Abstract
Virus-induced gene silencing (VIGS) is a widely used, powerful technique for reverse genetics. VIGS vectors derived from the Tobacco rattle virus (TRV) are among the most popular for VIGS. We have developed a TRV RNA2 vector that allows the insertion of gene silencing fragments by ligation-independent cloning (LIC). This new vector has several advantages over previous vectors, particularly for applications involving the analysis of large numbers of sequences, since TRV-LIC vectors containing the desired insert are obtained with 100% efficiency. Importantly, this vector allows the high-throughput cloning of silencing fragments without the use of costly enzymes required for recombination, as is the case with GATEWAY-based vectors. We generated a collection of silencing vectors based on 400 tomato (Solanum lycopersicum) expressed sequence tags in this TRV-LIC background. We have used this vector to identify roles for SlMADS1 and its Nicotiana benthamiana homologs, NbMADS4-1 and -2 in flowering. We find that NbMADS4-1 and -2 act nonredundantly in floral development and silencing of either gene results in loss of organ identity. This TRV-LIC vector should be a valuable resource to the plant community.
The last decade has seen an explosion in the availability of plant gene sequences. The genomes of the model species Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) have both been sequenced, while those of tomato (Solanum lycopersicum) and maize (Zea mays) are currently being sequenced (Mueller et al., 2005; http://www.sgn.cornell.edu/about/tomato_project/;http://www.maizesequence.org/index.html). Large collections of ESTs have also been generated for a variety of species that are widely used for research purposes. Concomitant with the availability of this sequence information, many important aspects of plant growth and development have been analyzed by DNA microarrays, leading to the identification of numerous genes potentially involved in these processes. At this time then, the challenge to most plant biologists is to effectively mine this data to identify and characterize the genes and gene products that are critical to the crucial processes that have been investigated. This calls for techniques that start with a known DNA sequence and allow the determination of biological function. This approach is called reverse genetics and some of the most common methods for performing reverse genetic studies are based on RNA silencing.
Although recently discovered, RNA silencing is a well-characterized, endogenous system for monitoring RNA inside a cell and eliminating foreign molecules or inhibiting mRNA translation (for review, see Brodersen and Voinnet, 2006). It is a homology-based process that uses small RNA fragments to identify targets for destruction or inhibition. RNA silencing is also indispensable for normal plant growth and development, regulating the expression of central genes in flowering, meristem identity, and other processes (Meins et al., 2005). In plants, RNA silencing plays critical roles in viral defense, generating small interfering RNA sequences that target the foreign viral RNA for degradation. It is this capability of RNA silencing in plants that has been harnessed for use as a tool in reverse genetics. Virus-induced gene silencing (VIGS) is an efficient tool based on RNA silencing. VIGS uses viral vectors to introduce gene fragments homologous to a gene of interest into a plant host (for review, see Lu et al., 2003; Burch-Smith et al., 2004). Endogenous RNA silencing machinery then acts to eliminate viral RNA sequences and inadvertently also targets the mRNA of the gene of interest for removal. In this way, the gene of interest is silenced, through the removal of its mRNA.
VIGS has been widely adapted for functional gene studies (Lu et al., 2003; Burch-Smith et al., 2004; Robertson, 2004). It was originally described for use in tobacco (Nicotiana tabacum) and its relative Nicotiana benthamiana (Kumagai et al., 1995; Ruiz et al., 1998), but VIGS has now been adopted for gene analysis in many dicotyledonous species including Arabidopsis (Burch-Smith et al., 2006; Wang et al., 2006), potato (Solanum tuberosum; Brigneti et al., 2004), tomato (Liu et al., 2002a), and pepper (Capsicum annum; Chung et al., 2004). Even plant species that are not core eudicots have been analyzed with VIGS, including opium poppy (Papaver somniferum; Hileman et al., 2005), and Aquilega (Gould and Kramer, 2007). VIGS has also been developed for use in monocots and it has been described in barley (Hordeum vulgare; Holzberg et al., 2002; Hein et al., 2005), wheat (Triticum aestivum; Scofield et al., 2005), and rice (Ding et al., 2006, 2007).
VIGS has become a widely used technique for reverse genetics because it is transient and does not require the generation of stable transgenics. In addition, it is rapid with phenotypes observed with 10 d to 3 weeks of silencing. Another advantage of VIGS is that it can be designed to silence either a single member or multiple members of a gene family, addressing the problem of functional redundancy. The most widely used VIGS vectors are derived from the Tobacco rattle virus (TRV). It has been used for VIGS in the Solanaceae (Ratcliff et al., 2001; Liu et al., 2002a, 2002b; Brigneti et al., 2004; Chung et al., 2004), Arabidopsis (Burch-Smith et al., 2006; Wang et al., 2006), opium poppy (Hileman et al., 2005), California poppy (Eschscolzia californica; Wege et al., 2007), and Aquilega (Gould and Kramer, 2007). TRV is a bipartite, single-stranded, positive sense RNA virus with a large host range (Hull, 2002). Some of the features of this virus that are believed to contribute to its success as a VIGS vector include its ability to enter growing parts of the plant, its toleration of DNA inserts up to about 1 kb in size, and mild symptoms of infection (Lu et al., 2003; Burch-Smith et al., 2004).
We have previously generated and described TRV-based VIGS vectors (Liu et al., 2002a, 2002b). Our first generation TRV VIGS vector used restriction digestion and ligation for cloning of inserts (Liu et al., 2002b). We then developed a second-generation vector that carried GATEWAY recombination sites (Liu et al., 2002a). However, prohibitive features of this vector include time-consuming and complicated cloning procedures. In addition, the comparatively high cost of the enzymes used for GATEWAY-based cloning and proprietary issues are also a consideration for large-scale gene cloning and VIGS screening. To overcome these limitations, we have adopted modified ligation-independent cloning (LIC; Aslanidis and de Jong, 1990; Dieckman et al., 2002) as an approach for high-throughput cloning into the TRV VIGS vector. LIC is much faster and more accurate than other cloning strategies as only a single transformation is required. We used this new TRV-LIC vector to clone 400 tomato ESTs and we assessed its effectiveness in N. benthamiana. Thus, the TRV-LIC vector is an improved vector for high-throughput VIGS.
In addition, we observed several interesting phenotypes and identified N. benthamiana MADS-box genes with important, nonredundant roles in flowering. MADS-box genes represent a large family of transcription factors with critical roles in floral and general development. In particular, they function in conferring floral organ identity, floral determinancy, and the timing of the vegetative-floral transition (Ng and Yanofsky, 2001; Henderson and Dean, 2004). We used our TRV-LIC vector to silence NbMADS4-1 and NbMADS4-2 simultaneously or individually. In all instances we observed a loss of floral organ identity and determinancy, resulting in green, bushy flowers. These data suggest that NbMADS4-1 and -2, despite their high homology, have critical, nonredundant roles in floral development.
RESULTS
Development of a LIC Strategy for TRV-Based VIGS
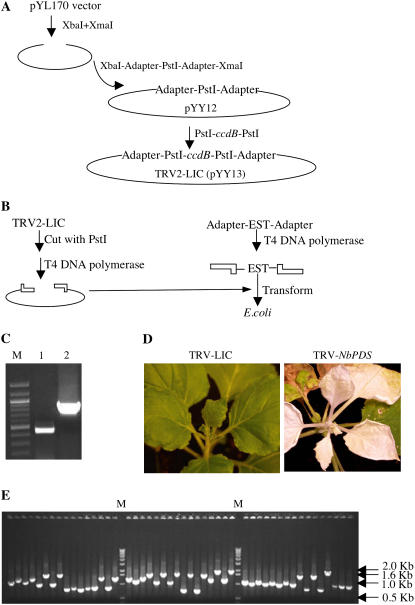
One limitation of currently available VIGS vectors is the multiple steps involved in the cloning of the silencing fragment into the vector. This can become quite expensive when using GATEWAY-based recombination vectors. To overcome these limitations, we adopted a modified LIC strategy (Aslanidis and de Jong, 1990; Dieckman et al., 2002) for the high-throughput cloning of inserts into our TRV VIGS vector. To do this we added two LIC adaptors to the TRV2 vector, pYL170 (Burch-Smith et al., 2006), to generate pYY12. These adaptors consist of 15 bp sequences containing central PstI restriction sites (Fig. 1A). We then inserted a ccdB gene between the two LIC sites by PstI digestion to create pYY13, referred to as TRV2-LIC (Fig. 1A). The ccdB gene allows us to screen putative recombinant colonies rapidly and accurately, a strategy employed by the widely used GATEWAY technology. Digestion of TRV2-LIC with PstI linearizes the vector, and subsequent treatment with T4 DNA polymerase generates sticky ends. PCR products carrying ends homologous to the adaptors and similarly treated with T4 DNA polymerase can form covalent bonds with ends of the linearized vector (see “Materials and Methods” for details). Our new TRV2 vector, TRV2-LIC, can thus be used for high-throughput cloning since any number of sequences carrying compatible ends can be combined with TRV2-LIC.
Figure 1.
TRV2-LIC vector. A, Construction of the TRV2-LIC vector. The TRV2 vector, pYL170, was used to generate the new TRV2-LIC vector by inserting a cassette containing adapters and two PstI sites in two digestion and ligation reactions. B, LIC cloning of inserts into TRV2-LIC. Briefly, the TRV2-LIC vector is digested with PstI and treated with T4 DNA polymerase. The EST carrying the relevant adapter sequences is generated by PCR and also treated with T4 DNA polymerase. The vector and insert are then mixed and used to transform Escherichia coli cells. C, TRV-LIC does not interfere with viral replication and spread as shown by the presence of TRV RNA1 (lane 1) and modified RNA2 (lane 2) in upper leaves of N. benthamiana plant. Lane M contains DNA size marker. D, TRV2-LIC allows efficient silencing of NbPDS as shown by the photobleaching of silenced plant (right). Control plant infiltrated with empty TRV-LIC vector shown for comparison (left). E, EST cloning efficiency into TRV-LIC is 100% as shown by PCR on colonies obtained from transformation with the vector and PCR product mixture. All 48 of the colonies tested here contain insert. Lane M contains DNA size marker.
We confirmed that the changes we had made to TRV RNA2 in generating TRV2-LIC did not compromise its infectivity by examining whether the virus was able to spread to the upper leaves of plants. We infected N. benthamiana plants with the TRV RNA1 and TRV2-LIC vectors only and collected upper leaves and inflorescence tissue 20 d later. The presence of TRV RNA2 in these tissues was confirmed by reverse transcription (RT)-PCR (Fig. 1C). Thus, the modifications made to TRV RNA2 to produce the TRV2-LIC plasmid does not hinder the ability of the virus to replicate and spread in plants. We also confirmed the ability of the new TRV-LIC vector to initiate gene silencing by inserting a fragment of the NbPHYTOENE DESATURASE (NbPDS) gene into this vector. We performed PDS silencing experiments according to standard protocols (Dinesh-Kumar et al., 2003) and found that after 10 d, we observed photobleaching of upper leaves in presumably silenced plants (Fig. 1D). The timing and appearance of photobleaching was indistinguishable from that observed with earlier versions of our TRV VIGS vectors (data not shown). Thus, our TRV2-LIC vector functions in VIGS in a manner similar to other TRV-based VIGS vectors.
High-Throughput Cloning of Tomato ESTs
While N. benthamiana has become extremely popular for gene studies, the limited availability of sequence collections hinders the study of gene function. In contrast, large EST collections from tomato are readily accessible for large-scale gene function analyses (D'Agostino et al., 2007). Tomato and N. benthamiana are both members of the Solanaceae family and there is usually a high degree of similarity in gene coding sequences. Indeed, tomato gene fragments have been successfully used to silence homologs in N. benthamiana (Liu et al., 2004b) or vice versa (Ekengren et al., 2003). To demonstrate the efficacy of the TRV-LIC vector for high-throughput screening for gene function, we cloned 400 tomato ESTs into this vector (Supplemental Table S3; see “Materials and Methods”). These ESTs predominantly belonged to one of the following three classes: kinases, transcription factors, and phosphatases. The efficiency of this large-scale cloning approach was confirmed by the presence of inserts in the vector (representative gel is shown in Fig. 1E). The presence of the desired insert was detected in 100% of all clones examined.
Developmental Phenotypes Revealed by Silencing 400 Tomato ESTs
Our TRV-LIC EST clone collection was then screened for silencing phenotypes in N. benthamiana plants. For this, we infiltrated four-leaf stage plants with a 1:1 mixture of TRV1 and TRV2-LIC-EST fragment for each of the clones in our collection, and monitored the infiltrated plants for 8 weeks. Eventually, we observed several developmental phenotypes in our silenced plants, and some examples are shown (Fig. 2). For each EST, we infiltrated four plants per trial and did at least three trials.
Figure 2.
Selected phenotypes observed with silencing of tomato ESTs. A, Silencing of SlRibosomal protein L11 like, Slα-tubulin, SlHSP70, and SlPolyubiquitin causes death. B, Silencing SlMAP3K epsilon disrupts leaf development resulting in severe crinkling. C, Reduction in SlGeranylgeranyl reductase levels causes yellowing of leaves. D, Reduced SlPlastidic aldolase causes photobleaching and a variegated appearance. E, Decreased SlCyclin-dependent protein kinase, p34cdc2, levels cause severe stunting. F, Homeotic transformations of floral tissue in SlTAG1-silenced plants. G, mRNA levels in control (left) versus silenced (right) tissue indicate effective silencing of genes indicated. Number of PCR cycles is indicated above lanes. Lane M is the DNA size marker. C is no RT control. Actin is a loading control.
One of the most common phenotypes resulting from silencing was lethality (Fig. 2A). This is exemplified by the silencing of SlRibosomal protein L11 like, Slα-tubulin, SlHSP70, and SlPolyubiquitin (Fig. 2A). Another phenotype we observed was the deformation of the upper, presumably silenced, leaves as shown for SlMAP3K epsilon (Fig. 2B). These plants were ultimately much shorter than control plants. Interestingly, SlMAP3K epsilon-silenced plants did not produce inflorescences, suggesting severe disruption of development beyond leaf structure (Fig. 2B, center section). The silencing of several constructs led to loss of pigmentation or chlorosis, as shown for silencing with SlGeranylgeranyl reductase (Fig. 2C) and SlPlastidic aldolase (Fig. 2D). The phenotypes that we observed in these two cases are in close agreement with previous reports on the effects of reduction of the levels of these gene products (Boldt et al., 1992; Tanaka et al., 1999).
Interestingly, silencing of some clones led to developmental defects in the meristems (Fig. 2E) or inflorescences (Fig. 2F) of plants. Silencing SlCyclin-dependent protein kinase, p34cdc2 (Fig. 2E), resulted in an attenuation of apical growth and severe stunting. The leaves of silenced plants also showed chlorosis. Silencing initiated by the tomato AGAMOUS homolog, SlAG1, produced phenotypes previously described for knock down in tomato (Pnueli et al., 1994). As in tomato, we observed the homeotic transformation of the third whorl stamens into petals, and the replacement of fourth whorl carpels with other flowers or inflorescences (Fig. 2F). These results reinforce our previous conclusion that TRV-mediated VIGS is an effective tool for assessing the functions of genes in the reproductive tissues of plants (Chen et al., 2004; Liu et al., 2004a).
For 21 EST clones representing 18 unique genes that showed interesting developmental phenotypes when silenced, we attempted to clone their N. benthamiana homologs into the TRV2-LIC silencing vector (Supplemental Table S1). We failed to obtain N. benthamiana homologs corresponding to four tomato ESTs. We repeated the silencing using 14 N. benthamiana sequences and observed similar developmental phenotypes as had been obtained with the tomato sequences (data not shown). We then determined the degree of silencing of each of the 14 genes by RT followed by semiquantitative RT-PCR using primers annealing outside the sequence used for silencing (Fig. 2G; Supplemental Table S2; “Materials and Methods”). In all cases we observed a greater than 60% reduction in transcript levels in the silenced plants. Thus, the approach used for the high-throughput cloning and silencing of tomato ESTs to assess their function using N. benthamiana as a heterologous system was successful.
Silencing SlMADS1 Produces Homeotic and Other Transformations in Flowers
Plants infiltrated with TRV carrying a fragment of SlMADS1 displayed very interesting developmental phenotypes, particularly in the floral inflorescences and individual flowers (Fig. 3). The altered development is first manifested as increased branching, first of the primary stem and then of the secondary stems, giving rise to a bushy plant compared to the control plants (Fig. 3A). Most of the branches of the SlMADS1-silenced plants went on to produce inflorescences, such that the final number of flowers in the silenced plants is almost double that of the control plants (Fig. 3A; data not shown). The timing of the floral transition did not differ between control and SlMADS1-silenced plants.
Figure 3.
Silencing SlMADS1. A, SlMADS1-silenced plants (right) show loss of apical dominance compared to control empty vector-treated plants (left). B, Flowers on control empty vector-treated plants show distinct white petals surrounded by sepals. C, Flowers on SlMADS1-silenced plants have enlarged sepals and white petals are largely absent. D, Sepals separated from inner whorls from control (left) and SlMADS1-silenced (right) flowers. E, Magnified view of inner whorls of flower dissected in D. Note reduced white petals and the presence of flower-like organs. F, Indeterminate flowers of SlMADS1-silenced plants. G, Magnified view of one of the flowers in F.
Wild-type N. benthamiana flowers typically possess a first whorl of five green sepals and a second whorl of five white petals that fuse to form a tube that surrounds the reproductive organs (Fig. 3B). The third whorl usually consists of four stamens that surround the central gynoecium of the fourth whorl (Fig. 2F, left section). SlMADS1-silenced flowers carried enlarged sepals that were about 5-times bigger than the sepals of control flowers (Fig. 3, C and in D compare left and right sections). Further, petals and stamens converted into green leaf-like structures (Fig. 3, C–E). Interestingly, in some cases the fourth whorl carpel was replaced by another flower or even inflorescence (Fig. 3, D–G), resulting in indeterminate flowers.
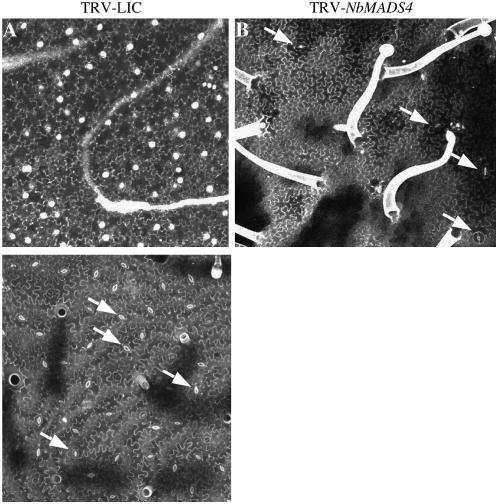
We examined the ultrastructure of these modified petals and stamens by confocal microscopy. Wild-type petals and stamens normally do not possess stomata and trichomes on their abaxial surface (Fig. 4, left section). Both these structures were present in the leaf-like structures of SlMADS1-silenced flowers (Fig. 4, right section). Thus, the cells on the leaf-like petals and stamens in SlMADS1-silenced plants looked very similar to pavement cells of leaf epidermis.
Figure 4.
NbMADS4-silenced petals compared to controls. A, Abaxial surface of control N. benthamiana petal. Note absence of trichomes. B, Abaxial surface of SlMADS1-silenced petal contains both trichomes and stomata (arrows). C, Abaxial surface of control N. benthamiana leaf contains many stomata. Scale bar represents 50 μm.
NbMADS4-1 and 4-2 Have Nonredundant, Critical Roles in Floral Development
To further characterize SlMADS1's role in plant development, we cloned the N. benthamiana sequences showing highest similarity to SlMADS1. A search of the GenBank database revealed that the tobacco sequence, NtMADS4, showed highest similarity (85%) to SlMADS1at the amino acid level (Supplemental Fig. S1). Using RT-PCR with primers based on NtMADS4, we obtained two sequences from N. benthamiana that we named NbMADS4-1 and NbMADS4-2 (Supplemental Fig. S2). These two genes are 82% identical to each other at the nucleotide and amino acid levels. NbMADS4-1 and NbMADS4-2 show 83% and 97% identity to NtMADS4, respectively (Supplemental Fig. S1).
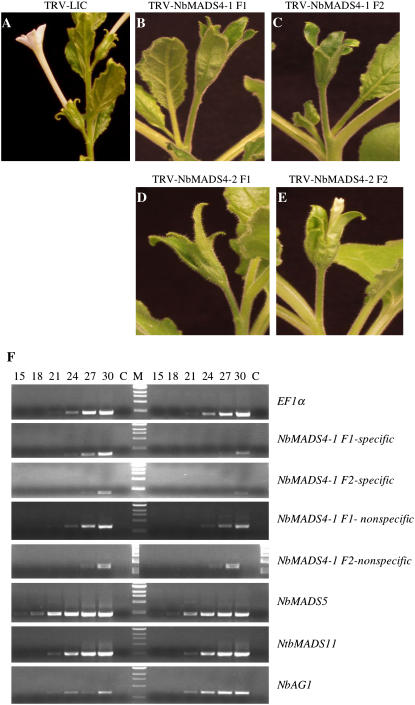
We then cloned two different fragments of each of these genes separately into the TRV2-LIC vector for silencing (Supplemental Figs. S3 and S4). The phenotypes obtained on silencing NbMADS4-1 and NbMADS4-2, each with either fragment, were identical to those observed on silencing SlMADS1 (Figs. 3 and 5). This suggests that NbMADS4-1 and NbMADS4-2 have closely related functions in flower development. To determine whether NbMADS4-1 and NbMADS4-2 were functionally redundant, we examined the mRNA levels of these genes in plants where silencing using the alternate gene had been initiated. In NbMADS4-1-silenced plants, NbMADS4-1 RNA transcript levels decreased by about 90% (Fig. 5F), while levels of NbMADS4-2 mRNA were virtually unaffected (Fig. 5F). Similarly, while NbMADS4-2 silencing produced a drastic reduction in NbMADS4-2 transcript levels, NbMADS4-1 was unaffected (Fig. 5F). We confirmed that the expression of other MADS-box genes was not affected when silencing NbMADS4-1 and -2 by monitoring levels of two closely related genes, NbMADS5 and NbMADS11 (Fig. 5F). We also confirmed that other genes involved in floral organ identity were not disrupted in NdMADS4-1 or -2-silenced tissue by determining that levels of NbAG1, the homolog of AtAGAMOUS, were unchanged (Fig. 5F). These data suggest that NbMADS4-1 and NbMADS4-2 have important, nonredundant roles in floral development in N. benthamiana.
Figure 5.
Silencing NbMADS4-1 and NbMADS4-2. A, Control N. benthamiana flower. B and C, Silencing NbMADS4-1 using fragment 1 (B) or fragment 2 (C) causes enlargement of sepals and the loss of large white sepals. D and E, Silencing NbMADS4-2 causes identical phenotypes to silencing with either NbMADS4-1 fragment. F, The reduction of NbMADS4-1 transcript levels was confirmed by semiquantitative RT-PCR in tissue that had been silenced with fragment 1 (row 2) or fragment 2 (row 3). The levels of NbMADS4-2 mRNA were not altered in NbMADS4-1 fragment 1 silenced plants (row 4) and NbMADS4-1 fragment 2 silenced plants (row 5). The transcript levels of the related genes NbMADS5 and NbMADS11 were also examined to confirm specificity of silencing (rows 6 and 7, respectively). The phenotypes shown are not due to the silencing of another MADS-box transcription factor NbAG1 (row 8). Lanes to the left of the marker (M) contain control tissue and those on the right contain tissue from silenced tissue. C is no RT control. Numbers at the top of each lane indicate number of PCR cycles.
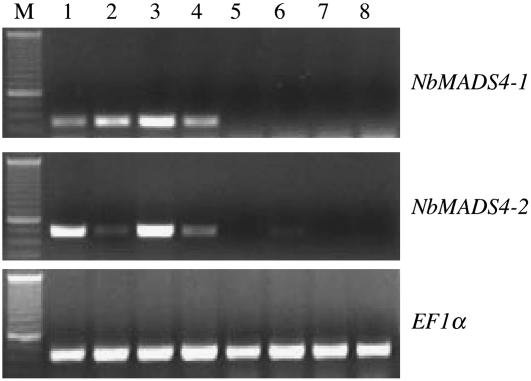
Given the effects on floral development we observed, we examined the expression of NbMADS4-1 and NbMADS4-2 in different organs. Both genes were expressed in all whorls of floral organs (Fig. 6), while neither was highly expressed in any of the vegetative organs sampled (Fig. 6).
Figure 6.
Analysis of NbMADS4 tissue-specific expression. NbMADS4-1 and NbMADS4-2 transcripts were detected by semiquantitative RT-PCR. Lane 1 is tissue from sepals, 2 is petals, 3 is petioles, 4 is carpels, 5 is roots, 6 is stems, 7 is leaves, and 8 is seedlings. Lane M is the size marker. EF1α was used as a loading control.
DISCUSSION
We report here the construction of a TRV-LIC vector that facilitates the high-throughput cloning of silencing fragments. This vector allows rapid, efficient cloning of inserts and has the added advantage of doing so at markedly reduced costs. While gene-specific primers are still required for each insert of interest, these are shorter than those used with other systems like GATEWAY, which have been adopted for cloning large numbers of inserts (Liu et al., 2002a). Further, there are fewer cloning steps and neither ligases nor recombinases are required, further reducing the tediousness and cost of the procedure. In addition, we have demonstrated that for the 400 tomato ESTs cloned here, cloning efficiency approaches 100%; thus, there is less need for screening colonies obtained after the cloning process to confirm the presence of inserts.
The TRV-LIC vector achieves silencing of target genes with the same timing and efficiency as previous TRV-based VIGS vectors. We observed greater than 60% reduction in target transcript levels in all cases examined. The TRV-LIC vector is also able to silence genes expressed in the growing parts of the plant as demonstrated by the flowering defects we observed in several instances (Figs. 2 and 3). This is consistent with other reports of VIGS of meristematic and flowering genes (Ratcliff et al., 2001; Chen et al., 2004; Liu et al., 2004a; Hileman et al., 2005; Gould and Kramer, 2007; Wege et al., 2007).
We have used the TRV-LIC vector to generate a collection based on 400 tomato ESTs. This collection includes kinases, phosphstases, and transcription factors. It should therefore prove to be a useful collection for researchers interested in investigating the function of members of these protein families. TRV-VIGS is effective in several Solanaceous species and our tomato collection could be used for VIGS in tomato, potato, pepper, or N. benthamiana. Here, we used tomato clones to silence several N. benthamiana genes. Based on the tomato sequence, we cloned the corresponding N. benthamiana sequence and examined whether the phenotypes we obtained with the tomato clone were an accurate representation of the endogenous gene function. In all cloned 14 homologs of N. benthamiana, we found that the results from tomato clones held true for the N. benthamiana sequence.
In the course of our studies with this silencing collection we observed a range of developmental phenotypes. Of particular interest was that generated by silencing SlMADS1. Sequence analysis shows that SlMADS1 belongs to the MADS transcription factor family. Members of this family have critical roles in flowering time and floral organ identity, in addition to roles in other important aspects of plant development (Ng and Yanofsky, 2001; Henderson and Dean, 2004). SlMADS1 belongs to the MIKC class of MADS transcription factors and its domain architecture consists of the MADS box (M) that is important for DNA binding and dimerization, the Intervening (I) domain for functional specialization, the Keratin-like (K) domain, and the C-terminal (C) domain for transcriptional activation (Kaufmann et al., 2005). The flowers of SlMADS1-silenced plants displayed enlarged sepals, the conversion of second-whorl petals to leaves in some cases, and the replacement of the fourth-whorl carpel with other flowers that also showed larger-than-normal sepals and petal conversion. Some of these phenotypes are reminiscent of the sepallata1 (sep1) sep2 sep3 triple mutant (Pelaz et al., 2000). However, SlMADS1-silenced plants also displayed a loss of apical dominance and the production of many secondary branches. SlMADS1 is most similar to NtMADS4, a member of the AGAMOUS-like2 subfamily of MADS transcription factors (Jang et al., 2002). NtMADS4 is known to have a role in flowering in tobacco, where ectopic overexpression leads to early flowering, although the flowers showed no defects in organization or tissue identity (Jang et al., 2002).
We cloned the N. benthamiana homolog of SlMADS1 and found two very similar sequences, NbMADS4-1 and NbMADS4-2. Both NbMADS4-1 and NbMADS4-2 show typical MIKC-type MADS transcription factor domain architecture (Supplemental Fig. S1) and they are highly similar to other MADS proteins, including Arabidopsis SEP proteins. Similar to NtMADS4 and other AGL2 family members, NbMADS4-1 and -2 were expressed almost exclusively in floral tissue (Jang et al., 2002). We used the TRV2-LIC vector to silence both NbMADS4-1 and NbMADS4-2, either simultaneously or individually. It should be noted that the sequences we used for specific silencing contained one or two regions of greater than 21 bp of homology between NbMADS4-1 and -2 (Supplemental Figs. S3 and S4). Despite this, we were able to generate specific silencing, as shown by our RT-PCR analysis. One possible explanation for this is that insufficient small interfering RNAs were derived from these regions to initiate silencing of the homolog not being targeted.
Silencing NbMADS4-1 and NbMADS4-2, either simultaneously or separately, resulted in a loss of apical dominance and defects in floral organ identity. However, we did not observe a change in the timing of floral initiation. Interestingly, a loss of apical dominance was also observed when NtMADS4 was overexpressed (Jang et al., 2002). Together, these phenotypes point to an important role for NtMADS4/NbMADS4-1/2 in flowering. Interestingly, NbMADS4-1 and -2 genes are not redundant as specific silencing of either homolog produced all the phenotypes observed when SlMADS1 was silenced. This is quite different from the situation with the AtSEP genes, which are functionally redundant (Pelaz et al., 2000). Thus while NbMADS4-1 and NbMADS4-2 function in the same process, they appear to have independent, essential roles. MADS proteins are known to function as dimers (Kaufmann et al., 2005). We speculate the NbMADS4-1 and -2 may act together as a dimer and loss of either protein results in loss of activity.
MATERIALS AND METHODS
Plasmid Construction
The VIGS vectors pTRV1 (pYL192) and pTRV2 (pYL170) have been described previously (Liu et al., 2002b; Burch-Smith et al., 2006). The TRV2-LIC vector (pYY13) was derived from pYL170 (Burch-Smith et al., 2006). Two adaptors with PstI restriction sites were inserted into pYL170 by XbaI and XmaI digestion and ligation to produce pYY12. The ccdB gene was amplified by primers 5′-GCTAAGGAAGCTAAACTTTTGCTGACGAGAACAGGGACTGG-3′ and 5′-CGCCTGCAGCTCGAGCAGACTGGCTGTGTATAAGGGAGCCTG-3′, and inserted into pYY12 at the PstI site to yield TRV2-LIC (pYY13; Fig. 1A).
Cloning Tomato EST VIGS Collection
Four hundred tomato (Solanum lycopersicum) ESTs were amplified with primers: 5′-CGACGACAAGACCCT-plasmid-specific sequence-3′ and 5′-GAGGAGAAGAGCCCT-plasmid-specific sequence-3′. The PCR products were purified with polyethylene glycol/MgCl2 to remove any nonspecific PCR products and primers. A total of 50 ng of purified PCR product was treated with T4 DNA polymerase (New England Biolabs) in 1× reaction buffer containing 5 mm dATP and dithiothreitol at 22°C for 30 min followed by 20 min of inactivation of T4 DNA polymerase at 70°C. The TRV2-LIC vector was then digested with PstI and similarly treated with T4 DNA polymerase but dTTP replaced dATP. A total of 50 ng of treated PCR product and TRV2-LIC vector were mixed and incubated at 65°C for 2 min and then 22°C for 10 min. Then 6 μL of the mixture was transformed into Escherichia coli DH10B competent cells (Fig. 1B). Transformants were tested by PCR amplification using primers 5′-TGTTACTCAAGGAAGCACGATGAGCT-3′ and 5′-GAGGAGAAGAGCCCTGCCGCTCTAGAACTAGTGGATCC-3′. Plasmids from positive clones were purified and sequenced. To generate TRV2-LIC-NbPDS, NbPDS was amplified from Nicotiana benthamiana genomic DNA using primers 5′-CGACGACAAGACCCTCGGTCTAGAGGCACTCAACTTTATAAACC-3′ and 5′-GAGGAGAAGAGCCCTTCCCTTCAGTTTTCTGTCAAACC-3′.
Plant Growth and Agroinfiltration
N. benthamiana plants were grown in pots at 25°C on light carts under continuous light. For VIGS TRV1 (Liu et al., 2002b), TRV2 or TRV2-LIC and its derivatives were introduced into Agrobacterium tumefaciens strain GV2260 by heat shock. For screening the TRV2-LIC tomato EST collection, 2 mL overnight cultures were grown at 28°C in the appropriate antibiotic selection medium in a 96-well culture box. The next day, cultures were spun down and cells were resuspended in infiltration medium (10 mm MES, 10 mm MgCl2, 200 μm acetosyringone), adjusted to OD600 of 1, and incubated at room temperature for 3 h. N. benthamiana infiltration was performed as previously reported (Liu et al., 2002b). Infiltrated plants were observed for 8 weeks.
RNA Extraction and RT-PCR Analysis
Total RNA was extracted from leaves or flowers of wild-type N. benthamiana plants using RNeasy plant minikit (Qiagen). First-strand cDNA was synthesized using 1 μg of total RNA, gene-specific primers, and SuperScript reverse transcriptase (Invitrogen) according to the manufacturer's protocol. Primers are listed in Supplemental Table S1. RT-PCR products were cloned into TRV-LIC vector (pYY13) and performed VIGS in N. benthamiana. The expression levels of 14 genes whose silencing resulted in severe developmental phenotypes were monitored by semiquantitative RT-PCR using gene-specific primers that anneal outside the region targeted for silencing (Supplemental Table S2).
In NbMADS4-silenced plants, expression levels of LeMADS5, NtMADS11, and NAG1 were examined by semiquantitative RT-PCR using primers 5′-ACCGAATATATCAACACGAGAAGCACTG-3′ and 5′-CTAGGCCCTGCTCCTCCTACTGTAATTG-3′, 5′-CTCATATGCTGAGAGGCAGCTTACTGCT-3′ and 5′-GATGGCGAAGCATCCATGGCGGCATTAC-3′, and 5′-CTCTCCACAAAGGAAACTGGGAAGAG-3′ and 5′-GACTAGTTGAAGAGATGGTTGGTC-3′, respectively.
Cloning NbMADS4-1 and NbMADS4-2 from N. benthamiana
NbMADS4-1 was amplified by RT-PCR from N. benthamiana flower RNA using primers 5′-CGACGACAAGACCCTGGGAAGAGGAAGAGTTGAACTTAAG-3′ and 5′-GACCACTTTGTACAAGAAAGCTGGG(T)25V. NbMADS4-2 was amplified by RT-PCR using primers 5′-ACGACAAGACCCTCTTTCCTTCTTCTGTATCTGTGAGAGAAAAGAAAG-3′ and 5′-GAGGAGAAGAGCCCTCATCATCGTCGTATTAGTTCATACAAGTAG-3′. RT-PCR products were cloned into pCR2.1-TOPO (Invitrogen). The cDNA sequences were aligned by ClustalW.
NbMADS4 Expression Analysis in Different Tissues
Total RNA was purified from sepals, petioles, petals, carpels, roots, stems, leaves, and seedlings of wild-type N. benthamiana, and 2 μg RNA was used for RT-PCR. Primers used to amplify NbMADS4-1 were 5′-CGACGACAAGACCCTGAAGATTTGGGGACATTAAGTAC-3′ and 5′-GAGGAGAAGAGCCCTCACCCATGGGACTATATCCAAATTGAGG-3′. Primers 5′-CAATGCAGCTACGCCTCTTTGGACCCAATG-3′ and 5′-CAGTTGCTGCTGCATTAACCTCGTTTCCAC-3′ were used for NbMADS4-2.
Silencing Specificity
Two cDNA fragments were amplified for each NbMADS4 gene by PCR. Primers for fragment 1 of NbMADS4-1 were 5′-CGACGACAAGACCCTGGGAAGAGGAAGAGTTGAACTTAAG-3′ and 5′-GAGGAGAAGAGCCCTCTTCCTTGACCTGATTTGCTTCAAGG-3′. Primers for fragment 2 were 5′-CGACGACAAGACCCTGAAGATTTGGGGACATTAAGTAC-3′ and 5′-GAGGAGAAGAGCCCTCACCCATGGGACTATATCCAAATTGAGG-3′. Primers for fragment 1 of NbMADS4-2 were 5′-CGACGACAAGACCCTATGGGAAGAGGAAGAGTTGAACTAAAG-3′ and 5′-GAGGAGAAGAGCCCTGTGTGCCCAAGTCCTCCCCAAGAAG-3′. Primers for fragment 2 were 5′-CGACGACAAGACCCTTCCAAGGAACTTGAGCATCTTGAG-3′ and 5′-GAGGAGAAGAGCCCTTTACAGCATCCATCCTGGAATAAATC-3′. cDNA fragments were cloned into TRV2-LIC vector. Gene expression levels were monitored by semiquantitative RT-PCR using specific primers that anneal outside the region targeted for silencing. In addition, NbMADS4-1-silenced plants were also tested with NbMADS4-2-specific primers 5′-CGACGACAAGACCCTGGGAAGAGGAAGAGTTGAACTTAAG-3′and 5′-GAGGAGAAGAGCCCTCACCCATGGGACTATATCCAAATTGAGG-3′.
Microscopy
Tissue was vacuum infiltrated with 0.1 mg/mL propidium iodide and incubated for 30 min at 4°C in the dark. Confocal images were acquired on a Zeiss Axiovert 200 m light microscope equipped with a Zeiss LSM 510 NLO laser scanning microscope using a 10× C-Apochromat (NA 1.2) water-corrected objective lens. Scale bar is 50 μm.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of amino acids of N. benthamiana MADS4, and their tobacco and tomato homologs.
Supplemental Figure S2. Alignment of cDNAs of NbMADS4-1 and NbMADS4-2.
Supplemental Figure S3. Nucleotide sequence comparison of fragment 1 of NbMADS4-1 (NbMADS4-1F1) and fragment 1 from NbMADS4-2 (NbMADS4-2F1) used for silencing.
Supplemental Figure S4. Nucleotide sequence comparison of fragment 2 of NbMADS4-1 (NbMADS4-1F2) and fragment 2 from NbMADS4-2 (NbMADS4-2F2) used for silencing.
Supplemental Table S1. Primers to amplify N. benthamiana cDNAs.
Supplemental Table S2. Primers for RT-PCR.
Supplemental Table S3. EST clones.
Supplementary Material
Acknowledgments
We thank Dr. Greg Martin of the Boyce Thompson Institute at Cornell University for his generous gift of a partial tomato EST collection. We thank Shawn Bachan of Yale University for helpful suggestions on sequencing of TRV-EST clones. We are also grateful to Dr. Kirk Czymmek of the Delaware Biotechnology Institute for his assistance with microscopy.
This work was supported by the National Science Foundation Plant Genome (grant no. DBI–0211872).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Savithramma P. Dinesh-Kumar (savithramma.dinesh-kumar@yale.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Aslanidis C, de Jong PJ (1990) Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res 18 6069–6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldt R, Borner T, Schnarrenberger C (1992) Repression of the plastidic isoenzymes of aldolase, 3-phosphoglycerate kinase, and triosephosphate isomerase in the barley mutant “albostrians”. Plant Physiol 99 895–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigneti G, Martin-Hernandez AM, Jin H, Chen J, Baulcombe DC, Baker B, Jones JD (2004) Virus-induced gene silencing in Solanum species. Plant J 39 264–272 [DOI] [PubMed] [Google Scholar]
- Brodersen P, Voinnet O (2006) The diversity of RNA silencing pathways in plants. Trends Genet 22 268–280 [DOI] [PubMed] [Google Scholar]
- Burch-Smith TM, Anderson JC, Martin GB, Dinesh-Kumar SP (2004) Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J 39 734–746 [DOI] [PubMed] [Google Scholar]
- Burch-Smith TM, Schiff M, Liu Y, Dinesh-Kumar SP (2006) Efficient virus-induced gene silencing in Arabidopsis. Plant Physiol 142 21–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Jiang CZ, Gookin TE, Hunter DA, Clark DG, Reid MS (2004) Chalcone synthase as a reporter in virus-induced gene silencing studies of flower senescence. Plant Mol Biol 55 521–530 [DOI] [PubMed] [Google Scholar]
- Chung E, Seong E, Kim YC, Chung EJ, Oh SK, Lee S, Park JM, Joung YH, Choi D (2004) A method of high frequency virus-induced gene silencing in chili pepper (Capsicum annuum L. cv. Bukang). Mol Cells 17 377–380 [PubMed] [Google Scholar]
- D'Agostino N, Traini A, Frusciante L, Chiusano ML (2007) Gene models from ESTs (GeneModelEST): an application on the Solanum lycopersicum genome. BMC Bioinformatics (Suppl 1) 8 S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckman L, Gu M, Stols L, Donnelly MI, Collart FR (2002) High throughput methods for gene cloning and expression. Protein Expr Purif 25 1–7 [DOI] [PubMed] [Google Scholar]
- Dinesh-Kumar SP, Anandalakshmi R, Marathe R, Schiff M, Liu Y (2003) Virus-induced gene silencing. Methods Mol Biol 236 287–294 [DOI] [PubMed] [Google Scholar]
- Ding XS, Rao CS, Nelson RS (2007) Analysis of gene function in rice through virus-induced gene silencing. Methods Mol Biol 354 145–160 [DOI] [PubMed] [Google Scholar]
- Ding XS, Schneider WL, Chaluvadi SR, Mian MA, Nelson RS (2006) Characterization of a Brome mosaic virus strain and its use as a vector for gene silencing in monocotyledonous hosts. Mol Plant Microbe Interact 19 1229–1239 [DOI] [PubMed] [Google Scholar]
- Ekengren SK, Liu Y, Schiff M, Dinesh-Kumar SP, Martin GB (2003) Two MAPK cascades, NPR1, and TGA transcription factors play a role in Pto-mediated disease resistance in tomato. Plant J 36 905–917 [DOI] [PubMed] [Google Scholar]
- Gould B, Kramer EM (2007) Virus-induced gene silencing as a tool for functional analyses in the emerging model plant Aquilegia (columbine, Ranunculaceae). Plant Methods 3 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein I, Barciszewska-Pacak M, Hrubikova K, Williamson S, Dinesen M, Soenderby IE, Sundar S, Jarmolowski A, Shirasu K, Lacomme C (2005) Virus-induced gene silencing-based functional characterization of genes associated with powdery mildew resistance in barley. Plant Physiol 138 2155–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Dean C (2004) Control of Arabidopsis flowering: the chill before the bloom. Development 131 3829–3838 [DOI] [PubMed] [Google Scholar]
- Hileman LC, Drea S, Martino G, Litt A, Irish VF (2005) Virus-induced gene silencing is an effective tool for assaying gene function in the basal eudicot species Papaver somniferum (opium poppy). Plant J 44 334–341 [DOI] [PubMed] [Google Scholar]
- Holzberg S, Brosio P, Gross C, Pogue GP (2002) Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J 30 315–327 [DOI] [PubMed] [Google Scholar]
- Hull R (2002) Matthews' Plant Virology, Ed 4. Academic Press, New York
- Jang S, An K, Lee S, An G (2002) Characterization of tobacco MADS-box genes involved in floral initiation. Plant Cell Physiol 43 230–238 [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Melzer R, Theissen G (2005) MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene 347 183–198 [DOI] [PubMed] [Google Scholar]
- Kumagai MH, Donson J, Della-Cioppa G, Harvey D, Hanley K, Grill LK (1995) Cytoplamic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc Natl Acad Sci USA 92 1679–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Nakayama N, Schiff M, Litt A, Irish VF, Dinesh-Kumar SP (2004. a) Virus induced gene silencing of a DEFICIENS ortholog in Nicotiana benthamiana. Plant Mol Biol 54 701–711 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP (2002. a) Virus-induced gene silencing in tomato. Plant J 31 777–786 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP (2004. b) Involvement of MEK1 MAPKK, NTF6 MAPK, WRKY/MYB transcription factors, COI1 and CTR1 in N-mediated resistance to tobacco mosaic virus. Plant J 38 800–809 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP (2002. b) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30 415–429 [DOI] [PubMed] [Google Scholar]
- Lu R, Martin-Hernandez AM, Peart JR, Malcuit I, Baulcombe DC (2003) Virus-induced gene silencing in plants. Methods 30 296–303 [DOI] [PubMed] [Google Scholar]
- Meins F Jr, Si-Ammour A, Blevins T (2005) RNA silencing systems and their relevance to plant development. Annu Rev Cell Dev Biol 21 297–318 [DOI] [PubMed] [Google Scholar]
- Mueller LA, Solow TH, Taylor N, Skwarecki B, Buels R, Binns J, Lin C, Wright MH, Ahrens R, Wang Y, et al (2005) The SOL Genomics Network: a comparative resource for Solanaceae biology and beyond. Plant Physiol 138 1310–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M, Yanofsky MF (2001) Function and evolution of the plant MADS-box gene family. Nat Rev Genet 2 186–195 [DOI] [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405 200–203 [DOI] [PubMed] [Google Scholar]
- Pnueli L, Hareven D, Rounsley SD, Yanofsky MF, Lifschitz E (1994) Isolation of the tomato AGAMOUS gene TAG1 and analysis of its homeotic role in transgenic plants. Plant Cell 6 163–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff F, Martin-Hernandez AM, Baulcombe DC (2001) Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J 25 237–245 [DOI] [PubMed] [Google Scholar]
- Robertson D (2004) VIGS vectors for gene silencing: many targets, many tools. Annu Rev Plant Biol 55 495–519 [DOI] [PubMed] [Google Scholar]
- Ruiz MT, Voinnet O, Baulcombe DC (1998) Initiation and maintenance of virus-induced gene silencing. Plant Cell 10 937–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield SR, Huang L, Brandt AS, Gill BS (2005) Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiol 138 2165–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R, Oster U, Kruse E, Rudiger W, Grimm B (1999) Reduced activity of geranylgeranyl reductase leads to loss of chlorophyll and tocopherol and to partially geranylgeranylated chlorophyll in transgenic tobacco plants expressing antisense RNA for geranylgeranyl reductase. Plant Physiol 120 695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Cai X, Wang X, Zheng Z (2006) Optimisation of tobacco rattle virus-induced gene silencing in Arabidopsis. Funct Plant Biol 33 347–355 [DOI] [PubMed] [Google Scholar]
- Wege S, Scholz A, Gleissberg S, Becker A (2007) Highly efficient virus-induced gene silencing (VIGS) in California poppy (Eschscholzia californica): an evaluation of VIGS as a strategy to obtain functional data from non-model plants. Ann Bot (Lond) 100 641–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.