The use of transgenic technologies for the genetic manipulation of plant species has had a profound impact on basic plant research and biotechnology. Overexpression of heterogonous genes, for example, is widely used for the introduction of novel traits into transgenic crop plants. Overexpression can also be used in combination with down-regulation and controlled expression (e.g. induced, developmentally regulated, or tissue specific) studies as a tool for basic plant research and for functional analysis of native genes in various model plants. Two crucial components are required for the transient and/or stable expression of foreign genes in plant cells. The first is a plasmid vector capable of carrying the foreign gene-encoding sequence and the regulatory elements, i.e. promoter and terminator sequences, needed for its expression in plant cells (thus collectively referred to as an expression cassette). The second is the biological or physical methods by which the vector can be delivered into the target plant cells. With the exception of virus-mediated gene expression, where the distinction between the plasmid vector and the vehicle itself is rather blurry, biolistics, polyethylene glycol, and Agrobacterium-mediated gene transfer all require plasmid vectors, some of which have been specifically designed for optimal use by a particular method. Thus, for example, small and multicopy plasmids are most useful vectors for biolistics and polyethylene glycol-mediated genetic transformation where large quantities of DNA are typically used in each experiment. On the other hand, single-copy plasmids carrying certain biological features have been specifically designed for use with Agrobacterium as the transformation vehicle.
In the past several decades, a remarkable variety of plasmids suitable for the cloning, transfer, and expression of foreign genes in plant cells has been developed (e.g. Simoens et al., 1986; Hajdukiewicz et al., 1994; Hamilton, 1997; Xiang et al., 1999; Hellens et al., 2000b; Earley et al., 2006; Coutu et al., 2007). While these plasmids have proven instrumental in plant biology research and biotechnology, the basic designs of many of these vectors are quite restrictive and rarely permit the cloning and transfer of more than a single target gene (excluding the selection gene, which may or may not be present on the transformation vector) as a single unit into plant cells. The growing interest in dissecting and analyzing complex metabolic pathways and the need to exploit the full potential of multigene traits for plant biotechnology (for review, see Halpin et al., 2001; Halpin and Boerjan, 2003; Capell and Christou, 2004; Tyo et al., 2007) mandate the development of new methods and tools for the delivery and stacking of multiple genes in plant cells. Here, we review some of the methods that can be used for the delivery of multiple genes into plant cells, while focusing mainly on systems that allow the assembly of multigene plant transformation vectors.
AGROBACTERIUM BINARY PLASMIDS FOR DELIVERY OF A SINGLE TRANSGENE
Being the dominant method for plant genetic transformation, Agrobacterium vectors (Hellens et al., 2000a) have been the target of constant improvements and modifications over the past several decades. (For a unique and interesting historical perspective on the genesis of Agrobacterium vectors, the reader is referred to the excellent chronicle written by Chilton in 2001.) Agrobacterium vectors, commonly referred to as binary vectors, must carry the target gene's expression cassettes in a specialized area, designated the transferred DNA (T-DNA) region. When delivered into Agrobacterium's cells, binary plasmids replicate as the bacterial cell divides, but it is the T-DNA region that is typically transferred into the plant cells (for review, see Gelvin, 2003). Thus, the gene of interest and its regulatory sequences need to be placed within the borders (designated as left and right borders) of the T-DNA. Some of the early features seen on binary plasmids (e.g. Bevan, 1984) were the introduction of a plant selection cassette, a simple and narrow multiple-cloning site (MCS) region suitable for limited cloning of target sequences directly into the T-DNA region, and the addition of an Escherichia coli origin of replication (ori), which made it possible to propagate and manipulate these vectors in a friendlier and simpler environment. A crucial improvement made to the first generation of binary plasmids was the introduction of an empty plant expression cassette (e.g. Velten and Schell, 1985), a feature that allowed the plant biologist a simple and more direct route for cloning his/her gene of interest under the control of a plant-expressing constitutive promoter. Nevertheless, these vectors did not offer much flexibility in terms of target gene cloning, as they were typically constructed with a very narrow MCS (e.g. the number of unique restriction sites on the binary vectors developed back in 1985 by Velten and Schell ranged from one to a maximum of four). These pioneered constructs were soon followed by many other vectors offering new and improved features that included different bacterial ori, extended MCSs and various bacterial and plant selection markers, and even plant reporter genes (e.g. An et al., 1985; Simoens et al., 1986; Becker et al., 1992). The constant improvements in binary vectors even included the most famous binary vector, one which has been dominating the landscape of binary plasmids for several decades: pBin19 (Komori et al., 2007). This plasmid, the first to be commercialized for use in plant genetic transformation, to our knowledge, offers several features, including incorporation of the LacZ gene into the MCS to facilitate identification of recombinant plasmids using a colorimetric assay, a bacterial kanamycin-resistance gene, an E. coli ori, a complete plant selection marker expression cassette, and an extended MCS. One modification to this plasmid, for example, was the creation of pBINPLUS, a pBin19 derivative capable of producing a much higher copy number in E. coli and with a plant-selectable marker located near the T-DNA's left border (van Engelen et al., 1995), a feature that was found useful for integrating complete T-DNA sequences into the plant genome. Another modification to pBin19 was the incorporation of the GUS reporter gene and the creation of the even more popular vector pBI121, which has been used in many transformation studies and has just recently been reconstructed and completely sequenced (Chen et al., 2003).
With the advancement of transgenic technologies into nearly every niche of basic plant research and the ever-growing number of reports on the genetic transformation of various plant species came further advancement in vector technologies. New vectors were designed and constructed to provide the users with a more specialized set of tools suitable for performing various tasks in plant cells (e.g. the transfer of extremely long DNA molecules [Hamilton, 1997], the expression of fluorescent protein fusions [Goodin et al., 2002], and the detection of protein-protein interactions [Bracha-Drori et al., 2004]), while others were specifically designed for versatility and simplicity, allowing plant biologists not only a choice, but also the ability to manipulate these vectors for their own needs. The latter group of vectors are typically constructed as families of plasmids and include, for example, the pCB minibinary vector series that featured a collection of extremely small pBin19-derivative vectors (Xiang et al., 1999), the pGreen series of plasmids featuring a versatile and flexible series of binary vectors (Hellens et al., 2000b), and the pCAMBIA vectors featuring plasmids for GUSintron and GFP reporting, promoter cloning, and much more (http://www.cambia.org/daisy/cambia/materials/vectors/). These and many other families of vectors (e.g. McCormac et al., 1997; Curtis and Grossniklaus, 2003; Bracha-Drori et al., 2004; Earley et al., 2006; Coutu et al., 2007) provide the plant research community with a vast number of versatile tools for various plant expression analyses. (For a recent update on binary vectors, the reader is referred to an update by Komori et al., 2007).
DELIVERY OF MULTIPLE GENES BY SINGLE-GENE VECTORS
Most of the plasmids and families of plasmids that exist today are limited to the expression of a single gene (excluding the selection marker, when present). Realizing that many agronomical traits are polygenic in nature, the use of such plasmids for the coordinated manipulation of multiple traits presents a unique challenge for the plant genetic engineer. Several approaches can be considered when using single-gene vectors for the delivery of multiple genes into plant cells (for review, see Halpin et al., 2001; Daniell and Dhingra, 2002; Halpin and Boerjan, 2003). Some of the approaches used for the production of transgenic plants carrying multiple new traits include (1) retransformation (e.g. Singla-Pareek et al., 2003; Seitz et al., 2007), the stacking of several transgenes by successive delivery of single genes into transgenic plants; (2) cotransformation (e.g. Li et al., 2003), the combined delivery of several transgenes in a single transformation experiment; and (3) sexual crosses (e.g. Ma et al., 1995; Zhao et al., 2003; Lucker et al., 2004) between transgenic plants carrying different transgenes.
The manipulation of wood composition in forest trees is just one example of a challenge faced by molecular biologists while planning the engineering of forest trees with altered lignin content (Halpin and Boerjan, 2003). Rationalized by the complex, costly, and polluting process of cellulose removal during the pulping process, metabolic engineering of lignin structure and content is expected to yield transgenic trees with improved lignin properties, and Li et al. (2003) chose a cotransformation approach for the delivery of two different transgenes for the combinatorial modification of several lignin genes in aspen (Populus spp.) plants. The transgenes were cloned on independent binary vectors and were shuttled by two different Agrobacterium strains. Transgenic aspen plants carrying a variety of transgene combinations were recovered and revealed the additive affect of the analyzed lignin-biosynthesis pathway genes in forest trees (Li et al., 2003). The authors also reported that transformation of tobacco (Nicotiana tabacum) plants with a combination of four independent Agrobacterium strains even resulted in the recovery of transgenic plants carrying all four transgene constructs (Li et al., 2003). They thus suggested that cotransformation can be a useful method for the delivery of multiple transgenes into plant cells using single or dual gene-transformation vectors. Cotransformation was also used in the development of golden rice (Oryza sativa), in which four different genes (including a single selectable marker) were delivered from two independent constructs (Ye et al., 2000), and in the metabolic engineering for poly(3-hydroxybutyrate-co-3-hydroxyvalerate) production in Arabidopsis (Arabidopsis thaliana), where six transgenes (which included two different selectable markers) were delivered by two independent T-DNA molecules (Slater et al., 1999).
For all the benefits and simplicity of combining cotransformation, retransformation, and crosses while using single-gene vectors for the delivery of multiple genes into plant species, these methods suffer from several drawbacks. These include the undesirable incorporation of a complex T-DNA integration pattern, often observed during integration of T-DNA molecules from multiple sources (De Neve et al., 1997; De Buck et al., 1999), and the time needed for retransformation or crosses between transgenic plants. More importantly, transgenes derived from different sources typically integrate at different locations in the plant genome, which may lead to various expression patterns and possible segregation of the transgenes in the offspring.
Interestingly, a record number of transgenes was actually introduced into plant cells by codelivery of multiple single-gene plasmids using the particle bombardment transformation method. A mixture of 12 different plasmids was used for the delivery of 12 different gene constructs into soybean (Glycine max) embryogenic suspension culture (Hadi et al., 1996), and the bombardment of rice embryogenic culture with a mixture of 14 different plasmids resulted in the production of transgenic lines with up to 11 different transgenes (Chen et al., 1998). While expression of the transgenes delivered by this method proved stable across several generations, the independent segregation of the transgenes that integrated at different genomic locations and the limited efficiency of particle bombardment for the production of transgenic plants are just two of the major drawbacks for the use of such methods in plant biotechnology and research.
METHODS FOR SINGLE-VECTOR-BASED DELIVERY OF MULTIPLE TRANSGENES INTO PLANT CELLS
Delivering multiple genes by a single vector has a profound advantage over the use of multiple vectors, because only a single DNA molecule needs to be transferred into the cells. Thus, a smaller number of plants typically need to be generated, as compared with the transformation methods discussed above. Furthermore, because a single DNA molecule will finally integrate into the plant genome, all genes cloned into that molecule will most likely be inherited together. When using Agrobacterium, there is an additional advantage for launching a single T-DNA molecule from a single Agrobacterium strain, because the delivery of multiple T-DNA molecules by several Agrobacterium strains is more prone to complex integration patterns (De Neve et al., 1997; De Buck et al., 1999). Indeed, several methods and various vectors have been designed to facilitate the transfer of multiple genes using a single vector. One such design calls for the assembly of a special expression cassette in which the coding sequences (CDSs) of several proteins are fused together to produce a single transcriptional unit. Upon translation, the coded polyprotein can be processed and cleaved by specific proteases, giving rise to individual proteins. This method was used, for example, to coordinate the expression of multiple enzymes into various subcellular compartments in transgenic plants (Dasgupta et al., 1998). This was achieved by fusing together the CDSs of two reporter genes and of the tobacco vein mottling virus (TVMV) NIa proteinase and expressing them as a single transcript in plant cells. The proteins' CDSs were separated by TVMV NIa recognition sequences that allowed the digestion of the polyprotein by TVMV NIa and the release of processed proteins, which were properly targeted to different subcellular compartments. In another approach, the CDSs of two proteinase inhibitors were linked together by a sequence known to be sensitive to an as yet unidentified plant proteinase, which cleaved the polyprotein and produced two distinct proteins conferring plant resistance to nematodes (Urwin et al., 1998).
Several reports have demonstrated the expression of multiple proteins via a polyprotein construct in various plant species (e.g. Marcos and Beachy, 1994; von Bodman et al., 1995; Halpin et al., 1999; El Amrani et al., 2004; Ralley et al., 2004). The main advantages of these approaches are that they allow the production of multiple proteins at a controlled molar ratio and require the transfer of only a single expression cassette into the plant cells. However, the assembly of a fused transcript (which may be tedious and somewhat complicated), the translation of a long polyprotein (which may fold improperly), and the reliance on the cleaving activity of specific proteinases (that need to be present and active at the time and/or location of polyprotein production) may hinder the use of this technology for the routine expression of multiple genes in plant cells.
ASSEMBLY OF MULTIPLE-GENE EXPRESSION CASSETTES
Various strategies have been employed for the delivery of several transgenes into plant cells using single- and double-cassette vectors, each with its own advantages and disadvantages. One obvious solution for the delivery of multigenes into plant cells is the assembly of large, multicassette, plant transformation vectors. Nevertheless, the rigid design of many binary-vector systems and the limitations of type II restriction enzyme-mediated cloning hinder the user's ability to redesign and reconstruct these plasmids to carry several genes as a single transformable unit. Thus, while one cannot rule out the possibility of using traditional cloning methods and existing sets of binary vectors for the assembly of multiple-gene transformation vectors (as exemplified below), novel vectors and construction methods would certainly facilitate this task, making the assembly of complex vectors simpler and more straightforward.
VECTOR ASSEMBLY BY TRADITIONAL CLONING METHODS
In a recent report, Zhong et al. (2007) chose the option of assembling a triple-cassette binary vector to analyze the role of two NAC domain transcription factors, SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN1 (SND1) and NAC SECONDARY WALL-THICKENING PROMOTING FACTOR1 (NST1), in the regulation of secondary wall synthesis. The vector was composed of a single selectable marker and two SND1 and NST1 RNA interference (RNAi) cassettes. The construct was then used for the production of transgenic Arabidopsis plants in which the simultaneous RNAi inhibition of SND1 and NST1 led to loss of secondary wall formation in stem fibers (Zhong et al., 2007). While the specific technical details on the construction of this ternary plasmid were not clear, it can be assumed that conventional type II restriction enzymes and regular cloning techniques were applied. Indeed, conventional enzymes and common recombinant DNA techniques have also proven instrumental in the assembly of large binary plasmids, carrying four (Bohmert et al., 2000) and even five (Bohmert et al., 2002) different expression cassettes. A quadruple vector was assembled using a combination of EcoRI and HindIII digestions, with or without filling in of the restriction site overhangs by T4 DNA polymerase, and by successive cloning of three phb (polyhydroxybutyrate biosynthesis) expression cassettes into an acceptor vector carrying a plant-selectable marker. Although all three phb genes were cloned under the control of the same regulatory elements (i.e. cauliflower mosaic virus [CaMV] 35S promoters and nopaline terminators) in each cassette, expression of all genes was stable in transgenic Arabidopsis plants, which exhibited an accumulation of polyhydroxybutyrate to up to 4% of their fresh weight (Bohmert et al., 2000). Using a slightly different strategy and with the help of PCR amplification of specific DNA fragments, the same group of researchers later described the assembly of several more quadruple binary plasmids and even the assembly of a quintuple binary plasmid. The latter plasmid carried, in addition to the plant selection marker (under the control of the nopaline synthase promoter), three phb genes and the phasing gene from Ralstonia eutropha, all driven by the CaMV 35S constitutive promoter (Bohmert et al., 2002). The assembly of these multigene expression cassette vectors was proven useful for the production of multitransgene plants and for the development of a strategy for overproducing polyhydroxybutyrate in transgenic plants. Interestingly, the authors reported that constitutive expression of the β-ketothiolase (phbA) gene may cause certain problems during the transformation of Arabidopsis, potato (Solanum tuberosum), and tobacco plants (Bohmert et al., 2002). They thus suggested an inducible promoter or a somatically activated expression system as a possible strategy to overcome the transformation issues (Bohmert et al., 2002), thus emphasizing the need for the development of easy-to-use, flexible, and modular systems for the assembly of multiple-gene expression cassettes with various regulatory elements, as described further on.
A CRE/LOXP RECOMBINATION SYSTEM FOR VECTOR ASSEMBLY
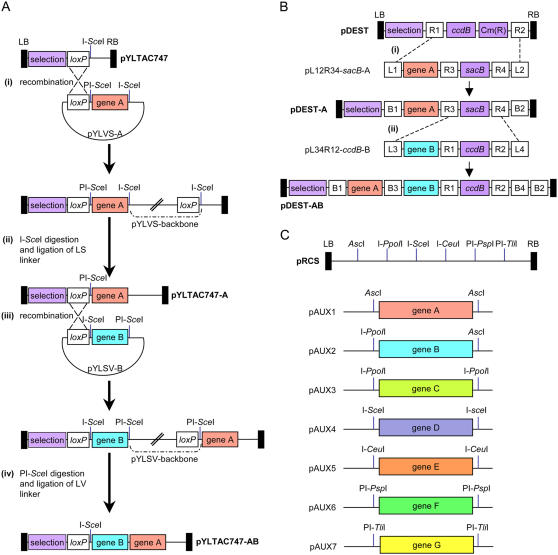
One of the major drawbacks of using conventional type II restriction enzymes for the cloning of large DNA fragments is the high occurrence of their restriction recognition sites within many DNA sequences. Thus, the use of these restriction enzymes for the assembly of multiple-gene expression cassettes can be rather tedious, limited, and sometimes even impossible. One interesting approach to overcoming these limitations was recently presented by Lin et al. (2003), who used a combination of the Cre/loxP recombination system and two rare-cutter (PI-SceI and I-SceI) endonucleases for the sequential assembly of multiple genes onto a transformation-competent artificial chromosome (TAC)-based vector. The system is based on the function of Cre as a reversible site-specific recombinase capable of recombining two loxP sites and on the idea that a pair of DNA sequences, located on two separate plasmids, can be linked together indefinitely by repeated recombination events. An acceptor, TAC-based binary vector, designated pYLTAC747, was constructed to contain loxP and I-SceI sites on the T-DNA region. The same loxP site was also placed next to the MCS of two small high-replicating donor vectors, pYLVS and pYLSV. Assembly of a multiple-gene binary vector begins with recombination between a pYLVS donor vector and pYLTAC747 and results in the incorporation of the entire donor plasmid onto the T-DNA region (Fig. 1A, step i). The unwanted donor plasmid backbone is removed by I-SceI digestion of the recombined plasmid (Fig. 1A, step ii), the cutter having been engineered to flank the MCS, together with PI-SceI, in both pYLVS and pYLSV. Ligation of the digested I-SceI with a suitable linker eliminates this site from the recombined plasmid, leaving it with the original loxP site and a new PI-SceI site, creating the binary plasmid pYLTAC747-A (Fig. 1A). These sites can now be used for new recombination and digestion steps using a pYLSV-based donor plasmid (Fig. 1A, step iii). Alternating between pYLVS and pYLSV and using multiple rounds of recombination and backbone removal, the authors assembled a binary vector carrying a set of 10 foreign or functional DNA sequences and used it for the successful transformation of rice plants.
Figure 1.
Methods for the assembly of multiple-gene binary plasmids. A, The Cre/loxP-mediated multigene assembly process. i, Cre/loxP recombination of the pYLVS-A plasmid into the pYLTAC747 acceptor binary plasmid. ii, Release of the pYLVS backbone by I-SceI digestion and ligation with a LS linker. This ligation abolishes the I-SceI site from pYLTAC747-A. iii, Cre/loxP recombination of the pYLSV-B plasmid into the pYLTAC747-A and release of the pYLSV backbone by PI-SceI digestion and ligation with a LV linker (iv). B, The MultiRound Gateway assembly process. i, Gateway recombination between attL1 and attR1 and between attL1 and attR1 sites by LR clonase and conversion of the ccdB-based binary pDEST vector into a sacB-based Destination vector. ii, Gateway recombination between attL3 and attR3 and between attL4 and attR4 sites by LR clonase and reconversion of the destination binary vector into a ccdB-based plasmid. C, The homing endonucleases pRCS/pAUX vector system. Assembly of a multigene binary plasmid is achieved by successive cloning of various gene expression cassettes using rare-cutting homing endonucleases.
GATEWAY-BASED METHODS FOR VECTOR ASSEMBLY
Gateway-mediated DNA cloning is another recombination-based system that can offer an alternative to the use of Cre/loxP and rare cutters for the assembly of multiple-gene vectors. Similar to Cre/loxP-mediated recombination, the Gateway technology is based on the action of recombinases that are capable of recombining compatible DNA sequences. Thus, for example, the LR clonase can recombine attL with attR sites in an irreversible fashion and is typically used for the delivery of DNA fragments from an Entry clone into a Destination vector (Walhout et al., 2000). The simplicity of the Gateway technology led to the development of various Gateway-compatible plant transformation vectors. While these vectors were designed to perform various tasks, such as protein localization studies, constitutive and inducible expression of foreign genes, and promoter analysis (e.g. Karimi et al., 2002; Curtis and Grossniklaus, 2003; Walter et al., 2004; Earley et al., 2006), they were not designed with the intention of allowing the stacking of several cassettes in a single plant transformation vector. The introduction of Multisite Gateway as a method for recombining multiple DNA segments in a single recombination step (Cheo et al., 2004; Sasaki et al., 2004; Karimi et al., 2005) led to the development of new multigene stacking systems. Using a set of Destination binary vectors that were specifically designed for the Multisite Gateway LR clonase reaction and a set of three Entry plasmids (each carrying a different att site), a four-expression-cassette vector was recently assembled (Wakasa et al., 2006). The vector carried, in addition to a plant selection marker, three independent modified glutelin gene cassettes, and its introduction into rice plants resulted in high-level accumulation of the hypocholesterolemic peptide lactostatin (IIAEK) in the seed endosperm (Wakasa et al., 2006). While useful for the delivery of several transgenes into plant cells, the use of Multisite Gateway for the stacking of multiple genes is still limited by the complexity of the LR reactions and the small number of available att sites.
Another Gateway-based system describes the assembly of multigene vectors through a series of recombination steps while alternating between two different Gateway Entry vectors (Chen et al., 2006). This MultiRound Gateway cloning strategy is based on two unique Gateway Entry vectors that can be used in conjunction with virtually any type of Gateway Destination vector that carries the attR1-attR2 or attR3-attR4 recombination sites (e.g. pMDC99, Curtis and Grossniklaus, 2003; pHWG, Karimi et al., 2002; or pHm43GW, Karimi et al., 2005). The two entry vectors, pL12R34-sacB and pL34R12-ccdB, were designed for a Gateway negative screening strategy using sacB and ccdB. (The SacB protein synthesizes a substrate, in the presence of Suc, that is toxic to gram-negative bacteria, while the product of the cddB gene is poisonous to most E. coli strains.) Each vector also featured a unique pair of attL and attR recombination sites: The pL12R34-sacB carried the attR3-attR4 recombination sites flanked by a pair of attL1-attL2 sites, while the pL34R12-ccdB carried attR1-attR2 recombination sites flanked by a pair of attL3-attL4 sites and several restriction sites for the cloning of target genes (Fig. 1B). Assembly of a multiple-gene vector begins with LR recombination between one of the pL Entry vectors and a Destination vector (exemplified by the introduction of pL12R34-sacB-A into the empty binary Destination vector pDEST; Fig. 1B, step i). This recombination step results in (1) transfer of the first target DNA sequence into the Destination vector; (2) replacement of the Destination vector's negative selection gene with that of the Entry vector (e.g. ccdB with sacB; Fig. 1B); and (3) the incorporation of a new pair of attR recombination sites (e.g. the insertion of attR3-attR4; Fig. 1B). A second recombination LR step can now take place between the second Entry vector and the recombinant Destination vector (exemplified by the introduction of pL34R12-ccdB-B into pDEST-A; Fig. 1B, step ii). This step leads to the transfer of the second target DNA sequence into the Destination vector. It also leads to the replacement of the negative selection gene (e.g. sacB with ccdB; Fig. 1B) and the incorporation of a new pair of attR recombination sites (e.g. the attR1-attR2; Fig. 1B) into the Destination vector. The resulting vector (pDEST-AB; Fig. 1B) can now be used for a third recombination step with the proper Entry vector (e.g. pL12R34-sacB-C). Using multiple recombination cycles, Chen et al. (2006) reported the assembly of a plant transformation vector containing seven functional DNA fragments and its use for the stable genetic transformation of Arabidopsis and tobacco plants. The authors also constructed pL12R34-Ap and pL34R12-Cm, a compatible pair for pL12R34-sacB and pL34R12-ccdB, which were designed for a positive screening strategy with chloramphenicol and ampicillin and an additional two pairs of plasmids suitable for negative and positive selection, but which also contained I-SceI sites. The latter sites can be used for linearizing Entry vectors prior to their use in a recombination experiment, a practice that is considered to assist in LR recombination efficiency.
UTILIZING RARE CUTTERS FOR VECTOR ASSEMBLY
The use of rare cutters for removal of the plasmid backbone following Cre/loxP-mediated recombination (Lin et al., 2003), or for plasmid linearization prior to Gateway recombination (Chen et al., 2006), is possible due to the low occurrence of their recognition sequences within natural and artificial DNA sequences. Indeed, rare cutter recognitions sites have been incorporated into other plant vectors, typically within their MCSs, where they assist with the cloning of large DNA fragments (e.g. Lonsdale et al., 1995; Uberlacker and Werr, 1996; de Maknik et al., 1997). In a unique approach, Goderis et al. (2002) addressed the issue of multigene stacking by constructing a system in which several expression cassettes can be mounted together into specifically designed binary vectors using a set of different rare-cutting enzymes. The system featured two binary vectors (designated pPZP-RCS1 and pPZP-RCS2) with a MCS consisting of 11 rare-cutting (six octanucleotide and five homing endonuclease) recognition sites (Fig. 1C) and the addition of 13 hexanucleotide restriction sites. It also included a set of auxiliary (pAUX) plasmids that allowed the cloning of DNA sequences between pairs of homing endonuclease recognition sites (Fig. 1C) and thus the stacking of up to seven different target DNA sequences into the MCS of the pPZP-RCS1 and pPZP-RCS2 binary plasmids. The authors next assembled six different expression cassettes carrying various genes into different pAUX plasmids and successively mounted all six expression cassettes onto pPZP-RCS2. Biochemical and molecular analyses confirmed that all genes were stably expressed in transgenic Arabidopsis plants obtained by Agrobacterium-mediated transformation of the recombinant pPZP-RCS2 plasmid (Goderis et al., 2002).
One of the main advantages of the pAUX system over recombinase-based vector systems is that it is somewhat modular and allows the replacement of expression cassettes without interfering with the other elements already present in the vector. Nevertheless, it still suffers from one crucial drawback: Its original design limits its capacity to seven pAUX plasmids, and while other expression cassettes and functional DNA fragments can still be cloned into the remaining octanucleotide and hexanucleotide restriction sites, these sites do not feature the flexibility provided by the homing endonucleases and their corresponding pAUX plasmids. Furthermore, the pAUX plasmids were originally designed with a rather limited MCS and did not contain any regulatory elements for plant expression. Thus, additional cloning and reconstruction of functional expression cassettes are required from those who plan to use this system for the assembly of multigene binary vectors.
THE pSAT FAMILY OF PLASMIDS
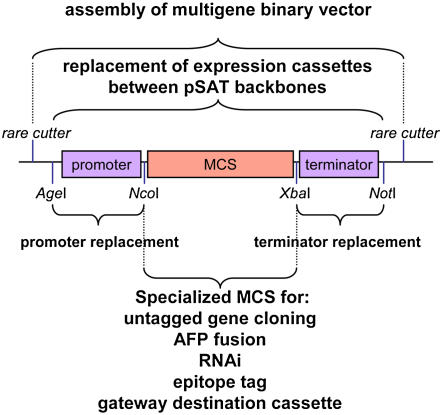
Appreciating the power of rare cutters for the assembly of binary vectors, several reports described the conversion of the pAUX series of plasmids into versatile and modular collections of plasmids useful for executing various tasks in plant cells (summarized in Table I). The first set of pAUX-based vectors, designated pSAT (satellite)-AFP plasmids (Tzfira et al., 2005), were constructed to support the N- and C-terminal fusion of five different autofluorescent tags. Developed with the intention of providing a flexible design (Fig. 2), the vectors carry expanded MCSs that allow easy exchange of target genes between different autofluorescent tags. Expression of the tagged proteins is controlled by tandem CaMV 35S promoter and terminator and each of these regulatory elements can be replaced using a single, simple cloning step. Finally, all expression cassettes can be mobilized between the seven different pAUX/pSAT plasmids using simple, one-step cloning by AgeI and NotI (Fig. 2). The pSAT-AFP series of plasmids was supplemented with a set of plasmids (pSAT-MCS) suitable for overexpression of untagged, free proteins under the control of the tandem CaMV 35S promoter. The flexibility of the pSAT-MCS design proved useful during the extension of the pSAT family of plasmids to carry various promoters and terminators (Chung et al., 2005). The pSAT.pro.MCS.ter series of plasmids supported the expression of untagged proteins under the control of five additional constitutive promoters (Table I). Furthermore, it also allowed easy modification and removal of the ATG-containing NcoI site from several pSAT-AFP and pSAT.pro.MCS.ter plasmids, allowing for greater versatility while cloning ATG-containing target genes into these vectors (Chung et al., 2005). Other modifications of the basic pSAT-AFP plasmids (Table I) included the introduction of the pSAT-BiFC family of plasmids, useful for the bimolecular fluorescence complementation assay (Citovsky et al., 2006), the construction of pSAT-red fluorescent protein (RFP), for the N- and C-terminal fusion of RFP (enhanced cyan variant of GFP-enhanced cyan fluorescent protein [ECFP], or the monomeric form of DsRed2; Chung et al., 2005; Citovsky et al., 2006), and the assembly of pSAT-RNAi, a set of vectors useful for the expression of hairpin structures for RNAi in plant cells (Dafny-Yelin et al., 2007). Additional plasmids, specifically designed to facilitate the expression of epitope-tagged proteins in plant cells, have also been constructed (T. Tzfira, unpublished data), as have two sets of plant selection-marker expression cassettes, controlled under two different constitutive promoters (Chung et al., 2005; Tzfira et al., 2005), allowing the user the freedom of choosing the preferred selection marker while assembling a multigene binary vector. Finally, to accommodate the need for Gateway-mediated gene cloning, the pSAT-DEST plasmids, which were originally designed to support N- and C-terminal fusion to enhanced GFP (EGFP) and promoter-expression analysis (Tzfira et al., 2005), were recently expanded by the introduction of the pSITE family of vectors (Chakrabarty et al., 2007). In this last expansion, the authors mounted the Gateway-ready expression cassettes onto a pRCS-based plasmid, making the use of these plasmids more straightforward. Future plans for the extension of the pSAT family of plasmids include the assembly of subcellular markers tagged with various autofluorescent tags, the construction of tissue-specific expression cassettes, and the assembly of whole-plant and tissue-specific inducible systems.
Table I.
pSAT family of plasmids
T, Tags; G, genes; P, promoters. EYFP, Enhanced yellow fluorescent protein; citrine-YFP, citrine yellow fluorescent protein; nEYFP, amino-terminal half of EYFP; cEYFP, carboxy-terminal half of EYFP; GST, glutathione S-transferase.
| Plasmid Series | Uses and Features | Tags, Genes, or Promoters | References |
|---|---|---|---|
| pAUX | Empty auxiliary plasmids | n/a | Goderis et al. (2002) |
| pSAT-AFP | N- and C-terminal fusion to autofluorescent tags | T: EGFP, EYFP, citrine-YFP, ECFP, and DsRed2 | Tzfira et al. (2005) |
| P: Tandem CaMV 35S with a translational enhancer | |||
| pSAT-MCS | Overexpression of untagged proteins | P: Tandem CaMV 35S with a translational enhancer | Tzfira et al. (2005) |
| pSAT-pro.MCS.ter | Overexpression of untagged proteins from various constitutive promoters | P: Actin, agropine synthase, manopine synthase, nopaline synthase, octopine synthase, Rubisco small subunit, tandem CaMV 35S with a translational enhancer | Chung et al. (2005) |
| pSAT-selection | Overexpression of plant selection markers | G: nptII, bar, hpt | Chung et al. (2005); Tzfira et al. (2005) |
| P: Tandem CaMV 35S with a translational enhancer; octopine synthase | |||
| pSAT-BiFC | Subcellular localization of interacting proteins by bimolecular fluorescence complementation | T: nEYFP, cEYFP | Citovsky et al. (2006) |
| P: Tandem CaMV 35S with a translational enhancer | |||
| pSAT-RFP | N- and C-terminal fusion to autofluorescent mRFP tag | T: RFP | Chung et al. (2005); Citovsky et al. (2006) |
| P: Tandem CaMV 35S with a translational enhancer | |||
| pSAT-RNAi | Expression of hairpin structures for RNAi; expression is controlled by various constitutive promoters | T: RNAi | Dafny-Yelin et al. (2007) |
| P: Manopine synthase, nopaline synthase, Rubisco small subunit, tandem CaMV 35S, superpromoter | |||
| pSAT-epitope | N- and C-terminal fusion to various epitope tags | T: 1xFLAG, 3xFLAG, His, HA, Xpress, MYC, GST | T. Tzfira, unpublished data |
| P: Tandem CaMV 35S with a translational enhancer | |||
| pSAT-DEST | Promoter analysis and N- and C-terminal fusion to EGFP, Gateway Destination vectors | T: EGFP | Tzfira et al. (2005) |
| P: Tandem CaMV 35S with a translational enhancer | |||
| pSITE | Gateway-mediated N- and C-terminal fusion to autofluorescent tags and expression of untagged proteins | T: EGFP, EYFP, ECFP, RFP | Chakrabarty et al. (2007) |
| P: Tandem CaMV 35S with a translational enhancer |
Figure 2.
The general structure of a pSAT-based plant expression vector. A typical vector is composed of promoter and terminator sequences, flanked by AgeI and NcoI and XbaI and NotI sites, respectively. This arrangement allows for easy replacement of these regulatory elements in nearly all of the pSAT-based vectors as well as the construction of various specialized tags and functional constructs between the NcoI and XbaI sites. Fully assembled expression cassettes can be mobilized between different pSAT backbones by AgeI-NotI and be cloned into pRCS-based vectors using various rare cutters. [See online article for color version of this figure.]
EXPANDING CLONING POSSIBILITIES WITH ARTIFICIAL RESTRICTION ENZYMES
While the expansion of the pSAT family of plasmids provides the user with a comprehensive and versatile collection of plant expression vectors, the original design of the pAUX and pRCS vectors was limited to only seven auxiliary plasmids. Several approaches can be taken to increase the capacity of the pAUX/pSAT cloning system. One of our recent approaches was to expand the pRCS-based binary plasmids' MCS cloning capacity by adding a new site for the PI-SceI homing endonuclease (V. Zeevi, R. Dafny, and T. Tzfira, unpublished data). In addition, we observed that we cannot obtain double digestion of the pRCS-based MCS using a pair of homing endonucleases, a problem that can be attributed to the lack of appropriate space between these enzymes' recognition sites on the T-DNA region to enable successive digestion by these enzymes. We thus began modifying the T-DNA region by incorporating long spacers between the I-PopI, I-SceI, I-CeuI, PI-PspI, PI-SceI, and PI-Tlil, and assembling pSAT vectors with compatible sites (V. Zeevi, R. Dafny, and T. Tzfira, unpublished data), which would increase the capacity of the modified pRCS binary plasmids to a total number of 13 expression cassettes. The constant search for new and modified homing endonucleases (e.g. Arnould et al., 2006; Smith et al., 2006) could potentially lead to the commercial release of new enzymes that can be harnessed to further expand the pRCS MCS with new and rare-cutting recognition sites.
A second, more revolutionary approach to increasing the cloning capacity of the pRCS/pSAT multisite vector system is the use of artificial restriction enzymes, i.e. zinc finger nucleases (ZFNs). ZFNs are hybrid synthetic restriction enzymes that can be specifically designed to bind and cleave long (typically 24–30 bp) stretches of DNA sequences (Mani et al., 2005). ZFNs have been successfully used for various purposes of genome engineering in various organisms (Durai et al., 2005), but their potential for DNA recombination was never explored. We have recently designed, assembled, and expressed several ZFNs and demonstrated their potential for cloning of long DNA molecules (V. Zeevi, A. Tovach, and T. Tzfira, unpublished data). Furthermore, we are currently exploring the possibilities of introducing novel sequences into the T-DNA region of pRCS-based vectors, assembling and expressing ZFNs to target these 24-bp-long DNA sequences, and constructing the pSAT plasmids to be used in conjunction with these enzymes. The ability to design ZFNs to bind and specifically cleave virtually any target DNA sequence makes these enzymes most powerful for DNA cloning purposes and they may hold great potential for the assembly of future multigene plant genetic transformation vectors.
FUTURE PROSPECTS
The simultaneous manipulation of multiple genes in transgenic plants presents a clear challenge for plant biologists and biotechnologists. Although several approaches can be used for the delivery of multiple genes into plant cells, the stacking of multiple expression cassettes onto a single binary plasmid seems to be the most straightforward method. The availability of the aforementioned multigene vector assembly systems will most likely facilitate the introduction of multiple genes into plant species. Nevertheless, several questions still remain open and technical issues need to be addressed before these systems, and others that may follow, can be routinely used for plant research and biotechnology. One of these questions, for example, is whether it is wise to use the same promoter and terminator sequences to control the expression of multiple genes in large vectors. While it has been shown that stable expression of four different genes can be achieved in transgenic plants, even when all genes are driven by the same constitutive promoter (Bohmert et al., 2002), further research is needed to determine whether such long constructs with repetitive elements remain stable in both bacteria and plant cells. Naturally, new strong constitutive promoters need to be identified to expand the user's choice of regulatory elements beyond the rather limited number of constitutive promoters that exist today.
The construction of large binary vectors also raises the issue of vector size limitations and the efficiency of delivering long T-DNA molecules into plant cells. These issues can be addressed, for example, by using vectors based on binary bacterial artificial chromosomes (Hamilton, 1997) or TAC (Liu et al., 1999), as these vectors possess the capacity to clone very large DNA fragments. Indeed, the Cre/LoxP-based vector system (Lin et al., 2003) utilizes TAC vectors as the acceptor plasmids for the plant expression cassettes and a Gateway-compatible TAC vector has also been constructed to support the transfer of long DNA fragments via the MultiRound Gateway cloning strategy (Chen et al., 2006). Similar vectors will need to be designed and constructed to support the use of rare-cutter cloning strategies for the assembly of large multigene plant transformation vectors.
Finally, the modularity of the rare-cutter-based cloning pSAT system is limited only by the small number of commercially available rare-cutting restriction enzymes. ZFNs, which can be artificially assembled and used for cloning long DNA molecules (V. Zeevi, A. Tovach, and T. Tzfira, unpublished data), can facilitate the expansion of the pSAT system beyond its current capacity of seven expression cassettes. Thus, further research should be directed toward the development of novel ZFNs, as well as toward the establishment of protocols for their assembly, purification, and use for DNA cloning purposes.
This work was supported by grants from the Biotechnology Research and Development Cooperation and the University of Michigan startup funds.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Tzvi Tzfira (ttzfira@umich.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
References
- An G, Watson BD, Stachel S, Gordon MP, Nester EW (1985) New cloning vehicles for transformation of higher plants. EMBO J 4 277–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnould S, Chames P, Perez C, Lacroix E, Duclert A, Epinat JC, Stricher F, Petit AS, Patin A, Guillier S, et al (2006) Engineering of large numbers of highly specific homing endonucleases that induce recombination on novel DNA targets. J Mol Biol 355 443–458 [DOI] [PubMed] [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson R (1992) New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol 20 1195–1197 [DOI] [PubMed] [Google Scholar]
- Bevan MW (1984) Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res 12 1811–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmert K, Balbo I, Kopka J, Mittendorf V, Nawrath C, Poirier Y, Tischendorf G, Trethewey RN, Willmitzer L (2000) Transgenic Arabidopsis plants can accumulate polyhydroxybutyrate to up to 4% of their fresh weight. Planta 211 841–845 [DOI] [PubMed] [Google Scholar]
- Bohmert K, Balbo I, Steinbuchel A, Tischendorf G, Willmitzer L (2002) Constitutive expression of the beta-ketothiolase gene in transgenic plants: a major obstacle for obtaining polyhydroxybutyrate-producing plants. Plant Physiol 128 1282–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha-Drori K, Shichrur K, Katz A, Oliva M, Angelovici R, Yalovsky S, Ohad N (2004) Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J 40 419–427 [DOI] [PubMed] [Google Scholar]
- Capell T, Christou P (2004) Progress in plant metabolic engineering. Curr Opin Biotechnol 15 148–154 [DOI] [PubMed] [Google Scholar]
- Chakrabarty R, Banerjee R, Chung S-M, Farman M, Citovsky V, Hogenhout SA, Tzfira T, Goodin M (2007) pSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: probing Nicotiana benthamiana-virus interactions. Mol Plant Microbe Interact 20 740–750 [DOI] [PubMed] [Google Scholar]
- Chen L, Marmey P, Taylor NJ, Brizard JP, Espinoza C, D'Cruz P, Huet H, Zhang S, de Kochko A, Beachy RN, et al (1998) Expression and inheritance of multiple transgenes in rice plants. Nat Biotechnol 16 1060–1064 [DOI] [PubMed] [Google Scholar]
- Chen P-Y, Wang C-K, Soong S-C, To K-Y (2003) Complete sequence of the binary vector pBI121 and its application in cloning T-DNA insertion from transgenic plants. Mol Breed 11 287–293 [Google Scholar]
- Chen QJ, Zhou HM, Chen J, Wang XC (2006) A Gateway-based platform for multigene plant transformation. Plant Mol Biol 62 927–936 [DOI] [PubMed] [Google Scholar]
- Cheo DL, Titus SA, Byrd DR, Hartley JL, Temple GF, Brasch MA (2004) Concerted assembly and cloning of multiple DNA segments using in vitro site-specific recombination: functional analysis of multi-segment expression clones. Genome Res 14 2111–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilton M-D (2001) Agrobacterium. A memoir. Plant Physiol 125 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SM, Frankman EL, Tzfira T (2005) A versatile vector system for multiple gene expression in plants. Trends Plant Sci 10 357–361 [DOI] [PubMed] [Google Scholar]
- Citovsky V, Lee LY, Vyas S, Glick E, Chen MH, Vainstein A, Gafni Y, Gelvin SB, Tzfira T (2006) Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J Mol Biol 362 1120–1131 [DOI] [PubMed] [Google Scholar]
- Coutu C, Brandle J, Brown D, Brown K, Miki B, Simmonds J, Hegedus DD (2007) pORE: a modular binary vector series suited for both monocot and dicot plant transformation. Transgenic Res (in press) [DOI] [PubMed]
- Curtis MD, Grossniklaus U (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dafny-Yelin M, Chung S-M, Frankman EL, Tzfira T (2007) pSAT RNA interference vectors: a modular series for multiple gene down-regulation in plants. Plant Physiol 145 1272–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Dhingra A (2002) Multigene engineering: dawn of an exciting new era in biotechnology. Curr Opin Biotechnol 13 136–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta S, Collins GB, Hunt AG (1998) Co-ordinated expression of multiple enzymes in different subcellular compartments in plants. Plant J 16 107–116 [DOI] [PubMed] [Google Scholar]
- De Buck S, Jacobs A, Van Montagu M, Depicker A (1999) The DNA sequences of T-DNA junctions suggest that complex T-DNA loci are formed by a recombination process resembling T-DNA integration. Plant J 20 295–304 [DOI] [PubMed] [Google Scholar]
- de Maknik J, Joseph RG, Tanner GJ, Larkin PJ, Djordjevic MA, Rolfe BG, Weinman JJ (1997) A convenient set of vectors for expression of multiple gene combinations in plants. Plant Mol Biol Rep 15 134–140 [Google Scholar]
- De Neve M, De Buck S, Jacobs A, Van Montagu M, Depicker A (1997) T-DNA integration patterns in co-transformed plant cells suggest that T-DNA repeats originate from co-integration of separate T-DNAs. Plant J 11 15–29 [DOI] [PubMed] [Google Scholar]
- Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, Chandrasegaran S (2005) Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res 33 5978–5990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45 616–629 [DOI] [PubMed] [Google Scholar]
- El Amrani A, Barakate A, Askari BM, Li X, Roberts AG, Ryan MD, Halpin C (2004) Coordinate expression and independent subcellular targeting of multiple proteins from a single transgene. Plant Physiol 135 16–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin SB (2003) Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev 67 16–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goderis IJ, De Bolle MF, Francois IE, Wouters PF, Broekaert WF, Cammue BP (2002) A set of modular plant transformation vectors allowing flexible insertion of up to six expression units. Plant Mol Biol 50 17–27 [DOI] [PubMed] [Google Scholar]
- Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO (2002) pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J 31 375–383 [DOI] [PubMed] [Google Scholar]
- Hadi MZ, McMullen MD, Finer JJ (1996) Transformation of 12 different plasmids into soybean via particle bombardment. Plant Cell Rep 15 500–505 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25 989–994 [DOI] [PubMed] [Google Scholar]
- Halpin C, Barakate A, Askari BM, Abbott JC, Ryan MD (2001) Enabling technologies for manipulating multiple genes on complex pathways. Plant Mol Biol 47 295–310 [PubMed] [Google Scholar]
- Halpin C, Boerjan W (2003) Stacking transgenes in forest trees. Trends Plant Sci 8 363–365 [DOI] [PubMed] [Google Scholar]
- Halpin C, Cooke SE, Barakate A, El Amrani A, Ryan MD (1999) Self-processing 2A-polyproteins: a system for co-ordinate expression of multiple proteins in transgenic plants. Plant J 17 453–459 [DOI] [PubMed] [Google Scholar]
- Hamilton CM (1997) A binary-BAC system for plant transformation with high-molecular-weight DNA. Gene 200 107–116 [DOI] [PubMed] [Google Scholar]
- Hellens R, Mullineaux P, Klee H (2000. a) Technical focus: a guide to Agrobacterium binary Ti vectors. Trends Plant Sci 5 446–451 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000. b) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42 819–832 [DOI] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P (2005) Modular cloning in plant cells. Trends Plant Sci 10 103–105 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7 193–195 [DOI] [PubMed] [Google Scholar]
- Komori T, Imayama T, Kato N, Ishida Y, Ueki J, Komari T (2007) Current status of binary vectors and superbinary vectors. Plant Physiol 145 1155–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhou Y, Cheng X, Sun J, Marita JM, Ralph J, Chiang VL (2003) Combinatorial modification of multiple lignin traits in trees through multigene cotransformation. Proc Natl Acad Sci USA 100 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Liu YG, Xu X, Li B (2003) Efficient linking and transfer of multiple genes by a multigene assembly and transformation vector system. Proc Natl Acad Sci USA 100 5962–5967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Shirano Y, Fukaki H, Yanai Y, Tasaka M, Tabata S, Shibata D (1999) Complementation of plant mutants with large genomic DNA fragments by a transformation-competent artificial chromosome vector accelerates positional cloning. Proc Natl Acad Sci USA 96 6535–6540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale DM, Moisan LJ, Harvey AK (1995) pFC1 to PFC7: a novel family of combinatorial cloning vectors. Plant Mol Biol Rep 13 343–345 [Google Scholar]
- Lucker J, Schwab W, van Hautum B, Blaas J, van der Plas LH, Bouwmeester HJ, Verhoeven HA (2004) Increased and altered fragrance of tobacco plants after metabolic engineering using three monoterpene synthases from lemon. Plant Physiol 134 510–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JK, Hiatt A, Hein M, Vine ND, Wang F, Stabila P, van Dolleweerd C, Mostov K, Lehner T (1995) Generation and assembly of secretory antibodies in plants. Science 268 716–719 [DOI] [PubMed] [Google Scholar]
- Mani M, Kandavelou K, Dy FJ, Durai S, Chandrasegaran S (2005) Design, engineering, and characterization of zinc finger nucleases. Biochem Biophys Res Commun 335 447–457 [DOI] [PubMed] [Google Scholar]
- Marcos JF, Beachy RN (1994) In vitro characterization of a cassette to accumulate multiple proteins through synthesis of a self-processing polypeptide. Plant Mol Biol 24 495–503 [DOI] [PubMed] [Google Scholar]
- McCormac AC, Elliott MC, Chen DF (1997) pBECKS: a flexible series of binary vectors for Agrobacterium-mediated plant transformation. Mol Biotechnol 8 199–213 [DOI] [PubMed] [Google Scholar]
- Ralley L, Enfissi EM, Misawa N, Schuch W, Bramley PM, Fraser PD (2004) Metabolic engineering of ketocarotenoid formation in higher plants. Plant J 39 477–486 [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Sone T, Yoshida S, Yahata K, Hotta J, Chesnut JD, Honda T, Imamoto F (2004) Evidence for high specificity and efficiency of multiple recombination signals in mixed DNA cloning by the Multisite Gateway system. J Biotechnol 107 233–243 [DOI] [PubMed] [Google Scholar]
- Seitz C, Vitten M, Steinbach P, Hartl S, Hirsche J, Rathje W, Treutter D, Forkmann G (2007) Redirection of anthocyanin synthesis in Osteospermum hybrida by a two-enzyme manipulation strategy. Phytochemistry 68 824–833 [DOI] [PubMed] [Google Scholar]
- Simoens C, Alliotte T, Mendel R, Muller A, Schiemann J, Van Lijsebettens M, Schell J, Van Montagu M, Inze D (1986) A binary vector for transferring genomic libraries to plants. Nucleic Acids Res 14 8073–8090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla-Pareek SL, Reddy MK, Sopory SK (2003) Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proc Natl Acad Sci USA 100 14672–14677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater S, Mitsky TA, Houmiel KL, Hao M, Reiser SE, Taylor NB, Tran M, Valentin HE, Rodriguez DJ, Stone DA, et al (1999) Metabolic engineering of Arabidopsis and Brassica for poly(3-hydroxybutyrate-co-3-hydroxyvalerate) copolymer production. Nat Biotechnol 17 1011–1016 [DOI] [PubMed] [Google Scholar]
- Smith J, Grizot S, Arnould S, Duclert A, Epinat JC, Chames P, Prieto J, Redondo P, Blanco FJ, Bravo J, et al (2006) A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res 34 e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyo KE, Alper HS, Stephanopoulos GN (2007) Expanding the metabolic engineering toolbox: more options to engineer cells. Trends Biotechnol 25 132–137 [DOI] [PubMed] [Google Scholar]
- Tzfira T, Tian G-W, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V (2005) pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol 57 503–516 [DOI] [PubMed] [Google Scholar]
- Uberlacker B, Werr W (1996) Vectors with rare-cutter restriction enzyme sites for expression of open reading frames in transgenic plants. Mol Breed 2 293–295 [Google Scholar]
- Urwin PE, McPherson MJ, Atkinson HJ (1998) Enhanced transgenic plant resistance to nematodes by dual proteinase inhibitor constructs. Planta 204 472–479 [DOI] [PubMed] [Google Scholar]
- van Engelen FA, Molthoff JW, Conner AJ, Nap JP, Pereira A, Stiekema WJ (1995) pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Res 4 288–290 [DOI] [PubMed] [Google Scholar]
- Velten J, Schell J (1985) Selection-expression plasmid vectors for use in genetic transformation of higher plants. Nucleic Acids Res 13 6981–6998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bodman SB, Domier LL, Farrand SK (1995) Expression of multiple eukaryotic genes from a single promoter in Nicotiana. Biotechnology (N Y) 13 587–591 [DOI] [PubMed] [Google Scholar]
- Wakasa Y, Yasuda H, Takaiwa F (2006) High accumulation of bioactive peptide in transgenic rice seeds by expression of introduced multiple genes. Plant Biotechnol J 4 499–510 [DOI] [PubMed] [Google Scholar]
- Walhout A, Temple G, Brasch M, Hartley J, Lorson M, van den Heuvel S, Vidal M (2000) GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol 328 575–592 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40 428–438 [DOI] [PubMed] [Google Scholar]
- Xiang C, Han P, Lutziger I, Wang K, Oliver DJ (1999) A mini binary vector series for plant transformation. Plant Mol Biol 40 711–717 [DOI] [PubMed] [Google Scholar]
- Ye X, Al-Babili S, Kloti A, Zhang J, Lucca P, Beyer P, Potrykus I (2000) Engineering the provitamin A (beta-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287 303–305 [DOI] [PubMed] [Google Scholar]
- Zhao JZ, Cao J, Li Y, Collins HL, Roush RT, Earle ED, Shelton AM (2003) Transgenic plants expressing two Bacillus thuringiensis toxins delay insect resistance evolution. Nat Biotechnol 21 1493–1497 [DOI] [PubMed] [Google Scholar]
- Zhong R, Richardson EA, Ye ZH (2007) Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 225 1603–1611 [DOI] [PubMed] [Google Scholar]