using anti-HA primary antibodies (Invitrogen). The results of this analysis demonstrated that different recombinant proteins accumulated to different levels in plants. Some (e.g. adenosine kinase) accumulated to greater levels than GFP. Other proteins accumulated at lower levels. The results also demonstrated that TRBO can be used to express His-6- and epitope-tagged recombinant proteins. Although several of the lesser accumulating proteins had His-6(HA)2 C-terminal amino acid tags, it is not proposed that this peptide tag was solely responsible for the lower accumulation levels. However, this tag may have an effect on the final level of accumulation on some proteins. For example, His-6(HA)2-tagged GFP (Fig. 7, lane 3) did accumulate to slightly lower levels than that of nontagged GFP.
Figure 7.
Expression of various proteins from the TRBO vector. N. benthamiana leaves were agroinoculated with pJL-TRBO vectors expressing various genes. Total soluble protein extracts were prepared approximately 5 dpi. Equal volumes of extract were loaded per lane. In some cases, proteins were expressed as fusions to a peptide tag of His-6-hemaglutinin peptide (duplicated), H6HA2. A, Coomassie Blue-stained SDS-PAGE gel of extracts. B, Western-blot (immunoblot) analysis of extracts using anti-HA peptide primary antibody. Lanes: M, molecular mass marker; M2, SeeBlue (Invitrogen) molecular mass marker; H, healthy plant extract. Extracts from tissue infected with JL-TRBO vector expressing the following genes: 1, Phytopthora infestans Avr3a (15 kD); 2, Aequorea victoria GFP (27 kD); 3, GFP:H6HA2 fusion (31 kD); 4, Arabidopsis adenosine kinase (38 kD); 5, 10th type III (FN10) domain from human fibronectin:H6HA2 (13 kD); 6, tomato RCR-3 proteinase:H6HA2 (42 kD); 7, tomato P69b proteinase:H6HA2 (73 kD). White and gray circles denote location of FN10 and RCR-3 proteins on Coomassie Blue-stained gel, respectively. [See online article for color version of this figure.]
DISCUSSION
We recently reported on the construction of a full-length TMV vector that could be efficiently delivered to cells by agroinfection provided a 35S-driven RNA-silencing suppressor gene was cointroduced at the same time (Lindbo, 2007). In this study, we describe a CP deletion mutant of that 35S-driven TMV vector. This vector was efficiently delivered to cells by agroinfection regardless of the coexpression of an RNA-silencing suppressor protein. These data demonstrated that the sequences in or around the TMV CP gene had a significant, negative impact on agroinfection efficiency. The negative effect of the TMV CP sequence could be at least partially neutralized by ectopic expression of a suppressor of RNA silencing. Therefore, it is hypothesized that the RNA sequence of the TMV CP subgenomic promoter and CP ORF may be a potent inducer of RNA silencing.
Since their inception, the majority of the published work on TMV vector development has focused on generating TMV expression vectors that expressed the TMV CP and therefore moved systemically in plants. This was in part because early TMV vectors were very inefficiently delivered to plants by hand rubbing of naked TMV vector RNAs (transcribed in vitro from TMV cDNA clones) onto plant leaves. As a result, the only way to obtain significant quantities of TMV vector-infected plant tissue was to allow the vector to move systemically in the plant. By dramatically improving the agroinfection rate of a TMV vector, we have made it possible to reliably infect large amounts of plant tissue with a TMV vector in the absence of systemic movement of the vector. This makes it possible to use TMV vectors that do not express the CP. This approach allows for several advantages, including higher protein expression levels from the TMV vector in a shorter time frame and the absence of large amounts of TMV CP in infected tissue. All of these advantages make it easier to produce and purify recombinant proteins of interest from plants with a TMV vector.
It is interesting that the agroinfectivity and expression capacity of the CP deletion vector described here are comparable to a TMV CP deletion vector described by Marillonnet et al. (2005). Their vector was capable of producing the equivalent of 4 g of recombinant GFP/kg fresh weight of infiltrated N. benthamiana tissue (Marillonnet et al., 2005), the highest protein expression levels reported for plants. The vector described here produced comparable levels of recombinant protein. TRBO-G expressed the equivalent of 3.3 to 5.5 g of recombinant GFP/kg of infiltrated N. benthamiana tissue (Fig. 5). Complete infections of agroinfiltrated leaves could be obtained when A. tumefaciens cells carrying the Marillonnet et al. vector were diluted up to 100-fold from an OD600 of 0.35 (Marillonnet et al., 2005). Our vector, when delivered by A. tumefaciens cultures diluted 100- to 300 -fold from an OD600 of 1.0, also gave complete infections of infiltrated leaves (Fig. 3). Two very significant differences between these two vectors are their size and composition. Marillonnet et al. achieved their high agroinfection rates by mutating cryptic introns in the TMV cDNA (making nearly 100 point mutations) and inserting 16 plant introns into the TMV cDNA. This significantly increased the size of the binary plasmid containing the intron-modified TMV cDNA. In contrast, we made no such modifications to our CP deletion vector, keeping the overall plasmid size smaller. The smaller size of this plasmid should make it easier to work with and clone into. The intron-modified vector of Marillonnet et al. is based on the crucifer strain of TMV (Gleba et al., 2004; Marillonnet et al., 2005). In contrast, our vector is based entirely on the TMV U1 strain. It may be that the crucifer strain of TMV has cryptic introns that do not exist in the U1 strain and therefore our vector did not require extensive sequence modifications.
The CP deletion vector TRBO described in this article had several advantages over a full-length TMV vector. The TRBO vector had a dramatically higher agroinfection rate. Because of this, plants could be efficiently inoculated even with very dilute suspensions of A. tumefaciens. This is important for two reasons. First, there are some plant species that demonstrate a hypersensitive response when infiltrated with high-density A. tumefaciens suspensions (Wroblewski et al., 2005). Being able to agroinoculate plants with lower density cell suspensions will reduce the chances of such negative responses of the plant to A. tumefaciens. Second, efficient inoculation with low-density A. tumefaciens cell suspensions will make it easier to obtain enough inoculum for infiltration of multiple leaves or plants. Another useful feature of TRBO was its remarkably high protein expression rate: For some proteins, gram quantities of recombinant protein were produced per kilogram of infiltrated tissue (equivalent to milligrams of recombinant protein per gram of infiltrated tissue). These levels are on par with the highest recombinant protein expression levels reported for plants (Marillonnet et al., 2005). High protein expression levels make it easier to obtain useful quantities of recombinant protein from less tissue in less time.
One of the challenges of working with plant virus expression vectors is the tendency of vector deletion mutants to appear in systemically infected portions of inoculated plants (Dawson et al., 1989; Beck and Dawson, 1990; Lehto and Dawson, 1990; Shivprasad et al., 1999). Although the phenomenon is not completely understood, it was demonstrated that a recombinant TMV vector with an insert moved more slowly than the same virus without an insert (Toth et al., 2002). This was especially true for systemic movement of recombinant viruses (Toth et al., 2002). Therefore, there is a selective advantage in movement for viruses that have lost their insert. As a result, a plant can be inoculated with a virus containing an insert, yet, when the virus appears in systemic (noninoculated) tissue, the insert may have been lost by recombination. The ability to synchronously inoculate large numbers of cells in a leaf and purify proteins from the inoculated leaf itself reduces the chances for insert loss from the virus. Therefore, TRBO is expected to be a more reliable expression vector because insert loss is less likely to be a problem than with full-length TMV vectors.
Another advantage of TRBO is that it does not produce TMV CP. Because TMV CP is required for systemic movement, TRBO is not capable of systemic movement in plants. It also will not produce virions in plants. This has definitive biocontainment and protein purification advantages. First, this feature reduces the chances for inadvertent plant-to-plant movement of the vector. Second, when extracting proteins from TRBO-infected tissue, the recombinant protein of interest does not need to be purified away from virion particles. If one is using a viral vector that does generate virus particles (such as JL24), efforts must be taken to both separate virion particles from the recombinant protein of interest and also to inactivate virus particles in any extracts of infected plant materials. These issues are not a concern with TRBO because it does not generate virus particles.
It is important to realize that not every protein expressed from a viral vector will necessarily accumulate to very high levels. The results in Figure 7 demonstrate that some proteins accumulate to levels greater than GFP. Other proteins accumulate to levels significantly lower than GFP. For example, in Figure 7, A and B, lane 7, the recombinant protein expressed could only be detected by immunoblotting (western-blotting) procedures and not by Coomassie Blue staining. There are no doubt effects of protein stability involved in the final accumulation level of any protein. Regardless, the high expression capacity of the TRBO expression vector provides an excellent opportunity to produce detectable levels of recombinant protein in plants in a very short time frame.
CONCLUSION
The TMV overexpression vector in pJL-TRBO, which lacks the TMV CP gene, has several useful advantages over a TMV vector that does express the CP gene. These advantages include higher agroinfection rates, easier scale up, higher protein expression levels, and biocontainment/protein purification advantages. The TRBO expression vector can express proteins at up to 100 times the level of the routinely used enhanced Agrobacterium transient expression method of cointroduction of a gene of interest and the P19 RNA-silencing suppressor gene into plants. It is proposed that this vector will be very useful for researchers interested in rapidly expressing recombinant proteins in plants. The ease of use of this expression vector system will make it accessible to a wide range of researchers in plant biology and biotechnology.
MATERIALS AND METHODS
Plasmid Construction
The construction of pJL24, pCB:GFP, and pJL3:P19 have been described previously (Lindbo, 2007). pJL24 is a 35S promoter-driven version of the TMV expression vector 30B (Shivprasad et al., 1999). The TMV vector 30B is a chimera of sequences from the U1 and U5 strains of TMV (the CP subgenomic promoter, CP ORF, and 3′-nontranslated sequences from the U5 strain of TMV, the remainder from the U1 strain). pJL-TRBO was constructed from pJL24 and a full-length cDNA clone of TMV U1 using standard cloning procedures. The final sequence of TRBO is as follows: TMV U1 nucleotides 1 to 5,756 (with the CP start codon, nucleotides 5,712–5,714, mutated from ATG to AGA); the polylinker sequence ttaattaacggcctagggcggccgc; then U1 nucleotides 6,177 to 6,395. Numbering of U1 nucleotide sequences was as according to Dawson et al. (1986). Immediately following U1 nucleotide 6,395 are a KpnI site, a ribozyme cDNA sequence (Turpen et al., 1993), and CaMV 3′ polyA signal/transcription terminator (Carrington and Freed, 1990). All plasmids had the mini binary plasmid pCB301 (Xiang et al., 1999) as their backbone, which can replicate in both Escherichia coli and Agrobacterium tumefaciens.
PCR products of the following genes were cloned into PacI- and AvrII-digested pJL-TRBO: (1) the cycle 3 mutant version of the gfp gene (gfpc3; Crameri et al., 1996); (2) gfpc3 with the coding sequence for a hexa-His and (HA)2 epitope tag fused to its 3′ end; (3) Arabidopsis (Arabidopsis thaliana) adenosine kinase gene (Wang et al., 2003); (4) 10th domain of the human fibronectin gene (Baron et al., 1991); (5) Phytophthora infestans avirulence protein gene avr3a (Armstrong et al., 2005); (6) tomato (Solanum lycopersicum) 69-kD proteinase gene (Tian et al., 2007); and (7) tomato Cys proteinase rcr-3 gene (Dixon et al., 2000; Tian et al., 2007). The adenosine kinase gene was a kind gift from Dave Bisaro. The fibronectin domain coding sequence was chemically synthesized. The Phytophthora and tomato genes were a kind gift from Sophien Kamoun. In all cases, the ATG start codon for the gene of interest was downstream of the PacI (ttaattaa) site, typically in the following context: ttaattaacATG.
Agroinfection
Binary plasmids purified from E. coli cultures were transformed into A. tumefaciens GV3101 using the freeze-thaw method (Chen et al., 1994). Transformed A. tumefaciens was plated on Luria-Bertani plates with 50 μg/mL kanamycin, 25 μg/mL gentamycin, and 10 μg/mL rifampicin for plasmid selection. For agroinfection, colonies of binary plasmid-transformed A. tumefaciens cells were grown (12–24 h) at 25°C to 28°C, 225 rpm in Luria-Bertani medium (Sambrook et al., 1989) supplemented with 10 mm MES, pH 5.7, 50 μg/mL kanamycin, 25 μg/mL gentamycin. Overnight cultures were diluted 1:10 in the same medium supplemented with 20 μm acetosyringone and grown as above to an OD600 of about 1.0. Cells were collected by centrifugation and resuspended in induction medium (Johansen and Carrington, 2001), 10 mm MES, pH 5.7, 10 mm MgCl2, 200 μm acetosyringone at an OD600 of 1.0. Cells sat at room temperature in induction medium for 2 to 24 h before infiltration into the abaxial surface of Nicotiana benthamiana leaves using a 1-mL syringe with no needle. When mixed cultures of A. tumefaciens were infiltrated into plants, bacterial cultures were prepared separately in induction medium and combined immediately before infiltration.
Plants and Photography
N. benthamiana plants were grown in a growth chamber with an 18-h photoperiod, 25°C to 27°C. For GFP photography, plants were photographed with a Cannon G6 digital camera equipped with a Tiffen Deep Yellow 15 filter. Plants were illuminated with a hand-held long-wave UV lamp (UVP Blak-Ray model UVL-56).
SDS-PAGE
Total soluble protein extracts of agroinfiltrated plant tissue were prepared by freezing tissue samples in liquid nitrogen and then grinding in the presence of 4 volumes/g fresh weight, 50 mm Tris, pH 7.5, 150 mm NaCl, 0.1% Tween 20, 0.1% β-mercaptoethanol. Extracts were clarified by centrifuging for 15 min at 12,000g to 15,000g at 4°C. Clarified supernatant was stored at −20°C. Equal volumes of clarified extract of each treatment were combined with SDS-PAGE loading dye (Laemmli, 1970) and analyzed on 4% to 20% SDS-PAGE gels. Gels were stained with Coomassie Blue to visualize proteins.
GFP Assay
Samples of clarified plant protein extracts, prepared as described above (or standards of purified GFP), were diluted in 50 mm carbonate buffer (pH 9.6). Protein samples in wells of a 96-well plate (Costar; white polystyrene) were read on a Perkin-Elmer HTS 7000 BioAssay reader with 405-nm excitation/535-nm emission filters.
Purification of His-6-Tagged GFP
His-6 C-terminally tagged GFP was expressed in plants from a TRBO replicon by agroinfection. Plant tissue was collected at approximately 5 dpi and ground in 4 volumes of extraction buffer (50 mm phosphate, pH 8.0, 10 mm Tris, pH 8.0, 500 mm NaCl, 0.1% Tween 20, 0.1% Nonidet P-40, 0.1% β-mercaptoethanol, 1 mm phenylmethylsulfonyl fluoride). Extract was filtered through cheesecloth, then centrifuged at 12,000g, 4°C for 20 min. Clarified supernatant was then passed through a −20°C freeze-thaw cycle. After thawing, samples were centrifuged (as before). Imidazole was added to supernatant for a final concentration of 10 mm. One-half milliliter of washed nickel nitilotriacetic acid agarose beads (Qiagen) was added to 8 to 10 mL of extract and incubated at 4°C on a rocker for 1 to 2 h. The column was washed in 20 to 25 column volumes of wash buffer (50 mm phosphate, pH 8.0, 500 mm NaCl, 0.1% Tween 20, 20 mm imidazole). Bound His-tagged GFP was eluted with 250 mm imidazole in 1× phosphate-buffered saline. The eluted fraction was dialyzed twice (6 h to overnight) into 1,000 volumes of 1× phosphate-buffered saline (11.9 mm phosphate, pH 7.4, 137 mm NaCl, 2.7 mm KCl) at 4°C. Protein concentration was estimated using bicinchoninic acid assay (Pierce) and bovine serum albumin as a standard.
Acknowledgments
We thank Shannon Woody for excellent technical assistance.
This work was supported by a grant from The Ohio State University/OARDC Research Enhancement Competitive Grants Program.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: John A. Lindbo (john_lindbo2003@yahoo.com).
Some figures in this article are displayed in color online but in black and white in the print edition.
Open Access articles can be viewed online without a subscription.
References
- Armstrong MR, Whisson SC, Pritchard L, Bos JI, Venter E, Avrova AO, Rehmany AP, Bohme U, Brooks K, Cherevach I, et al (2005) An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc Natl Acad Sci USA 102 7766–7771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron M, Norman DG, Campbell ID (1991) Protein modules. Trends Biochem Sci 16 13–17 [DOI] [PubMed] [Google Scholar]
- Beck DL, Dawson WO (1990) Deletion of repeated sequences from tobacco mosaic virus mutants with two coat protein genes. Virology 177 462–469 [DOI] [PubMed] [Google Scholar]
- Carrington JC, Freed DD (1990) Cap-independent enhancement of translation by a plant potyvirus 5′ nontranslated region. J Virol 64 1590–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M (1995) Green fluorescent protein. Photochem Photobiol 62 651–656 [DOI] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC (1994) Green fluorescent protein as a marker for gene expression. Science 263 802–805 [DOI] [PubMed] [Google Scholar]
- Chen H, Nelson RS, Sherwood JL (1994) Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques 16 664–668, 670 [PubMed] [Google Scholar]
- Crameri A, Whitehorn EA, Tate E, Stemmer WP (1996) Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat Biotechnol 14 315–319 [DOI] [PubMed] [Google Scholar]
- Creager AN, Scholthof KB, Citovsky V, Scholthof HB (1999) Tobacco mosaic virus. Pioneering research for a century. Plant Cell 11 301–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver JN, Lehto K, Close SM, Hilf ME, Dawson WO (1993) Genomic position affects the expression of tobacco mosaic virus movement and coat protein genes. Proc Natl Acad Sci USA 90 2055–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson WO, Beck DL, Knorr DA, Grantham GL (1986) cDNA cloning of the complete genome of tobacco mosaic virus and production of infectious transcripts. Proc Natl Acad Sci USA 83 1832–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson WO, Lewandowski DJ, Hilf ME, Bubrick P, Raffo AJ, Shaw JJ, Grantham GL, Desjardins PR (1989) A tobacco mosaic virus-hybrid expresses and loses an added gene. Virology 172 285–292 [DOI] [PubMed] [Google Scholar]
- Dixon MS, Golstein C, Thomas CM, van Der Biezen EA, Jones JD (2000) Genetic complexity of pathogen perception by plants: the example of Rcr3, a tomato gene required specifically by Cf-2. Proc Natl Acad Sci USA 97 8807–8814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donson J, Kearney CM, Hilf ME, Dawson WO (1991) Systemic expression of a bacterial gene by a tobacco mosaic virus-based vector. Proc Natl Acad Sci USA 88 7204–7208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleba Y, Klimyuk V, Marillonnet S (2007) Viral vectors for the expression of proteins in plants. Curr Opin Biotechnol 18 134–141 [DOI] [PubMed] [Google Scholar]
- Gleba Y, Marillonnet S, Klimyuk V (2004) Engineering viral expression vectors for plants: the ‘full virus’ and the ‘deconstructed virus’ strategies. Curr Opin Plant Biol 7 182–188 [DOI] [PubMed] [Google Scholar]
- Grimsley N (1995) Agroinfection. Methods Mol Biol 44 325–342 [DOI] [PubMed] [Google Scholar]
- Grimsley N, Hohn B, Hohn T, Walden R (1986) “Agroinfection,” an alternative route for viral infection of plants by using the Ti plasmid. Proc Natl Acad Sci USA 83 3282–3286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen LK, Carrington JC (2001) Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol 126 930–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Lehto K, Dawson WO (1990) Replication, stability, and gene expression of tobacco mosaic virus mutants with a second 30K ORF. Virology 175 30–40 [DOI] [PubMed] [Google Scholar]
- Lindbo JA (2007) High-efficiency protein expression in plants from agroinfection-compatible Tobacco mosaic virus expression vectors. BMC Biotechnol 7 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man M, Epel BL (2006) Assessment of the effectiveness of a nuclear-launched TMV-based replicon as a tool for foreign gene expression in plants in comparison to direct gene expression from a nuclear promoter. Transgenic Res 15 107–113 [DOI] [PubMed] [Google Scholar]
- Marillonnet S, Thoeringer C, Kandzia R, Klimyuk V, Gleba Y (2005) Systemic Agrobacterium tumefaciens-mediated transfection of viral replicons for efficient transient expression in plants. Nat Biotechnol 23 718–723 [DOI] [PubMed] [Google Scholar]
- Pogue GP, Lindbo JA, Dawson WO, Turpen TH, editors (1998) Tobamovirus Transient Expression Vectors: Tools for Plant Biology and High-Level Expression of Foreign Proteins in Plants. Kluwer Academic Publishers, Dordrecht, The Netherlands
- Pogue GP, Lindbo JA, Garger SJ, Fitzmaurice WP (2002) Making an ally from an enemy: plant virology and the new agriculture. Annu Rev Phytopathol 40 45–74 [DOI] [PubMed] [Google Scholar]
- Popescu SC, Popescu GV, Bachan S, Zhang Z, Seay M, Gerstein M, Snyder M, Dinesh-Kumar SP (2007) Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc Natl Acad Sci USA 104 4730–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Scholthof HB, Scholthof KB, Jackson AO (1996) Plant virus gene vectors for transient expression of foreign proteins in plants. Annu Rev Phytopathol 34 299–323 [DOI] [PubMed] [Google Scholar]
- Scholthof KB (2004) Tobacco mosaic virus: a model system for plant biology. Annu Rev Phytopathol 42 13–34 [DOI] [PubMed] [Google Scholar]
- Shivprasad S, Pogue GP, Lewandowski DJ, Hidalgo J, Donson J, Grill LK, Dawson WO (1999) Heterologous sequences greatly affect foreign gene expression in tobacco mosaic virus-based vectors. Virology 255 312–323 [DOI] [PubMed] [Google Scholar]
- Tian M, Win J, Song J, van der Hoorn R, van der Knaap E, Kamoun S (2007) A Phytophthora infestans cystatin-like protein targets a novel tomato papain-like apoplastic protease. Plant Physiol 143 364–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth RL, Pogue GP, Chapman S (2002) Improvement of the movement and host range properties of a plant virus vector through DNA shuffling. Plant J 30 593–600 [DOI] [PubMed] [Google Scholar]
- Turpen TH, Turpen AM, Weinzettl N, Kumagai MH, Dawson WO (1993) Transfection of whole plants from wounds inoculated with Agrobacterium tumefaciens containing cDNA of tobacco mosaic virus. J Virol Methods 42 227–239 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33 949–956 [DOI] [PubMed] [Google Scholar]
- Wang H, Hao L, Shung CY, Sunter G, Bisaro DM (2003) Adenosine kinase is inactivated by geminivirus AL2 and L2 proteins. Plant Cell 15 3020–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroblewski T, Tomczak A, Michelmore R (2005) Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnol J 3 259–273 [DOI] [PubMed] [Google Scholar]
- Xiang C, Han P, Lutziger I, Wang K, Oliver DJ (1999) A mini binary vector series for plant transformation. Plant Mol Biol 40 711–717 [DOI] [PubMed] [Google Scholar]