Dr. Pangloss was right, at least for live-cell imaging in plants, in his contention that this is “the best of all possible worlds” (Voltaire, 1759). We are now able to look inside cells in detailed ways that the fathers of microscopy, Antoni van Leeuwenhoek and Robert Hook, could not have possibly imagined (Gest, 2004). Current state-of-the-art instrumentation has made routine an array of imaging techniques that were inaccessible to most researchers just a few years ago. Additionally, the number of autofluorescent proteins (AFPs) with which we can color intercellular structures increases almost daily, thus providing ever greater flexibility in the types of experiments that can be performed. Moreover, the completion of large-scale genome sequencing projects has sparked many derivative microscopy-based projects, which are essential to put the function into functional genomics and to realize the emergent field of systems biology (Murphy, 2005; Li et al., 2006; Pepperkok and Ellenberg, 2006). For indeed, if a picture is still worth a thousand words (Orenstein, 2000), consider all that will be learned when, by microscopy en masse, we visualize the thousands of proteins predicted to be encoded in the genomes of eukaryotes in general (Brasch et al., 2004; Rual et al., 2004; Matsuyama et al., 2006) and plants in particular (Sterck et al., 2007).
This review is designed primarily to introduce researchers to the myriad of factors, beyond vector selection, that should be considered prior to embarking on protein localization experiments (Table I). To begin, we will review some of the most recently published vector systems for the expression of AFPs in plant cells, with particular emphasis on the pSAT (modular satellite plasmid) and pSITE (stable integration and transient expression plasmid) vectors (Tzfira et al., 2005; Chakrabarty et al., 2007; Goodin et al., 2007). Using the pSITE vectors, we have developed several transgenic lines to support cell biology studies conducted with Nicotiana benthamiana, a model host essential for the study of plant-pathogen interactions, which is being utilized increasingly in plant biology research, primarily due to the facile manner by which large populations of cells can be transfected. Next, we will compare protein localization data produced using transient assays versus transgenic plants. We will also discuss recent results from our lab supporting that protein localization data obtained using transient assays is comparable to that obtained from transgenic plants (Chakrabarty et al., 2007; Goodin et al., 2007). Finally, we discuss how advances in AFP-vector development will realize their greatest utility when used in conjunction with state-of-the-art imaging systems. Therefore, we conclude this review by examining the application of total internal reflectance fluorescence microscopy (TIRFM) as an adjunct to confocal imaging with a study of endoplasmic reticulum (ER) dynamics as an example.
Table I.
Key points for live-cell imaging in plants
| High-throughput protein localization is critical to the success of comprehensive functional genomics projects (Tian et al., 2004; Koroleva et al., 2005; Bhat et al., 2006; Matsuyama et al., 2006; Pepperkok and Ellenberg, 2006). |
| Current algorithms for predicting protein localization are often incapable of making accurate determinations (Fig. 2B). Thus live-cell imaging of chimeric AFP protein fusions are essential to characterize proteins whose function or localization cannot be predicted using computational methods (Li et al., 2006). |
| Gateway-cloning technology is currently the most effective means to integrate high-throughput proteomics, localization, and gene-silencing projects since the same set of entry clones could be used in all studies (Brasch et al., 2004; Earley et al., 2006). Many new binary vectors are being constructed using Gateway technology, thereby enhancing exchange of resources between labs (Chung et al., 2005; Earley et al., 2006; Chakrabarty et al., 2007). |
| Of any comparable system, the pSAT vectors offer the widest choices of (1) AFPs, (2) promoters to drive expression, and (3) flexibility in constructing binary vectors containing more than one expression cassette (Chung et al., 2005; Tzfira et al., 2005). |
| Due to the inability to efficiently predict the effect of AFPs on the stability of fusion partners, high-throughput protein localization should be performed using both N- and C-terminal fusions (Simpson et al., 2001; Pepperkok and Ellenberg, 2006). Alternatively, cloning of AFPs into internal sites can be performed (Tian et al., 2004; Li et al., 2006). |
| More than simply markers for protein localization, next generation AFPs (1) provide a greater diversity of colors, (2) can be photoactivated or converted facilitating protein tracking studies, (3) have been optimized for several advanced imaging techniques, and (4) can be used for characterizing protein-protein interactions (Patterson and Lippincott- Schwartz, 2004; Habuchi et al., 2005; Chudakov et al., 2006; Gurskaya et al., 2006). |
| A. tumefaciens (agroinfiltration) can be used to transiently express proteins in virus-infected plants to provide data similar to that generated with transgenic plants. |
A BIGGER BOX OF CRAYONS
“In living color” is somewhat of a holy grail for cell biologists. Consider what it has meant to be able to paint cellular loci in living cells at will. First there was the green fluorescent protein (GFP; Chalfie et al., 1994) from which blue fluorescent protein, cyan fluorescent protein (CFP), and yellow fluorescent protein (YFP) forms were subsequently derived (Zhang et al., 2002). After the discovery of red fluorescent protein (RFP; Matz et al., 1999), colors such as banana, orange, cherry, tomato, and plum were produced (Shaner et al., 2004). This salad of mFruits (Shaner et al., 2004) has been followed by a palette with an ever-increasing number of colors (Brandizzi et al., 2004; Shaner et al., 2005; Giepmans et al., 2006; Stewart, 2006). To be more specific, the demonstration that the GFP isolated from the jellyfish Aqueora victoriae could be linked to proteins of interest to allow in vivo examination of protein localization and dynamics in real time has transformed cell biology in a manner similar to the effect of the PCR on molecular biology. Subsequent mutagenesis of GFP led to new spectral variants, which opened doors to powerful techniques to study protein dynamics and interactions in vivo, such as fluorescence recovery after photobleaching (Ward and Brandizzi, 2004; Bates et al., 2006), fluorescence resonance energy transfer (FRET; Hink et al., 2002; Bhat et al., 2006), and bimolecular fluorescence complementation (Bhat et al., 2006; Citovsky et al., 2006; Dixit et al., 2006). The impact of GFP and its many spectral variants ushered in the search for similar proteins, DsRed from coral being the first of major significance (Matz et al., 1999). Derivatives of DsRed, which is a tetramer in its native state, led to the isolation of monomeric forms displaying different colors, collectively called mFruits (Shaner et al., 2004). Most recently, a novel monomeric RFP, TagRFP, has been described (Merzlyak et al., 2007). This protein is brighter and more resistant to photobleaching than mRFP and can be used in combination with GFP in FRET experiments. This is a particularly exciting result as GFP, despite its overwhelming popularity as fusion partner for localization studies, has been of relatively little utility in FRET-based experiments, which are typically performed with the spectral variants CFP and YFP (Jares-Erijman and Jovin, 2006).
To date, more than 35 AFPs that span an emission range from blue to red of the visible spectrum (460–595 nm) have been described (Stewart, 2006). Among these, GFP, DsRed, and most of their derivatives share a common property, namely that their fluorescence cannot be regulated easily. However, new AFPs have now been discovered and developed that are photoactivatible (PA-AFPs), photoconvertible, or photoswitchable. Briefly, the fluorescence from PA-AFPs, such as PA-GFP (Patterson and Lippincott-Schwartz, 2004; Runions et al., 2006) or DRONPA (Habuchi et al., 2005) is very low, but the AFP can be activated by a brief but intense pulse of excitation at a particular wavelength. Similarly, the fluorescence of photoconvertibles (Gurskaya et al., 2006) and photoswitchables (Chudakov et al., 2006) can be changed from one color to another, typically green to red or cyan to green, respectively, by a brief pulse of light of a particular wavelength. These novel AFPs, therefore, facilitate tracking experiments in which the movement of a tagged protein from one subcellular locus to another can be monitored following activation. A further innovation is that of chromophore assisted laser inactivation (Vitriol et al., 2007), which is a light-mediated technique that can be used for AFP-tagged protein inactivation. Incorporation of these new AFPs into the vector systems described below is under way, which should be of great benefit to plant biologists seeking precise spatiotemporal control of their AFP fusion.
As the number of AFPs increase, there is something of a revolution afoot with respect to microscopes by which these proteins can be detected. In the past 5 years all the major manufacturers of laser scanning confocal microscopes have commercialized instruments that permit high resolution images to be acquired at greater speed (e.g. Zeiss Live5), spectral resolution (e.g. Leica SP5), or laser synchronization (e.g. Olympus FV1000). Additionally, nonconfocal systems such as total internal fluorescence microscopes or specialized multiphoton lasers can be purchased as add-on units that are integrated into laser scanning confocal microscopes, thereby dramatically expanding their functionality. Furthermore, despite the high image quality of confocal laser scanning microscopy, the imaging frame rate is slow (typically <3 frames/s); therefore, addition of a spinning-disc unit on the confocal microscope can facilitate video-rate imaging of protein dynamics (Nakano, 2002; Wang et al., 2005).
BEGINNING, MIDDLE, OR END—WHERE TO FUSE YOUR AFP?
Prior to embarking on a study involving the use of AFP fusions, researchers should address two fundamental questions: (1) Where to attach the AFP to a protein of interest?, and (2) What promoter should be used to drive expression? With respect to the first question, the usual approach taken is that AFPs are fused to the amino (N) or carboxy (C) terminus. However, some proteins may not tolerate AFPs fused to one or either termini. So, it may be necessary to insert the AFP into an internal site (Tian et al., 2004; Li et al., 2006). Importantly, the use of AFPs should by no means be considered a replacement for alternative, and at times more appropriate, methods such as immunolocalization (Ruzin, 1999; Sauer et al., 2006). Additionally, proteins that contain N-terminal signal peptides may mislocalize if expressed as fusions to the C termini of AFPs (Simpson et al., 2001; Li et al., 2006). Moreover, if one is interested in studying a single protein and there is a requirement for the AFP fusion to function as closely as possible to the native protein with respect to localization, tissue specificity, timing, and level of expression, then it may be necessary to express the fusion in transgenic plants under control of the native promoter. At the other extreme, if there is a need to localize thousands of fusions in a relatively short period of time, then transient expression may be the most cost effective. It is currently impossible to use computational methods to accurately predict the effect of an AFP on a particular fusion (Li et al., 2006). Therefore, many of the current expression systems have been developed so that N- or C-terminal fusions can be tested (Tzfira et al., 2005; Earley et al., 2006; Chakrabarty et al., 2007). We consider it good general practice to test both types of fusions in preliminary studies prior to embarking on more intensive research. For example, our attempts to express the sonchus yellow net virus (SYNV) glycoprotein by fusing RFP to its C terminus were unsuccessful. Despite the fact that SYNV glycoprotein contains a predicted N-terminal signal peptide (Goldberg et al., 1991), the correctly targeted fusion was that in which RFP was placed in frame ahead of the signal peptide (Goodin et al., 2007). In a more detailed study, Simpson et al. (2001) found that approximately 50% of human proteins, including those targeted to the ER or mitochondria, localized to the same subcellular locus when expressed as N-terminal fusions to CFP or C-terminal fusions to YFP (Pepperkok and Ellenberg, 2006).
While we prefer the simplicity of constructing N- or C-terminal fusions, Tian et al. (2004) have reported a highly successful breakthrough technology called fluorescent tagging of full-length proteins. Best suited to protein localization studies for plants with sequenced genomes, the fluorescent tagging of full-length proteins technique requires a triple-PCR procedure to incorporate an AFP into an internal locus near the C terminus of proteins of interest. To avoid effects of the AFP tag on native subcellular localization, the location of the tag relative to the target sequence has to be determined for each individual protein, based on computer-assisted predictions of protein folding and functional domains (Li et al., 2006). According to the authors the AFP tag should be placed within a stretch of hydrophilic residues, outside of any specific protein domain, and near the C terminus. Use of this location minimizes disturbances of the contiguous protein sequence and protects the activity of membrane anchoring signals typically found within a few C-terminal amino acid residues (Casey, 1995; Zhang and Casey, 1996).
After deciding the best manner in which to construct AFP fusions for a specific set of experiments, the next critical choice is to determine the promoter to drive expression. Some consider that expression under the control of native promoters will always be superior to that of employing constitutive promoters, most commonly the constitutive cauliflower mosaic virus 35S promoter. However, this opinion ignores the fact that simply using the native promoter, or more often 1 to 2 kb of upstream sequence, does not take into account that promoter/gene duplication may affect expression levels, as will genomic context since the AFP fusion is unlikely to be expressed from the same genetic locus as the native gene (Bhat et al., 2006). Another common misconception is that expression from 35S, or even double-35S promoters, necessarily results in accumulation of fusion proteins at levels higher than would be achieved were the native promoter used. However, all such results are highly dependent upon the protein under investigation. Fusion of an AFP to a protein may stabilize it or make it less so. In systems such as Arabidopsis (Arabidopsis thaliana), where it is straightforward to obtain T-DNA insertional knockout alleles for particular genes of interest (Alonso et al., 2003), single gene complementation by AFP-fusion proteins implies their correct functionality in planta.
NEW GATEWAY-COMPATIBLE VECTORS FOR EXPRESSION OF AUTOFLUORESCENT FUSIONS IN PLANTS
Gateway-compatible binary vectors have greatly improved the cloning efficiency of AFP-tagging projects (Curtis and Grossniklaus, 2003; Earley et al., 2006). Briefly, Gateway cloning utilizes the site-specific recombination system utilized by λ phage to transfer DNA fragments between plasmids containing compatible recombination sites (Walhout et al., 2000). What makes this strategy so attractive is that once DNA clones of interest are captured into an entry vector (pDONR), they can be mobilized into a plethora of vectors that permit expression in bacteria, insect cells, yeast (Saccharomyces cerevisiae), animal, or plant cells. This avoids the frustrating situation commonly encountered with ligase-mediated cloning, namely that there are often no compatible restriction sites to permit easy mobilization from one vector to the next. This results in the need to reclone and resequence DNA fragments of interest, which may be cost and time prohibitive when done on a large scale. Once pDONR constructs are generated and validated, all downstream expression systems can be utilized, which increases the efficiency of sharing clones for genomics research (Hilson et al., 2003, 2004).
Ultimately, once genome sequencing and assembly projects are complete, characterization in vivo is essential to establish function of constituents of the ORFeome, particularly for those genes whose function cannot be predicted in silico. Therefore, plant functional proteomics research is increasingly dependent upon vectors that facilitate high-throughput gene cloning and expression of fusions to AFPs. Our personal approach has been first to transiently express proteins as either C- or N-terminal fusions. Second, for proteins of significant interest, we have conducted expression in the context of transgenic plants. To facilitate such research we have developed the pSITE family of plasmids, a new set of Agrobacterium binary vectors, suitable for the stable integration or transient expression of various AFPs in plant cells (Fig. 1). It should be noted that the pSITE vectors were derived from the modular pSAT system (Chung et al., 2005; Tzfira et al., 2005). Specifically, the pSAT6 series and was subcloned into an RCS2 derivative containing the nopaline phosphotransferase (nptII) gene, which confers resistance to kanamycin in transformed plant tissue. Conversion of the remaining pSAT series to pSITE equivalents is already under way and pSAT derivatives for bimolecular fluorescence complementation are now available (Citovsky et al., 2006). Also, in development are pSAT and pSITE vectors that express alternative AFPs to those shown below (K. Martin and M.M. Goodin, unpublished data). We are also developing modified pMDC (Curtis and Grossniklaus, 2003) and pSITE vectors to contain PA-AFPs (S. DeBolt, J. Estevez, and M.M. Goodin unpublished data).
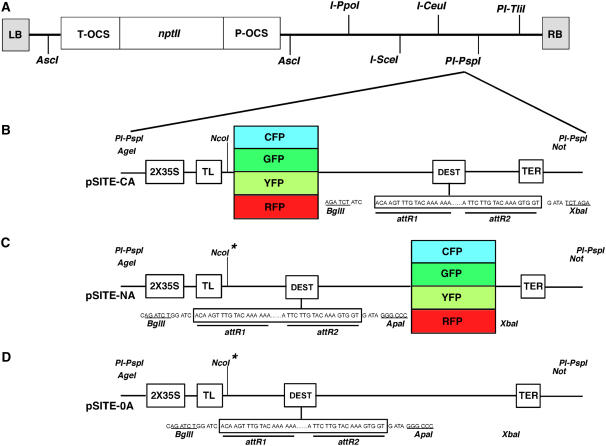
Figure 1.
Schematic representations of pSITE vectors. A, All modified pSAT6 cassettes were cloned into the pRCS2-ocs-nptII binary vector at the PI-Psp1 site. The ability to select transgenic plant cells is conferred by the nptII gene, the expression of which is controlled by the ocs promoter (P-OCS) and terminator (T-OCS). B, C-series pSITE vectors for Gateway recombination-mediated construction of binary vectors for expression of proteins of interest fused to the carboxy termini of AFPs. C, N-series pSITE vectors for Gateway recombination-mediated construction of binary vectors for expression of proteins of interest fused to the amino termini of AFPs. D, 0-series pSITE vectors for Gateway recombination-mediated construction of binary vectors for expression of native proteins. Protein expression is controlled by a duplicated CaMV 35S promoter (2X35S) and a tobacco etch virus translational leader (TL). All vectors employ the CaMV35S transcriptional terminator (TER). Nco1*, This restriction site was deleted to create the pSITE-NB and pSITE-0B vectors, thereby allowing translation to initiate at the native start codon on the gene of interest. This figure is a revision of that appearing in Chakrabarty et al. (2007). [See online article for color version of this figure.]
The current set of pSITE vectors can be used to express native proteins or enhanced CFP, GFP, YFP, or RFP fusions to either the C or N termini of proteins of interest (Fig. 1). These vectors were validated with respect to five criteria essential for high-throughput protein localization studies associated with the study of plant-pathogen interactions. Such experiments require facile vector systems that permit (1) high-throughput construction of recombinant expression vectors, (2) protein expression in either transient assays or transgenic plants without the need for subcloning into different vectors, (3) the ability to efficiently deliver proteins and their interacting targets or substrates to the same cell, (4) expression of proteins in pathogen-infected cells and, (5) the ability to monitor membrane or protein dynamics in a large number of cells so as to permit rigorous statistical analyses (Chakrabarty et al., 2007).
With respect to the first criterion, the pSITE vectors permit single-step Gateway-mediated recombination cloning for construction of binary vectors that can be used directly in transient expression studies or for the selection of transgenic plants on media containing kanamycin. Thus, following high-throughput protein localization, any construct of interest can be used to generate transgenic plants if necessary, without the need for subcloning into alternate vectors. Additionally, the ability to clone directly into a complete binary vector eliminates the two-step cloning procedure required for assembly of pSAT derivatives.
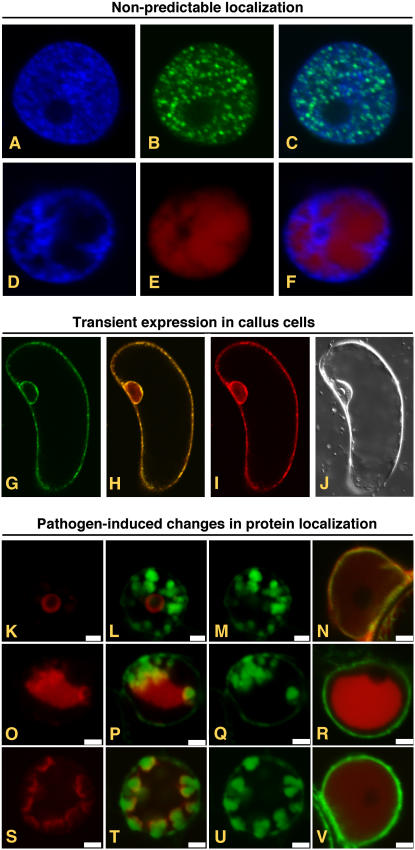
The pSITE vectors have proven to be useful in a diversity of applications (Fig. 2). For example, we have recently cloned the nucleocapsid and phosphoprotein genes of Potato yellow dwarf virus (D. Ghosh and M.M. Goodin, unpublished data). Computational analyses failed to identify karyophillic sequences in either of these proteins; however, AFP fusions of both proteins localize entirely to the nucleus (Fig. 2). Thus, protein localization is useful for characterization of proteins whose function or localization cannot be predicted using computational methods. When considering whole plant genomes, we are very much in the dark about protein localization. Li et al. (2006) make an excellent case for the need for large-scale plant protein localization studies given that, for Arabidopsis, only 48% of the genes have been assigned putative molecular functions, 30% have no predicted molecular functions, and 22% have not yet been annotated. For the majority of these genes that are assigned putative molecular functions, function was predicted from sequence similarity to other genes (Wortman et al., 2003). Worse yet, only 3.5% (917) of all predicted Arabidopsis proteins have had their molecular functions elucidated empirically, while only 5% (1,300) of all predicted Arabidopsis proteins have had their subcellular locations determined empirically. Clearly, as it is the paradigm for plant molecular genetics, there is a great need for Arabidopsis protein localization projects to be on par with those of other model systems (Wiemann et al., 2004; Li et al., 2006; Matsuyama et al., 2006).
Figure 2.
Transient expression of AFPs from pSITE vectors. A to F, Expression of proteins whose subcellular localization cannot be determined in silico. Fluorescence from DAPI (A) and GFP (B) in the nucleus of a cell expressing a GFP:PYDV-P protein. The overlay of A and B is shown in C. Fluorescence from DAPI (D) and RFP (E) in the nucleus of a cell expressing a RFP:PYDV-N protein. The overlay of D and E is shown in F. Although both PYDV-N and -P proteins are entirely localized to the nucleus, analysis of their primary structure failed to identify karyophillic domains. G to J, Expression of AFP fusions in callus cells of N. benthamiana. G, GFP fluorescence of transgenic callus cell expressing mGFP5-ER. H, Overlay of G and I. I, RFP fluorescence following agromediated expression of RFP-SYNV-P from a pSITE vector. J, Differential interference contrast image of cell shown in G to I. K to V, Expression to study differential protein localization in pathogen-infected cells. Shown are confocal micrographs of RFP fusions of SYNV proteins expressed in SYNV-infected and mock-inoculated mGFP5-ER transgenic N. benthamiana plants. Fluorescence images for GFP, RFP, and the corresponding overlay are shown for each fusion expressed in SYNV-infected cells. Only the overlay is shown for fusions expressed in mock-inoculated leaves. Sections from top to bottom show localization of RFP:P (K–N), RFP:N (O–R), and RFP:M (S–V). Sections K to V are reprinted from Goodin et al. (2007).
N. BENTHAMIANA: SERVING TO INTEGRATE PLANT OMICS PROJECTS
Like Vero cells, which served to accelerate localization of the human proteome, comparable projects in plants require a cell culture, or a similarly manipulatable system (Simpson et al., 2001). Thus, suspension cell cultures have been used for medium-throughput localization of Arabidopsis proteins (Koroleva et al., 2005; Pendle et al., 2005). In a similar manner, callus cells derived from N. benthamiana can be used (Fig. 2, G–J; S. Yelton and M.M. Goodin, unpublished data). However, an increasingly attractive alternative is to use agroinfiltration, a highly facile means to express proteins transiently in plants, which simply involves infiltrating leaves with suspension of virulent Agrobacterium tumefaciens transformed with binary vectors of interest (Schob et al., 1997; Goodin et al., 2002; Fig. 2). When using vectors, such as pSITE, that contain selectable markers, any constructs of interest identified in transient expression assays can be used to generate transgenic plants for further study. In addition to speed, another great advantage of agroinfiltration in N. benthamiana is the ease by which dyes such as 4′,6-diamidino-2-phenylindole (DAPI), to stain DNA (Chakrabarty et al., 2007; Goodin et al., 2007; Fig. 2, A–F), or BODIPY-TR, to stain endomembranes (Goodin et al., 2005), can be infiltrated into leaves prior to examination of tissue samples by microscopy. This approach serves to improve both the quality and accuracy of interpretation of micrographs, thus avoiding the low quality green dots on black images that lack any frame of reference to assist interpretation of the data. Agroinfiltration and facile counterstaining techniques are not easily conducted in Arabidopsis or other plants. Therefore, although N. benthamiana has been used traditionally in the context of host-pathogen interactions, it is rapidly being adopted in a myriad of studies in plant biology, particularly in cases where localization is being linked to protein-protein interactions (Tardif et al., 2007), or complementation of studies initiated in Arabidopsis (Levy et al., 2007).
TRANSGENIC PLANTS AND FLUORESCENT MARKER PROTEINS FOR PLANT CELL BIOLOGY RESEARCH
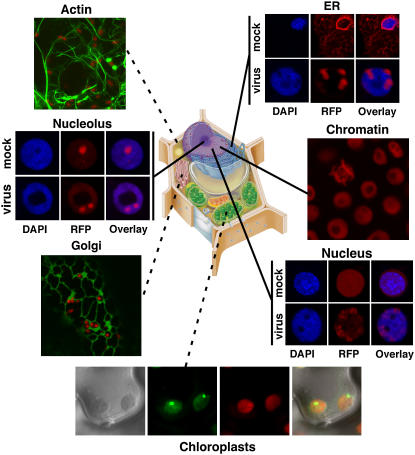
To support our own research and that of other plant viruses that cannot replicate in Arabidopsis, we have produced a series of pSITE derivatives and transgenic plant lines in N. benthamiana that express markers for fluorescent highlighting of actin filaments, chromatin, ER, and nucleoli (Fig. 3). We anticipate that these plants will be of general utility for the plant biology community, for example the transgenic line expressing Histone 2B fused to RFP (Fig. 3) will eliminate time consuming steps of counterstaining with DAPI, or similar dyes for the localization of nuclear proteins. We note that for confocal microscopy studies requiring a nuclear marker that the transgenic RFP:NbH2B lines can be imaged using the 543 nm laser line of the common He-Ne lasers. This should, therefore, provide an exceptional alternative to the use of propidium iodide which, being highly cell impermeant, requires incubation of plant tissue in harsh buffers when it is used as a nuclear counterstain (Kumar et al., 2006). Additionally, the use of the cell-permeant, DNA-selective dye DAPI requires a UV or near-UV laser, which, if unavailable, forces the use of propidium iodide. Thus, the RFP:NbH2B lines may circumvent two major technical limitations that often reduce the quality of micrographs of nuclear-localized proteins in plant cells.
Figure 3.
Confocal micrographs showing localization of AFPs targeted to a variety of subcellular loci. All markers are expressed from pSITE vectors in N. benthamiana. Micrographs marked with dashed lines represent transient expression while solid lines represent stable expression in transgenic plants. Plants expressing a RFP:SYNV-M fusion were not resistant to infection. Instead, RFP:SYNV-M was relocalized from the nucleoplasm to foci consistent with sites where intranuclear membranes accumulate (see Fig. 2). Clockwise, cell marker proteins used were the Rubisco small subunit (chloroplast), soybean (Glycine max) mannosidase (Golgi), Fib1 (nucleolus), RFP-HDEL (ER), Histone 2B (chromatin), and SYNV:M protein (nuclelus). Drawing of a plant cell courtesy of http://www.ualr.edu/botany/plantcelldiagram.jpg.
ACCURACY OF PROTEIN LOCALIZATION: TRANSIENT EXPRESSION COMPARES FAVORABLY TO TRANSGENIC PLANTS
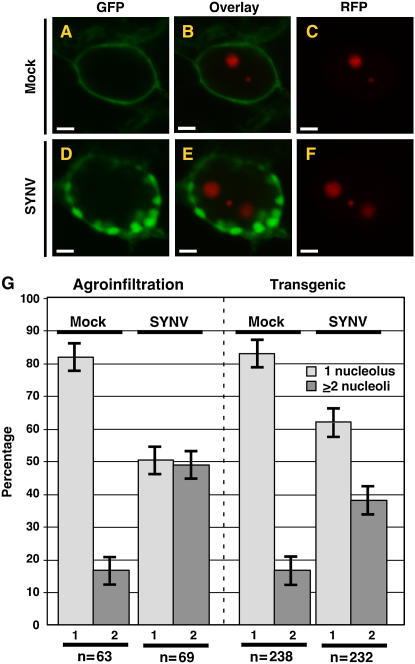
As noted above, the decision to use transient versus stable expression is not a trivial one. Of necessity, proteome-scale projects will rely heavily upon transient assays because of the number of proteins that must be examined. Protein localization studies conducted in the context of pathogens, or other situations associated with stress physiology face an additional challenge, namely, how to determine accurately protein localization in live cells in different physiological states. For example, we have previously shown that RFP fusions of SYNV-encoded proteins have radically different localization patterns in epidermal cells of mock-inoculated or SYNV-infected leaves (Chakrabarty et al., 2007; Goodin et al., 2007; Fig. 2, K–V). While this might be expected for virus-encoded proteins, similar pathogen-induced changes in host proteins have been observed, such as changes in the pattern of accumulation of the Arabidopsis nucleolar marker Fibrillarin1 (Fib1; Fig. 4; Chakrabarty et al., 2007). Our initial experiments were conducted using agroinfiltration to express an RFP-AtFib1 (RFP fusion of Arabidopsis nucleolar marker Fib1) fusion in mock-inoculated or SYNV-infected cells. Interestingly, we noted a statistically significant increase in nuclei with multiple nucleoli in virus-infected cells (Fig. 4; Chakrabarty et al., 2007). This raised the question as to whether a plant pathogen, A. tumefaciens, could be used to study protein localization in plant cells already infected with a pathogen, in this case a virus. To assess this, we generated transgenic plants expressing RFP-AtFib1. We could therefore determine SYNV-induced changes in Fib1 localization without potential artifacts introduced by A. tumefaciens in infiltrated leaves. Consistent with the finding that the process of agroinfiltration per se does not affect protein localization, the variation in nucleoli per nucleus was found to be identical in agroinfiltrated leaves of mock-inoculated plants and transgenic plants expressing RFP-AtFib1 (Fig. 4G). Similarly, statistically significant increases in the number of nucleoli per nucleus were observed in SYNV-infected plants in which RFP-AtFib1 was delivered via a stable transgene or agroinfiltration (Fig. 4G). However, these results were not identical to those obtained in mock-inoculated plants, with agroinfiltrated leaves showing at 51% and 49% of nuclei having 1 or ≥2 nucleoli per nucleus, respectively. In transgenic plants this ratio was 60% to 40%, respectively. This discrepancy can be explained in part due to the fact that in agroinfiltrated leaves, nucleoli were scored only in nuclei containing intranuclear membranes that accumulated GFP, which is an excellent marker for scoring virus-infected cells (Goodin et al., 2005). In contrast, in transgenic plants RFP-AtFib1 was the only fluorescent marker, thus reducing the bias of scoring only virus-infected cells per se. The significance of these data with respect to SYNV biology is, as yet, unclear. However, it has recently been shown that the OPEN READING FRAME3 (ORF3) protein of Groundnut rosette virus (Umbravirus) induces cajal bodies to fuse with nucleoli, resulting in changes in the localization of Arabidopsis Fib2 and coilin in cells of N. benthamiana (Kim et al., 2007). These authors present a model whereby ORF3 relocalizes coilin and Fib1 to the cytoplasm where all three proteins associated with groundnut rosette virus RNA to form ribonucleoprotein complexes that are involved in systemic movement through the phloem (Kim et al., 2007).
Figure 4.
A to F, Laser scanning confocal micrographs of N. benthamiana leaf epidermal cells showing shift in the localization of expression patterns of the RFP:AtFib1 nucleolar marker from two loci in mock-inoculated cells (A–C; mock) to three in SYNV-infected cells (D–F; SYNV). Transient expression of RFP:AtFib1 was conducted in mGFP5-ER plants. G, Quantitative comparison of RFP:AtFib1 expression patterns in mock-inoculated and SYNV-infected cells. Expression patterns were divided into two categories: nuclei with one nucleolus (1, light gray), or two nucleoli (2, dark gray). Results obtained using agroinfiltration are shown on the left while those obtained with transgenic plants are on the right. The numbers of nuclei examined (n) are shown at the bottom of the graph. Sections A to F and agroinfiltration data in G are reprinted from Chakrabarty et al. (2007).
The effects of viral infection on the localization of nucleolar proteins discussed above are but few small examples of a growing field of research that seeks to determine the effect of pathogens on the localization of host proteins (Gedge et al., 2005; Hiscox, 2006; Kim et al., 2007). The examples discussed above provide great confidence that agroinfiltration-mediated transient expression of proteins provides data comparable to that obtained using transgenic plants. Therefore, in addition to the essential task of localizing the proteome, transient expression assays in plants will continue to reveal the subtleties of proteins and membrane dynamics in pathogen-infected cells which, ultimately, will aid in establishing the relationship between pathogen-induced changes in protein localization and gene expression (Goodin et al., 2005).
NEW TIRF FOR PLANT CELL BIOLOGY
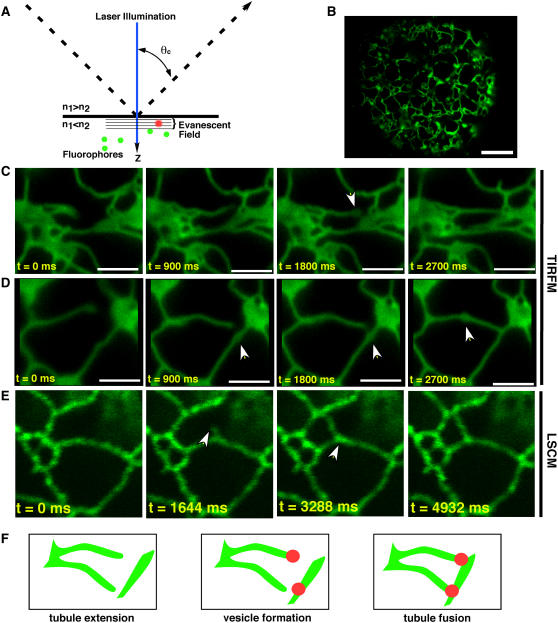
At present, protein localization studies are heavily reliant on wide-field or confocal microscopy. Clearly, the instrumentation in popular use is returning novel information at exponentially increasing rates. Yet, the question arises—are we seeing all that is possible? For this reason we have explored the use of TIRFM (for review, see Schneckenburger, 2005; Jaiswal and Simon, 2007) for monitoring fusion events of cortical ER tubules in plant protoplasts (Fig. 5). The underlying principle of TIRFM is that incident light rays that strike the interface of two media of differing refractive indices at greater than the critical angle result in the rays being totally internally reflected instead of passing through the second medium (Fig. 5A). At the point of reflection an evanescent wave is produced in the medium of lower refractive index (Fig. 5A). Fluorophores entering the evanescent wave, which is in the order of 100 nm, are excited, resulting in fluorescence detection with exceedingly high signal to noise ratios. Thus, TIRFM is well suited for monitoring events occurring at the cell surface. Additionally, as TIRFM instruments use CCD detectors to capture fluorescence emission, molecular events occurring on a rapid time scale can be captured, whereas the same events are missed when using point-detection systems in confocal microscopes (Fig. 5, D and E). We were able to observe that ER tubules appear to fuse at foci, where punctae accumulate either at the tip (Fig. 5C) of tubules or in internal sites of the extended tubule (Fig. 5D). In contrast to the results obtained with TIRFM, the resolution of confocal imaging was unable to capture tubule extension and fusion events (Fig. 5E).
Figure 5.
A, The underlying principle of TIRFM is that incident light rays (dashed line) that strike the interface of two media of differing refractive indices (n) at greater than the critical angle (θc) result in the rays being totally internally reflected instead of passing through the second medium. At the point of reflection an evanescent wave is produced in the medium of lower refractive index. Fluorophores (green dots) entering the evanescent wave, which is in the order of 100 nm, are excited (red dots), resulting in fluorescence detection with exceedingly high signal to noise ratios. B, TIRF micrograph of an N. benthamiana protoplast expressing m5GFP-ER. C and D, TIRF micrographs showing time series of ER tubule extension and fusion. Note that the point of fusion occurs at loci where puncta form at the termini (C) or within (D) tubules. E, Confocal micrographs showing ER tubule extension and fusion lack the speed and resolution of TIRFM. F, Model suggesting that puncta form and define the sites of ER tubule fusion.
While these data are insufficient to develop a significant model of ER-tubule fusion, they do serve to remind students to be cognizant of, and to seek out, new imaging techniques and instruments. Indeed, even if this is not the best of all possible worlds, our science demands the best possible micrographs.
Acknowledgments
Given the limitations on the length of this article and the breadth of live-cell imaging in plants, we apologize to colleagues whose excellent work was not cited in this review. We thank Ryan Gutierrez for critically reading the manuscript prior to submission. This manuscript is published with the approval of the Director of the Kentucky Agricultural Experiment Station as journal article number 07–12–097.
This work was supported by U.S. Department of Agriculture and Kentucky Tobacco Research and Development Center awards (to M.G.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Michael M. Goodin (mgoodin@uky.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Bates IR, Wiseman PW, Hanrahan JW (2006) Investigating membrane protein dynamics in living cells. Biochem Cell Biol 84 825–831 [DOI] [PubMed] [Google Scholar]
- Bhat RA, Lahaye T, Panstruga R (2006) The visible touch: in planta visualization of protein-protein interactions by fluorophore-based methods. Plant Methods 2 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi F, Irons SL, Johansen J, Kotzer A, Neumann U (2004) GFP is the way to glow: bioimaging of the plant endomembrane system. J Microsc 214 138–158 [DOI] [PubMed] [Google Scholar]
- Brasch MA, Hartley JL, Vidal M (2004) ORFeome cloning and systems biology: standardized mass production of the parts from the parts-list. Genome Res 14 2001–2009 [DOI] [PubMed] [Google Scholar]
- Casey PJ (1995) Protein lipidation in cell signaling. Science 268 221–225 [DOI] [PubMed] [Google Scholar]
- Chakrabarty R, Banerjee R, Chung SM, Farman M, Citovsky V, Hogenhout SA, Tzfira T, Goodin M (2007) pSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: probing Nicotiana benthamiana-virus interactions. Mol Plant Microbe Interact 20 740–750 [DOI] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC (1994) Green fluorescent protein as a marker for gene expression. Science 263 802–805 [DOI] [PubMed] [Google Scholar]
- Chudakov DM, Chepurnykh TV, Belousov VV, Lukyanov S, Lukyanov KA (2006) Fast and precise protein tracking using repeated reversible photoactivation. Traffic 10 1304–1310 [DOI] [PubMed] [Google Scholar]
- Chung SM, Frankman EL, Tzfira T (2005) A versatile vector system for multiple gene expression in plants. Trends Plant Sci 10 357–361 [DOI] [PubMed] [Google Scholar]
- Citovsky V, Lee LY, Vyas S, Glick E, Chen MH, Vainstein A, Gafni Y, Gelvin SB, Tzfira T (2006) Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J Mol Biol 362 1120–1131 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R, Cyr R, Gilroy S (2006) Using intrinsically fluorescent proteins for plant cell imaging. Plant J 45 599–615 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45 616–629 [DOI] [PubMed] [Google Scholar]
- Gedge LJ, Morrison EE, Blair GE, Walker JH (2005) Nuclear actin is partially associated with Cajal bodies in human cells in culture and relocates to the nuclear periphery after infection of cells by adenovirus 5. Exp Cell Res 303 229–239 [DOI] [PubMed] [Google Scholar]
- Gest H (2004) The discovery of microorganisms by Robert Hooke and Antoni Van Leeuwenhoek, fellows of the Royal Society. Notes Rec R Soc Lond 58 187–201 [DOI] [PubMed] [Google Scholar]
- Giepmans BN, Adams SR, Ellisman MH, Tsien RY (2006) The fluorescent toolbox for assessing protein location and function. Science 312 217–224 [DOI] [PubMed] [Google Scholar]
- Goldberg KB, Modrell B, Hillman BI, Heaton LA, Choi TJ, Jackson AO (1991) Structure of the glycoprotein gene of sonchus yellow net virus, a plant rhabdovirus. Virology 185 32–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodin M, Yelton S, Ghosh D, Mathews S, Lesnaw J (2005) Live-cell imaging of rhabdovirus-induced morphological changes in plant nuclear membranes. Mol Plant Microbe Interact 18 703–709 [DOI] [PubMed] [Google Scholar]
- Goodin MM, Chakrabarty R, Yelton S, Martin K, Clark A, Brooks R (2007) Membrane and protein dynamics in live plant nuclei infected with Sonchus yellow net virus, a plant-adapted rhabdovirus. J Gen Virol 88 1810–1820 [DOI] [PubMed] [Google Scholar]
- Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO (2002) pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J 3 375–383 [DOI] [PubMed] [Google Scholar]
- Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, Lukyanov KA (2006) Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol 24 461–465 [DOI] [PubMed] [Google Scholar]
- Habuchi S, Ando R, Dedecker P, Verheijen W, Mizuno H, Miyawaki A, Hofkens J (2005) Reversible single-molecule photoswitching in the GFP-like fluorescent protein Dronpa. Proc Natl Acad Sci USA 102 9511–9516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilson P, Allemeersch J, Altmann T, Aubourg S, Avon A, Beynon J, Bhalerao RP, Bitton F, Caboche M, Cannoot B, et al (2004) Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res 14 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilson P, Small I, Kuiper MT (2003) European consortia building integrated resources for Arabidopsis functional genomics. Curr Opin Plant Biol 6 426–429 [DOI] [PubMed] [Google Scholar]
- Hink MA, Bisselin T, Visser AJ (2002) Imaging protein-protein interactions in living cells. Plant Mol Biol 50 871–883 [DOI] [PubMed] [Google Scholar]
- Hiscox J, editor (2006) Viruses and the Nucleus. John Wiley & Sons, West Sussex, UK
- Jaiswal JK, Simon SM (2007) Imaging single events at the cell membrane. Nat Chem Biol 3 92–98 [DOI] [PubMed] [Google Scholar]
- Jares-Erijman EA, Jovin TM (2006) Imaging molecular interactions in living cells by FRET microscopy. Curr Opin Chem Biol 10 409–416 [DOI] [PubMed] [Google Scholar]
- Kim SH, Ryabov EV, Kalinina NO, Rakitina DV, Gillespie T, MacFarlane S, Haupt S, Brown JW, Taliansky M (2007) Cajal bodies and the nucleolus are required for a plant virus systemic infection. EMBO J 26 2169–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroleva OA, Tomlinson ML, Leader D, Shaw P, Doonan JH (2005) High-throughput protein localization in Arabidopsis using Agrobacterium-mediated transient expression of GFP-ORF fusions. Plant J 41 162–174 [DOI] [PubMed] [Google Scholar]
- Kumar PP, Usha R, Zrachya A, Levy Y, Spanov H, Gafni Y (2006) Protein-protein interactions and nuclear trafficking of coat protein and betaC1 protein associated with Bhendi yellow vein mosaic disease. Virus Res 122 127–136 [DOI] [PubMed] [Google Scholar]
- Levy A, Erlanger M, Rosenthal M, Epel BL (2007) A plasmodesmata-associated beta-1,3-glucanase in Arabidopsis. Plant J 49 669–682 [DOI] [PubMed] [Google Scholar]
- Li S, Ehrhardt DW, Rhee SY (2006) Systematic analysis of Arabidopsis organelles and a protein localization database for facilitating fluorescent tagging of full-length Arabidopsis proteins. Plant Physiol 141 527–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y, et al (2006) ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol 24 841–847 [DOI] [PubMed] [Google Scholar]
- Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA (1999) Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol 17 969–973 [DOI] [PubMed] [Google Scholar]
- Merzlyak EM, Goedhart J, Shcherbo D, Bulina ME, Shcheglov AS, Fradkov AF, Gaintzeva A, Lukyanov KA, Lukyanov S, Gadella TW, et al (2007) Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat Methods 4 555–557 [DOI] [PubMed] [Google Scholar]
- Murphy RF (2005) Location proteomics: a systems approach to subcellular location. Biochem Soc Trans 33 535–538 [DOI] [PubMed] [Google Scholar]
- Nakano A (2002) Spinning-disk confocal microscopy—a cutting-edge tool for imaging of membrane traffic. Cell Struct Funct 27 349–355 [DOI] [PubMed] [Google Scholar]
- Orenstein JM (2000) Isn't a picture still worth a thousand words? Ultrastruct Pathol 24 67–74 [DOI] [PubMed] [Google Scholar]
- Patterson GH, Lippincott-Schwartz J (2004) Selective photolabeling of proteins using photoactivatable GFP. Methods 32 445–450 [DOI] [PubMed] [Google Scholar]
- Pendle AF, Clark GP, Boon R, Lewandowska D, Lam YW, Andersen J, Mann M, Lamond AI, Brown JW, Shaw PJ (2005) Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Biol Cell 16 260–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepperkok R, Ellenberg J (2006) High-throughput fluorescence microscopy for systems biology. Nat Rev Mol Cell Biol 7 690–696 [DOI] [PubMed] [Google Scholar]
- Rual JF, Hill DE, Vidal M (2004) ORFeome projects: gateway between genomics and omics. Curr Opin Chem Biol 8 20–25 [DOI] [PubMed] [Google Scholar]
- Runions J, Brach T, Külner S, Hawes C (2006) Photoactivation of GFP reveals protein dynamics within the endoplasmic reticulum membrane. J Exp Bot 57 43–50 [DOI] [PubMed] [Google Scholar]
- Ruzin SE (1999) Plant Microtechnique and Microscopy. Oxford University Press, New York
- Sauer M, Paciorek T, Benkova E, Friml J (2006) Immunocytochemical techniques for whole-mount in situ protein localization in plants. Nat Protocols 1 98–103 [DOI] [PubMed] [Google Scholar]
- Schneckenburger H (2005) Total internal reflection fluorescence microscopy: technical innovations and novel applications. Curr Opin Biotechnol 16 13–18 [DOI] [PubMed] [Google Scholar]
- Schob H, Kunz C, Meins F Jr (1997) Silencing of transgenes introduced into leaves by agroinfiltration: a simple, rapid method for investigating sequence requirements for gene silencing. Mol Gen Genet 256 581–585 [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22 1567–1572 [DOI] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY (2005) A guide to choosing fluorescent proteins. Nat Methods 2 905–909 [DOI] [PubMed] [Google Scholar]
- Simpson JC, Neubrand VE, Wiemann S, Pepperkok R (2001) Illuminating the human genome. Histochem Cell Biol 115 23–29 [DOI] [PubMed] [Google Scholar]
- Sterck L, Rombauts S, Vandepoele K, Rouze P, Van de Peer Y (2007) How many genes are there in plants (… and why are they there)? Curr Opin Plant Biol 10 199–203 [DOI] [PubMed] [Google Scholar]
- Stewart CN Jr (2006) Go with the glow: fluorescent proteins to light transgenic organisms. Trends Biotechnol 24 155–162 [DOI] [PubMed] [Google Scholar]
- Tardif G, Kane NA, Adam H, Labrie L, Major G, Gulick P, Sarhan F, Laliberte JF (2007) Interaction network of proteins associated with abiotic stress response and development in wheat. Plant Mol Biol 63 703–718 [DOI] [PubMed] [Google Scholar]
- Tian GW, Mohanty A, Chary SN, Li S, Paap B, Drakakaki G, Kopec CD, Li J, Ehrhardt D, Jackson D, et al (2004) High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol 135 25–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Tian GW, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V (2005) pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol 57 503–516 [DOI] [PubMed] [Google Scholar]
- Vitriol EA, Uetrecht AC, Shen F, Jacobson K, Bear JE (2007) Enhanced EGFP-chromophore-assisted laser inactivation using deficient cells rescued with functional EGFP-fusion proteins. Proc Natl Acad Sci USA 104 6702–6707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voltaire F (1759) Candide. Translated by John Butt (1947). Penguin Classics; Deluxe edition (October 25, 2005). Penguin Putnam, New York
- Walhout AJ, Temple GF, Brasch MA, Hartley JL, Lorson MA, van den Heuvel S, Vidal M (2000) GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol 328 575–592 [DOI] [PubMed] [Google Scholar]
- Wang E, Babbey CM, Dunn KW (2005) Performance comparison between the high-speed Yokogawa spinning disc confocal system and single-point scanning confocal systems. J Microsc 218 148–159 [DOI] [PubMed] [Google Scholar]
- Ward TH, Brandizzi F (2004) Dynamics of proteins in Golgi membranes: comparisons between mammalian and plant cells highlighted by photobleaching techniques. Cell Mol Life Sci 61 172–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemann S, Arlt D, Huber W, Wellenreuther R, Schleeger S, Mehrle A, Bechtel S, Sauermann M, Korf U, Pepperkok R, et al (2004) From ORFeome to biology: a functional genomics pipeline. Genome Res 14 2136–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortman JR, Haas BJ, Hannick LI, Smith RK Jr, Maiti R, Ronning CM, Chan AP, Yu C, Ayele M, Whitelaw CA, et al (2003) Annotation of the Arabidopsis genome. Plant Physiol 132 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Campbell RE, Ting AY, Tsien RY (2002) Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol 3 906–918 [DOI] [PubMed] [Google Scholar]
- Zhang FL, Casey PJ (1996) Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem 65 241–269 [DOI] [PubMed] [Google Scholar]