Abstract
Plants use light as a major source of information for optimizing growth and development. The photoreceptor phytochrome A (phyA) mediates various far-red light induced responses. Here, we show that Arabidopsis FHY3 and FAR1, which encode two proteins related to Mutator-like transposases, act together to modulate phyA signaling by directly activating the transcription of FHY1 and FHL, whose products are essential for light-induced phyA nuclear accumulation and subsequent light responses. FHY3 and FAR1 possess separable DNA-binding and transcriptional activation domains that are highly conserved in Mutator-like transposases. Further, expression of FHY3 and FAR1 is negatively regulated by phyA signaling. We propose that FHY3 and FAR1 define a novel class of transcription factors co-opted from an ancient Mutator-like transposase(s) to modulate phyA signaling homeostasis in higher plants.
Plants constantly monitor their light environment in order to grow and develop optimally, using a battery of photoreceptors. Phytochromes are a family of photoreceptors that monitors the incident red (R, 600-700 nm) and far-red (FR, 700-750 nm) light wavelengths by switching reversibly between the R-absorbing, biologically inactive Pr form and the FR-absorbing, biologically active Pfr form (1, 2). Upon photoactivation, phyA, the primary photoreceptor for FR light, is translocated from the cytoplasm into the nucleus to induce FR-responsive gene expression required for various photoresponses, such as seed germination, seedling de-etiolation, FR-preconditioned blocking of greening, and flowering (3). Genetic studies have identified two pairs of homologous genes essential for phyA signaling: FAR1 (far-red-impaired response 1) and FHY3 (far-red elongated hypocotyl 3); FHY1 (far-red elongated hypocotyl 1) and FHL (FHY1-like) (4-7). FHY1 and FHL have been implicated in mediating the light-dependent nuclear accumulation of phyA (8, 9). However, the biochemical function of FHY3 and FAR1 remains to be elucidated.
FHY3 and FAR1 share extensive sequence homology with MURA, the transposase encoded by the maize Mutator element, and the predicted transposase of the maize mobile element Jittery (10, 11). Both of these transposons are members of the superfamily of Mutator-like elements (MULEs) (12). Database mining and phylogenetic analysis revealed that FHY3/FAR1-like sequences are present in various angiosperms and fall into several phylogenetic clusters intermingled with MULE transposases (13, table S1 and fig. S1). These proteins share an N-terminal C2H2-type zinc-chelating motif of the WRKY-GCM1 family, a central putative core transposase domain, and a C-terminal SWIM motif (14, 15), with highly conserved predicted secondary/tertiary structures (fig. S2 and S3). To investigate the molecular function of FHY3 and FAR1, we generated transgenic plants expressing FHY3 and FAR1 proteins fused with a glucocorticoid receptor (GR) to control their nuclear localization (16). Both the FHY3p::FHY3-GR and FAR1p::FAR1-GR transgenes conferred a dexamethasone (DEX)-dependent rescue of the respective mutant phenotype (fig. S4), indicating that FHY3 and FAR1 act in the nucleus.
Previous studies suggested that FHY3 is required for maintaining proper expression levels of FHY1 and FHL in a light-independent manner (7, 10). Semi-quantitative reverse transcriptase (RT)-PCR analysis showed that DEX or DEX plus cycloheximide (CHX, a protein synthesis inhibitor, 17, fig. S5) treatment, but not mock treatment, restored the expression levels of FHY1 and FHL in both dark- and FR-light grown FHY3p::FHY3-GR/fhy3-4 transgenic seedlings (Fig. 1A). This observation suggests that FHY3 activates FHY1 and FHL expression. In addition, quantitative RT-PCR showed that FHY1 expression was also reduced in the far1-2 mutant, and was much further reduced in the fhy3 far1 double mutant, compared to the wild-type seedlings (Fig. 1B). This result suggests that FHY3 and FAR1 act together to up-regulate FHY1 expression.
Fig. 1. FHY3 and FAR1 directly up-regulate FHY1 and FHL expression.
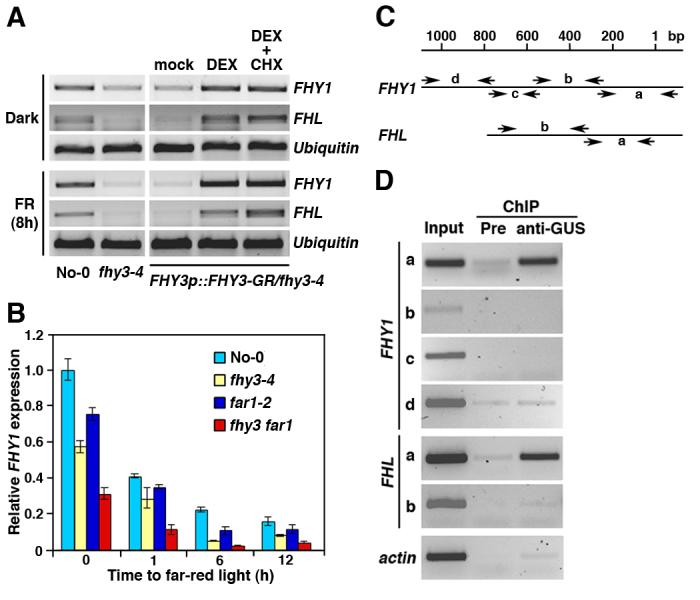
(A) Both FHY1 and FHL are up-regulated by DEX or DEX plus CHX treatment in the FHY3p::FHY3-GR/fhy3-4 transgenic plants. Seedlings were grown in darkness for 4 days then kept in darkness or transferred to FR light for 8 hours (h) before analysis. Expression of an Ubiquitin gene was shown as a control. No-0: Wild-type No-0 ecotype. (B) Reduced expression of FHY1 in the fhy3-4, far1-2, and fhy3 far1 double mutants, compared to the wild-type No-0 plants. Bars represent standard deviations of triplicate experiments. (C) Diagram of the promoter fragments of FHY1 and FHL. The locations of PCR primers used for enrichment test are indicated. “1” indicates the putative transcription initiation site. (D) Enrichment of the “a” fragments of the FHY1 and FHL promoters from the anti-GUS ChIP samples. Pre: Preimmune.
We next performed a chromatin immunoprecipitation (ChIP) assay to test for a direct interaction of FHY3 with the FHY1 and FHL promoters in vivo, using the 35S::GUS-FHY3/fhy3-4 transgenic plants (5). Multiplex PCR revealed enrichment for the “a” fragments (365-bp and 353-bp, respectively) of the FHY1 and FHL promoters in the anti-GUS ChIP samples, compared to the ChIP samples prepared with preimmune anti-sera and the Actin gene control (Fig.1, C and D). This result indicates that FHY3 directly occupies the FHY1 and FHL promoters in vivo. Moreover, constitutive overexpression of FHY1 suppressed the phenotypes of the fhy3-4, far1-2, and fhy3 far1 double mutants (fig. S6). Further, in response to FR treatment, the nuclear accumulation of phyA-GFP (Green Fluorescence Protein) is modestly reduced in the fhy3-4 mutant (reduced to about 60% of the wild-type levels), but is essentially abolished in the fhy3 far1 double mutant (fig. S7). Together, these findings suggest that FHY3 and FAR1 act together to regulate phyA nuclear accumulation through direct activation of FHY1 and FHL expression.
We next used a yeast one-hybrid assay to delineate the DNA sequences to which FHY3 and FAR1 bind. GAD-FHY3 or GAD-FAR1 fusion proteins (GAD, GAL4 transcriptional activation domain), but not GAD alone, activated the LacZ reporter genes driven by the FHY1 and FHL promoters (Fig. 2A). Deletion analysis narrowed down the FHY3/FAR1 binding site to a 39-bp promoter subfragment located on the “a” fragment for both FHY1 and FHL (Fig. 2B). Notably, these subfragments share a stretch of consensus sequence, 5′-TTCACGCGCC-3′ (Fig. 2C). Mutating the core sequence “CACGCGC” of this motif (m2 and m3 for FHY1, m5 for FHL) abolished the reporter gene activation by both GAD-FHY3 and GAD-FAR1. Mutating the flanking sequences (m1 and m4) did not obviously affect the reporter gene activation by GAD-FAR1, but clearly reduced activation by GAD-FHY3 (Fig. 2B). Thus, “CACGCGC” likely defines a cis-element that confers specific binding for FHY3 and FAR1 and is named FBS for FHY3-FAR1 binding site.
Fig. 2. FHY3 and FAR1 directly bind to the FBS motif present in the FHY1 and FHL promoters via the N-terminal zinc-finger motif.
(A) GAD-FHY3 and GAD-FAR1, but not GAD itself, strongly activate expression of the LacZ reporter genes driven by the FHY1 and FHL promoters in yeast. (B) GAD-FHY3 and GAD-FAR1 activate the LacZ reporter genes driven by the wild-type 39-bp subfragments of FHY1 and FHL promoters (wt::LacZ) in yeast. Mutations in the FBS motif (m2, m3, and m5) abolish activation of the LacZ reporter gene expression. (C) Diagram of the wild-type and mutant FHY1 and FHL subfragments used to drive the LacZ reporter gene expression and as probes in electrophoretic mobility shift assay (EMSA). The wild-type FBS motif is shown in red. Nucleotide substitutions in the mutant fragments are underlined. (D) and (E) EMSA assay showing that GST-FHY3N protein, but not GST by itself, specifically binds to the FHY1 and FHL wild-type probes (D), but not the m2, m3, and m5 mutant probes (E). Arrowheads indicate the up-shifted bands. Triangle indicates the super-shifted DNA-protein-antibody complex when incubated with anti-GST antibodies. Asterisks indicate non-specific binding. FP: free probe.
Domain deletion analysis revealed that the N-terminal fragments of FHY3 and FAR1 are necessary and sufficient for activating the LacZ reporter genes driven by the FHY1 and FHL promoters (fig. S8). Consistent with this, electrophoretic mobility shift assay (EMSA) showed that recombinant GST-FHY3N fusion protein (glutathione-S-transferase fused with the first 200 amino acids of FHY3, including the zinc finger motif) caused an up-shift of the radiolabeled wild-type FHY1 and FHL probes (Fig. 2D), but not the m2, m3, and m5 mutant probes (Fig. 2E). Moreover, addition of anti-GST antibodies caused a super-shift of the wild-type probes (Fig. 2D). Further, pre-incubation of the GST-FHY3N fusion proteins with two metal chelators, 1, 10-o-phenanthroline or EDTA, effectively reduced DNA-binding activity (fig. S9). Thus, we conclude that FHY3 binds directly to the FBS motif using the N-terminal zinc-finger motif. Genome wide analysis using the PatMatch program (18) against an Arabidopsis promoter database (http://stan.cropsci.uiuc.edu/sift/index.php) revealed that the FBS motif is also present in the promoters of hundreds of other genes, including the red light photoreceptor PHYTOCHROME B (PHYB), CIRCADIAN CLOCK-ASSOCIATED1 (CCA1) and EARLY FLOWERING4 (ELF4). Yeast one-hybrid assay showed that GAD-FHY3 and GAD-FAR1 are capable of activating the LacZ reporter genes driven by PHYB, ELF4 and CCA1 promoter fragments containing the wild-type FBS motif, but not a mutated FBS motif (fig. S10). This observation is consistent with a reported role of FHY3 in gating red light signaling to the circadian clock (19).
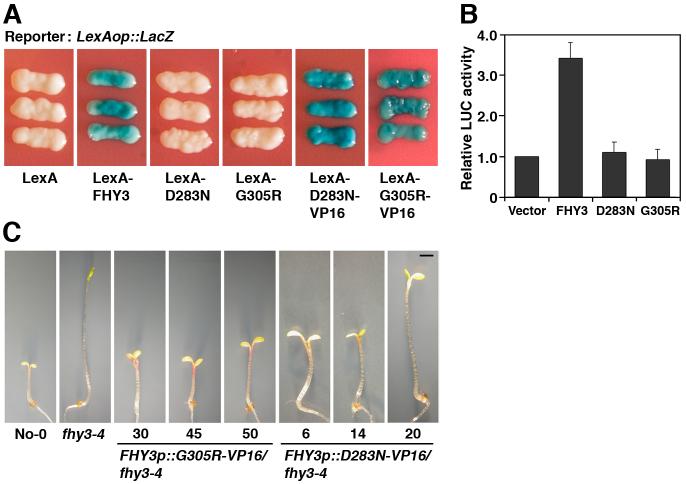
To test whether FHY3 possesses an intrinsic transcriptional regulatory activity, we fused a full-length FHY3 with the LexA DNA-binding domain. The LexA-FHY3 fusion protein, but not LexA alone, activated a LacZ reporter gene driven by the LexA operator (Fig. 3A). Two amino acid-substituted FHY3 proteins corresponding to the fhy3-9 (G305R) and fhy3-10 (D283N) mutant alleles (5) failed to activate the LacZ reporter gene (Fig. 3A), despite comparable levels of expression for the wild-type and mutant FHY3 fusion proteins. In addition, wild-type FHY3 protein, but not the G305R or D283N mutant proteins, activated a luciferase reporter gene driven by the FHY1 promoter in Arabidopsis protoplasts (Fig. 3B). Further, fusion with the VP16 activation domain of herpes simplex virus restored the transcriptional activation activity of G305R and D283N (Fig. 3A), and the fusion proteins conferred a complete or partial rescue of the fhy3-4 mutant phenotype (Fig. 3C). These results suggest that the intrinsic transcriptional activation activity of FHY3 is essential for its biological function. Domain deletion analysis revealed that the C-terminal region of FHY3 and FAR1 that lacks the N-terminal zinc-finger motif is necessary and fully capable of activating the reporter gene expression in yeast, whereas their N-terminal DNA-binding domains are unable to activate the reporter gene (fig. S11). These observations suggest that FHY3 and FAR1 possess separable DNA-binding and transcriptional activation domains.
Fig. 3. FHY3 possesses an intrinsic transcriptional activation activity.
(A) LexA-FHY3, LexA-D283N-VP16, and LexA-G305R-VP16 fusion proteins activate the LexAop::LacZ reporter gene expression in yeast, whereas LexA by itself, LexA-D283N or LexA-G305R fail to activate the reporter gene expression. (B) Wild-type FHY3, but not D283N and G305R, activates the FHY1p::Luc reporter gene expression in Arabidopsis protoplasts. Bars stand for standard deviations of triplicate experiments. (C) Images of 4-day old FR-grown seedlings of multiple independent lines showing that the FHY3p::D283N-VP16 and FHY3p::G305R-VP16 fusion genes confer complete or partial rescue of the fhy3-4 mutant phenotype. Bar: 2 mm.
Finally, we examined how FR light regulates the expression of FHY3 and FAR1 using quantitative RT-PCR. In a wild-type background, the transcript levels of FHY3 declined rapidly after exposure to FR light. Expression of FAR1 was also down-regulated by FR light, although with slower kinetics and to a lesser degree (Fig. 4A). Expression of FHY1 and FHL displayed a pattern similar to that of FHY3 (Fig. 4B), consistent with their being the direct target genes of FHY3 and FAR1. In contrast, expression of FHY3 and FAR1 remained high in the phyA-211 mutant under FR light (Fig. 4, C and D). These results indicate that expression of FHY3 and FAR1 is subject to a negative feedback regulation by phyA signaling and suggest that FHY3 and FAR1 act at a focal point of a feedback loop that maintains the homeostasis of phyA signaling (fig. S12).
Fig. 4. Down-regulation of FHY3, FAR1, FHY1 and FHL by phyA signaling.
(A) and (B) The transcript levels of FHY3 and FAR1 (A), FHY1 and FHL (B) are down-regulated by FR light. (C) and (D) The transcript levels of FHY3 (C) and FAR1 (D) remain relatively stable in the phyA-211 mutant, compared to the Columbia wild-type (Col) background. The expression levels in dark-grown wild-type plants were set as 1. Bars indicate standard deviations of triplicate experiments.
Our phylogenetic and functional analyses support a scenario whereby one or several related MULE transposases gave rise to the FHY3/FAR1-related genes during the evolution of angiosperms through a process termed “molecular domestication” (20, 21), with concomitant loss of the ability to transpose (fig. S13). Similar to this, DAYSLEEPER, an Arabidopsis hAT-like transposase, has been shown to act as a DNA-binding protein and is essential for plant development (22). However, it is not known whether this protein can directly regulate gene expression. Our results demonstrate that a transposase-derived protein can bind a promoter region and directly stimulate the transcription of that gene. Innovation of phyA, which occurred prior to the origin of angiosperms, has been hypothesized to confer adaptive advantage to the successful colonization of the first angiosperms on the earth (23). The domestication of FHY3 and FAR1 from an ancient transposase(s) might mark an event in the evolution of angiosperms to meet the challenges of changing light environments. Our results also provide functional evidence to support the proposition that transposable elements, which are prevalent throughout the genomes of many plants and animals, can serve as a source of new transcription factors that allow populations to adapt and species to evolve (24).
Supplementary Material
References and Notes
- 1.Neff MM, Fankhauser C, Chory J. Genes Dev. 2000;14:257. [PubMed] [Google Scholar]
- 2.Quail PH. Nat. Rev. Mol. Cell. Biol. 2002;3:85. doi: 10.1038/nrm728. [DOI] [PubMed] [Google Scholar]
- 3.Nagy F, Kircher S, Schäfer E. Semin. Cell Dev. Biol. 2000;11:505. doi: 10.1006/scdb.2000.0202. [DOI] [PubMed] [Google Scholar]
- 4.Hudson M, Ringli C, Boylan MT, Quail PH. Genes Dev. 1999;13:2017. doi: 10.1101/gad.13.15.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H, Deng XW. EMBO J. 2002;21:1339. doi: 10.1093/emboj/21.6.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desnos T, Puente P, Whitelam GC, Harberd NP. Genes Dev. 2001;15:2980. [Google Scholar]
- 7.Zhou Q, et al. Plant J. 2005;43:356. doi: 10.1111/j.1365-313X.2005.02453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiltbrunner A, et al. Curr. Biol. 2005;15:2125. doi: 10.1016/j.cub.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 9.Hiltbrunner A, et al. Plant Cell Physiol. 2006;47:1023. doi: 10.1093/pcp/pcj087. [DOI] [PubMed] [Google Scholar]
- 10.Hudson ME, Lisch DR, Quail PH. Plant J. 2003;34:453. doi: 10.1046/j.1365-313x.2003.01741.x. [DOI] [PubMed] [Google Scholar]
- 11.Lin R, Wang H. Plant Physiol. 2004;136:4010. doi: 10.1104/pp.104.052191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lisch D. Trends Plant Sci. 2002;7:498. doi: 10.1016/s1360-1385(02)02347-6. [DOI] [PubMed] [Google Scholar]
- 13.The accession numbers for Arabidopsis PHYA, FHY3, FAR1, FHY1, FHL, and the maize MURA and the predicted transposase of Jittery are NP_172428, NP_188856, NP_567455, NP_181304, AAC23638, AAA21566 and AAF66982, respectively.
- 14.Babu MM, Iyer LM, Balaji S, Aravind L. Nucleic Acids Res. 2006;34:6505. doi: 10.1093/nar/gkl888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makarova KS, Aravind L, Koonin EV. Trends Biochem. Sci. 2002;27:384. doi: 10.1016/s0968-0004(02)02140-0. [DOI] [PubMed] [Google Scholar]
- 16.Wagner D, Sablowski RWM, Meyerowitz EM. Science. 1999;285:582. doi: 10.1126/science.285.5427.582. [DOI] [PubMed] [Google Scholar]
- 17.The effectiveness of CHX treatment in inhibiting protein synthesis was shown by blocking HY5 protein accumulation in FR light-treated Arabidopsis seedlings (fig. S5). A full description of the Materials and Methods are available as supporting material on Science Online.
- 18.Yan T, et al. Nucleic Acids Res. 2005;33:W262. doi: 10.1093/nar/gki368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allen T, et al. Plant Cell. 2006;18:2506. doi: 10.1105/tpc.105.037358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller WJ, McDonald JF, Nouaud D, Anxolabéhère D. Genetica. 1999;107:197. [PubMed] [Google Scholar]
- 21.We observed partial synteny conservation of Arabidopsis FHY3 and FAR1 with their orthologs in Brassica and Populus, suggesting that the genomic locations of these genes have been fixed in the eudicots (fig. S13).
- 22.Bundock P, Hooykaas P. Nature. 2005;436:282. doi: 10.1038/nature03667. [DOI] [PubMed] [Google Scholar]
- 23.Mathews S. J. Hered. 2005;96:197. doi: 10.1093/jhered/esi032. [DOI] [PubMed] [Google Scholar]
- 24.Biémont C, Vieira C. Nature. 2006;443:521. doi: 10.1038/443521a. [DOI] [PubMed] [Google Scholar]
- 25.We thank F. Nagy, X. Dong, and P. Fobert for sharing the PHYAp::AtphyA-GFP4 transgenic line, a GR construct and a VP16 template, respectively. We thank X.W. Deng, G. Martin, J. Nasrallah, D. Stern and S. Mathews for commenting on the manuscript, Z. Fei for helping promoter analysis. This work was supported by funds from Boyce Thompson Institute, Triad Foundation, and National Science Foundation (IOS-0641639) (to H.W.), UT Arlington and National Institute of Health (to C. F.), and Microsoft Corporation to CBSU (D. R. R.).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.