Abstract
A dnaB gene encoding a homologue of the Escherichia coli DNA helicase DnaB was cloned and sequenced in the thermophilic eubacterium Rhodothermus marinus, predicting a DnaB protein that harbors an intein. This DnaB intein is 428 amino acid residues long, has several putative intein sequence motifs (including two putative endonuclease motifs), and is capable of protein splicing when produced in E. coli cells. The R. marinus DnaB intein is a close homologue of a DnaB intein in the cyanobacterium Synechocystis sp. strain PCC6803. The two inteins are positioned identically in their respective DnaB proteins. They also share a 54% sequence identity (74% sequence similarity) that is markedly higher than the 37% sequence identity shared by the extein sequences of the two DnaB proteins. Horizontal intein transfer (homing) is therefore invoked to relate these two DnaB inteins. The codon usage of R. marinus DnaB intein coding sequence differs markedly from the codon usages of its flanking extein coding sequences and other genes in the same genome, suggesting more recent acquisition of the DnaB intein in this organism.
Keywords: protein splicing, mobile genetic element, DNA helicase
An intein is a protein sequence embedded in-frame within a precursor protein sequence and excised during maturation (1, 2). This maturation process is termed “protein splicing” and involves precise excision of the intein sequence and joining of the flanking extein sequences. Protein splicing is therefore considered the protein equivalent of RNA splicing and adds another layer of complexity to the Central Dogma of molecular biology. Intein coding sequences have been found in eukaryote nuclear and organelle genomes (3–6), in archaebacteria (Archaea) (1, 7–9), and in eubacteria (5, 10–13), suggesting a wide distribution. Several inteins have been characterized for protein splicing activity, and some intein sequences appear to contain all cis information required for autocatalytic protein splicing (e.g., see refs. 3, 7, and 10). The chemistry of protein splicing has been discussed in several models, and some steps have been investigated experimentally (8, 14–17). They generally involve N → S acyl shift (or N → O shift) at the splice sites (18), formation of a branched intermediate (19), and cyclization of an invariant Asn residue at the C terminus of intein to succinimide (20), leading to excision of the intein and ligation of the exteins.
Little is known about the origin and evolution of inteins. Among the more than 30 intein or intein-like sequences reported, more than 20 of them are in archaebacteria (1, 7–9), 8 are in mycobacteria (5, 10–12), and the remaining few are in other organisms (3–6, 13). There is little overall sequence similarity among different inteins, unless comparison is made between inteins found at the same position in homologous proteins of related organisms (2). Nevertheless, a number of short sequence blocks have been recognized that show a significant degree of conservation among inteins (2, 5), which suggests common or similar origins of known inteins. Some conserved intein sequence blocks were also found at or near cleavage sites of some animal virus polyproteins (5, 15) and hedgehog family polyproteins (21), although the evolutionary implication of these findings is not clear. Many inteins have sequence blocks resembling the LAGLI-DADG motifs of many intron-encoded endonucleases (22, 23), leading to the suggestion that inteins may have arisen from an intron endonuclease that acquired protein splicing activity (22).
Like many introns (24), inteins are thought to be mobile genetic elements that can potentially be transmitted through horizontal intein transfer (homing) in addition to vertical inheritance (normal chromosomal transmission) (22, 25, 26). The yeast VMA intein was shown experimentally to “home” into an allele of the same gene lacking the intein (27). The distribution of a GyrA intein among several mycobacterium species was also interpreted to suggest intein homing within mycobacteria (12). Here we report the finding and characterization of a DnaB intein in Rhodothermus marinus that suggests intein transfer (homing) across remotely related organisms. DnaB is a replicative DNA helicase whose Escherichia coli homologue is crucial for DNA replication (28). Intein coding sequences have been found recently in dnaB genes of a cyanobacterium and a chloroplast (5, 13, 29), although protein splicing was not demonstrated for the predicted inteins. These cyanobacterial and chloroplast DnaB inteins are apparently related, but it could not be determined whether they are related through vertical inheritance or through intein transfer (13). We have now found a DnaB intein in the thermophilic eubacterium R. marinus and demonstrated its protein splicing activity in E. coli cells. More importantly, our analysis suggested that the R. marinus DnaB intein is a recent acquisition in this organism and is related to the cyanobacterial DnaB intein through horizontal intein transfer (homing).
EXPERIMENTAL PROCEDURES
Gene Cloning and Sequence Analysis.
Genomic DNA was isolated from R. marinus strain R-10 (ATCC 43812) grown at 75°C in ATCC medium 1599. A 600-bp DNA probe was prepared from the genomic DNA by PCR amplification, using a pair of oligonucleotide primers: 5′-GATCTTTTGCGCTTTGTAGAAAAATCGGT-3′ and 5′-GTTATGAGCGATAATGTCGTT-3′. In cloning the dnaB gene, genomic DNA was first digested with restriction enzyme BspEI and electrophoresed in an agarose gel. DNA fragments in the 4.6-kbp region were extracted from the agarose gel and randomly inserted into plastid vector pUC118. Clones containing the dnaB gene were identified by hybridizing to the 600-bp DNA probe described above. DNA sequence was determined on both strands using the dideoxynucleotide termination method on a LiCor 4000L automated sequencer.
GenBank searches were performed using the blast search program (30). Protein sequence alignment was carried out using the Clustal W program (31). Codon usage was analyzed by the technique of Gribskov et al. (32) using the GCG CodonPreference program with a window of 75 codons. Frequency of dG + dC present in the third position of codons was analyzed using the CodonPreference program with a window of 25 codons. A codon frequency table of R. marinus was generated from three genes—a β-mannanase gene, a DNA ligase gene, and a β-glucanase gene—which totaled 1,969 codons and represented all sequenced R. marinus genes. A codon frequency table of Synechocystis sp. PCC6803 was generated from 20 known genes surrounding the dnaB gene, which totaled 4,684 codons.
Protein Production and Splicing in E. coli Cells.
Recombinant plasmid pMR1 was constructed by cloning the 2.9-kbp NcoI–BsiW I DNA fragment (blunt ended) into the expression plasmid vector pMAL-c2 (New England Biolabs) at its SalI site (blunt ended). Recombinant plasmid pMR2 was constructed by removing a 346-bp AatII–HindIII DNA fragment from pMR1. Recombinant plasmid pMR3 was constructed by removing a 1059-bp SalI DNA fragment from pMR1. E. coli cells transformed with individual recombinant plasmids were grown in liquid Luria–Bertani medium at 37°C to late logarithmic phase (OD600 = 0.5). Isopropyl β-d-thiogalactoside (IPTG) was added at this time to a final concentration of 1 mM to induce production of recombinant proteins, and the induction was continued for 3 hr at 37°C. After induction, cells were lysed in SDS-containing gel loading buffer in a boiling water bath. Cellular proteins were resolved by SDS/polyacrylamide gel electrophoresis and visualized by staining with Coomassie blue R-250.
RESULTS
Cloning and Sequencing of the R. marinus dnaB Gene.
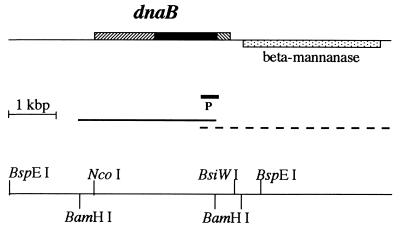
In a blast search (30) of GenBank for homologues of the Porphyra purpurea chloroplast DnaB protein, we noticed a short 3′ flanking sequence of the R. marinus β-mannanase gene (GenBank accession no. X90947). Our analysis of this sequence suggested a previously unrecognized potential coding region for a short polypeptide that has significant similarity to a 60-aa C-terminal sequence of DnaB proteins. In addition, we recognized in this sequence a putative intein motif, suggesting the presence of an intein-encoding dnaB gene in R. marinus. On the basis of this initial observation, we set out to clone the complete dnaB gene from this organism. R. marinus genomic DNA was digested with various restriction enzymes and analyzed on Southern blots by hybridizing to a 600-bp DNA probe derived from the 3′ flanking sequence of the β-mannanase gene (see Fig. 1). For each restriction enzyme that did not cut within the 600-bp probe DNA, a single DNA band was detected (data not shown), suggesting a single copy of the dnaB gene. Digesting R. marinus DNA with BspEI produced a single 4.6-kbp DNA band that hybridized to the probe DNA. DNA from this 4.6-kbp band was extracted from an agarose gel and randomly inserted into plasmid vector pUC118. Positive clones were identified by hybridizing to the same 600-bp probe DNA. Sequence determination of this cloned DNA fragment revealed a 2823-bp complete dnaB coding sequence (Fig. 1). The 3′ portion of this sequence overlaps and agrees with the previously reported 3′ flanking sequence of the β-mannanase gene, which is encoded on the opposite DNA strand and located 201 bp downstream of the dnaB gene (Fig. 1).
Figure 1.
Physical maps of R. marinus dnaB gene. On the top line, the dnaB gene is transcribed from left to right, while the neighboring β-mannanase gene (stippled box) is transcribed from right to left. The dnaB gene consists of extein coding sequences (hatched boxes) and intein coding sequence (solid box). In the middle, a solid line marks DNA sequence determined in this study, a dashed line marks DNA sequence determined previously by others, and a short thick line (P) marks a 600-bp DNA probe that was used in cloning the dnaB gene. The lower line shows restriction sites used in this study.
Identification and Sequence Analysis of the R. marinus dnaB Gene.
The R. marinus dnaB gene was identified by comparing its predicted protein sequence with sequences of known DnaB proteins (Fig. 2). This comparison revealed that the predicted R. marinus DnaB protein contains a 428-aa intein sequence (described later) bounded by a 421-aa N-extein and a 92-aa C-extein sequence (Fig. 2A). When only the extein sequences are considered, the R. marinus DnaB protein is 36%, 32%, and 35% identical to DnaB proteins of Synechocystis sp. PCC6803, P. purpurea chloroplast, and E. coli, respectively (Fig. 2B). These levels of sequence identity are comparable to the 36% sequence identity between DnaB proteins of Synechocystis sp. PCC6803 and E. coli. In addition, several sequence blocks that are highly conserved in known DnaB proteins are also present in the R. marinus DnaB. These include a putative ATP-binding motif, a putative DNA-binding motif, and a putative leucine-zipper motif (Fig. 2B). Together, these sequence features suggest strongly that the predicted R. marinus DnaB protein is a homologue of known DnaB proteins and possesses DNA helicase and ATPase activities.
Figure 2.
Sequence analysis of R. marinus DnaB protein. (A) Schematic illustration and comparison of DnaB proteins of R. marinus (Rma), Synechocystis sp. PCC6803 (Ssp), Porphyra purpurea chloroplast (Ppu), and E. coli (Eco). Hatched box and solid box represent extein and intein, respectively. Numbers of residues are shown for the R. marinus DnaB extein and intein sequences. (B) Comparison of extein sequences. Sequence names are the same as above. The R. marinus (Rma) sequence is numbered throughout, while total numbers of residues in other sequences are shown at the end of the corresponding sequences. An intein is marked by a big letter I in parentheses (e.g., at position 421 in the Rma sequence). Putative sequence motifs are marked by lines, including an ATP-binding motif (a), a DNA-binding motif (b), and a leucine-zipper motif (c). Hyphens represent gaps introduced to optimize the alignment; ∗ and . mark positions of identical and similar amino acids, respectively. (C) Comparison of intein sequences. Sequence names are the same as above. The R. marinus (Rma) sequence is numbered throughout, while the total number of residues in each sequence is shown at the end of that sequence. Putative intein motifs (blocks A to H) are marked. Hyphens represent gaps introduced to optimize the alignment; | and : mark positions of identical and similar amino acids, respectively.
Identification and Sequence Analysis of the R. marinus DnaB Intein.
The predicted R. marinus DnaB protein clearly contains an intein. The intein interrupts a 14-aa stretch of sequence that is extremely conserved among DnaB proteins with or without intein, so that the intein sequence boundaries are easily defined (Fig. 2B). The predicted intein boundaries also agree with other intein-defining features, including a nucleophilic residue (Cys) at the intein N terminus, a His-Asn dipeptide at the intein C terminus, and another nucleophilic residue (Ser) at the beginning of C-extein. These four residues and their positions are highly conserved among inteins and are known to be critical for the chemistry of protein splicing (18–20). In addition, the R. marinus DnaB intein contains putative intein sequence motifs (blocks A to H in Fig. 2C) that are significantly conserved among known inteins (2, 5). The R. marinus DnaB intein is similar in size to the 429-aa DnaB intein of Synechocystis sp. PCC6803 but much larger than the 150-aa DnaB intein of P. purpurea chloroplast. Nevertheless, all three DnaB inteins are positioned identically in their respective DnaB proteins.
Sequence comparison between DnaB inteins of R. marinus and Synechocystis sp. PCC6803 revealed a 54% sequence identity and a 74% sequence similarity (Fig. 2C). The much shorter DnaB intein sequence of P. purpurea chloroplast corresponded mostly to the two terminal regions of the R. marinus DnaB intein, and its 150-aa sequence is 26% and 29% identical to corresponding DnaB intein sequences of R. marinus and Synechocystis sp. PCC6803, respectively. Over the same 150-aa sequence region, a 66% sequence identity is shared between DnaB inteins of R. marinus and Synechocystis sp. PCC6803. Comparison between R. marinus DnaB intein and other known inteins revealed only low or very low levels of sequence similarity, with the highest being a 19% sequence identity to the RecA intein of Mycobacterium tuberculosis (10).
Codon Usage of R. marinus dnaB Gene.
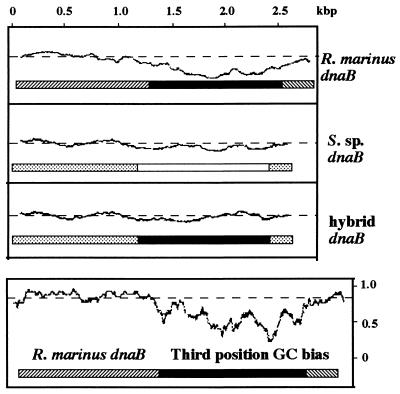
The R. marinus dnaB gene was examined for codon usage by comparing it to other known genes of the same genome. These other known genes have a total of 1,969 codons and do not encode intein. Codon usage of the R. marinus DnaB extein coding regions matched closely with codon usage of the other genes. In contrast, codon usage of the DnaB intein coding region exhibited marked deviation from the norm set by the other genes (Fig. 3). The dG+dC content of the DnaB intein coding region is 55%, which is markedly lower than the dG+dC contents of the DnaB extein coding regions (68%) and the other known genes (63–68%) in the same genome. The frequency of dG and dC at the third position of each codon was also examined, to avoid effects of amino acid composition bias. Marked deviation from the mean was observed in the intein coding region but not in the extein coding regions (Fig. 3). The average frequency of dG and dC at the third position of each codon was calculated to be 53% for the DnaB intein coding region, 87% for the DnaB extein coding regions, and 76–85% for other known genes in the same genome. Taken together, these observations suggested that the R. marinus DnaB intein coding sequence may be of foreign origin.
Figure 3.
Codon usage of the dnaB gene. (Upper) Pattern of codon usage determined by the technique of Gribskov et al. (32), using the GCG CodonPreference program with a window of 75 codons. Codon frequency tables used in the program were generated from 3 and 20 known genes for R. marinus and Synechocystis sp. PCC6803 (S. sp.), respectively. For each sequence, only the coding frame of the three possible reading frames is shown. Solid and open boxes correspond to intein coding regions of R. marinus dnaB and S. sp. dnaB, respectively. Hatched and stippled boxes correspond to extein coding regions of R. marinus dnaB and Synechocystis sp. dnaB, respectively. Hybrid dnaB is a hypothetical construct where the R. marinus DnaB intein is placed between Synechocystis sp. DnaB exteins and analyzed in the context of Synechocystis sp. genome. (Lower) Frequency of dG+dC present at the third position of each codon, which was determined using the CodonPreference program with a window of 25 codons. In each graph, codon usage typical of that organism scores close to the straight dashed line, as seen in the extein coding regions. Deviations from the norm result in troughs, which are readily apparent in the intein coding region of R. marinus dnaB.
For comparison, the dnaB gene of Synechocystis sp. PCC6803 was also examined for codon usage by comparing it to other genes of the same genome. No marked deviation from the norm was observed in the DnaB intein coding region (Fig. 3, S. sp. dnaB). The dG+dC content of the DnaB intein coding region was calculated to be 38%, which is close to the 41% average for other genes of the same genome. In a hypothetical analysis of codon usage, the R. marinus DnaB intein coding region was arbitrarily placed between DnaB extein coding regions of Synechocystis sp. PCC6803 and examined in the context of codon usage in Synechocystis sp. PCC6803. Under this hypothetical condition, the R. marinus DnaB intein coding region displayed a codon usage that is not markedly different from the norm of Synechocystis sp. PCC6803 (Fig. 3, hybrid dnaB).
Demonstration of Protein Splicing with the R. marinus DnaB Intein.
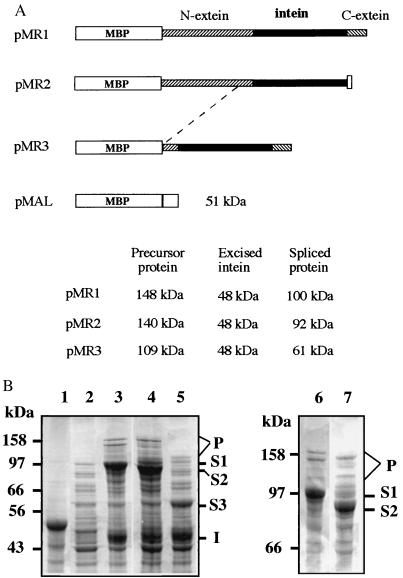
The R. marinus DnaB intein was tested for protein splicing in E. coli cells. Three recombinant plasmids (pMR1 to pMR3) were constructed to encode fusion proteins consisting of R. marinus DnaB sequence and a vector-encoded maltose-binding protein (MBP) sequence (Fig. 4A). Each fusion protein consists of the complete intein sequence flanked by various amounts of the extein sequences. pMR1 encodes the complete N- and C-extein sequences. pMR2 encodes the complete N-extein sequence and a 6-aa C-extein sequence closest to the intein, plus a vector-encoded 11-aa sequence at the C terminus. pMR3 encodes the complete C-extein sequence and a 68-aa N-extein sequence closest to the intein.
Figure 4.
Protein splicing of R. marinus DnaB. The R. marinus dnaB gene (complete or partial) was inserted into the expression plasmid vector pMAL-c2 to produce corresponding recombinant fusion proteins and to observe protein splicing. (A) Illustration of recombinant fusion proteins. Each fusion protein consists of the maltose-binding protein (MBP), the DnaB intein (solid box), and the DnaB exteins (hatched boxes) of different lengths, and some vector-encoded sequences (open boxs). Calculated molecular masses for the predicted protein products are listed. (B) Production and splicing of recombinant DnaB proteins. E. coli cells containing individual recombinant plasmids described above were induced by IPTG to produce the corresponding protein products. Total cellular proteins from induced cells were resolved by electrophoresis on SDS/polyacrylamide gels and visualized by Coomassie blue staining. Lane 1, cells transformed with pMAL and producing a 51-kDa protein, as a control. Lane 2, cells transformed with pMR1, but before IPTG induction. Lanes 3, 4, and 5, cells transformed with pMR1, pMR2, and pMR3, respectively, after IPTG induction. Lanes 6 and 7, same as lanes 3 and 4, respectively, but electrophoresed for a longer period of time. Letters S1, S2, and S3 mark positions of putative spliced proteins produced from pMR1, pMR2, and pMR3, respectively. Letter I marks position of putative excised intein. Letter P marks protein bands that may include precursor proteins and protein splicing intermediates.
Each recombinant plasmid was introduced into E. coli cells to produce the corresponding fusion protein and to observe possible protein splicing products (Fig. 4B). Two major protein products were observed in cells containing plasmid pMR1, and their sizes corresponded well with the predicted sizes of a spliced protein (100 kDa) and an excised intein (48 kDa), respectively. Two major protein products were also observed in cells containing plasmid pMR2, and their sizes corresponded well with the predicted sizes of a spliced protein (92 kDa), and an excised intein (48 kDa), respectively. Similarly, two major protein products were observed in cells containing plasmid pMR3, and their sizes again corresponded well with the predicted sizes of a spliced protein (61 kDa) and an excised intein (48 kDa), respectively. The putative spliced protein bands were identified not only by their individual sizes but also by their size changes, which corresponded well with alterations of either the N-extein or the C-extein sequences. The putative intein band was identified by its size as well as by the fact that its size was not affected by changing either one of the two extein sequences. In each case, one or more minor protein bands migrating near the top of the gel were also observably induced. In addition to expected precursor proteins, some of them may be protein splicing intermediates. Protein splicing has been known to produce a branched intermediate that migrates slower than the precursor protein in gel electrophoresis (14). Together, these results demonstrated a protein splicing activity of the R. marinus DnaB intein in E. coli cells.
DISCUSSION
The R. marinus dnaB Gene Encodes a Functional Intein.
The R. marinus dnaB gene was clearly identified by the protein sequence it encodes, despite the presence of an intein. The R. marinus DnaB intein was identified by several criteria: (i) it exists as a large intervening sequence interrupting a highly conserved region of the DnaB protein; (ii) it contains all of the previously recognized structural features of intein, including seven putative intein sequence blocks and several amino acid residues that are critical for protein splicing; and (iii) it exhibited protein splicing activity when produced in E. coli cells. Thus we have found an intein in a thermophilic eubacterium. Complete extein sequences appear not to be required for the R. marinus DnaB intein to undergo protein splicing (Fig. 4). This may be similar to some other intein sequences that are sufficient for protein splicing when placed within a foreign or target protein immediately before a nucleophilic residue (10, 14, 33), suggesting that all cis information required for protein splicing is contained within the intein. It was not determined whether the R. marinus DnaB protein undergoes protein splicing in vivo, but it is expected to do so, because the intein is clearly functional in E. coli cells.
Recent Intein Homing Relating DnaB Inteins of R. marinus and Synechocystis sp.
PCC6803? The DnaB inteins of R. marinus and the cyanobacterium Synechocystis sp. PCC6803 are clearly homologous, because of their sequence similarity and their identical position in DnaB protein. R. marinus is a Gram-negative heterotrophic marine thermophile that diverges deeply from cyanobacteria and is most closely allied to the Flexibacter–Cytophaga–Bacteroides group (34). It is therefore necessary to invoke horizontal intein transfer to explain the unusually high sequence identity (54%) between the two inteins, which is significantly higher than the 37% sequence identity between their exteins. If two homologous inteins are not related through recent horizontal transfer, the intein sequences would have diverged much more than their extein sequences; this possibility is similar to the intron situation, where intron sequences typically diverge much faster than exon sequences. As a known example, the homologous VMA inteins of Saccharomyces cerevisiae and another yeast, Candida tropicalis, share only 34% sequence identity, while their extein sequences are still 87% identical (3, 4). In another example, the homologous GyrA inteins of several mycobacterium species share only 66–74% sequence identity, while their extein sequences are almost 100% identical (12). The R. marinus DnaB intein and the much smaller DnaB intein of P. purpurea chloroplast are less likely related through recent intein transfer, because the two inteins share only 26% sequence identity compared with a 31% sequence identity shared by their exteins. On the basis of all these considerations, it appears most likely that the DnaB inteins of R. marinus and Synechocystis sp. PCC6803 are related through recent horizontal intein transfer, not through vertical inheritance. Horizontal intein transfer (intein homing) has frequently been suggested (1, 2, 12, 22, 25–27). Inteins present in the same location within extein homologues from different organisms have been called intein alleles (2). Some allelic inteins exhibited similar endonuclease target site specificities (2), which may explain “intein homing” to the same location of homologous genes.
The R. marinus DnaB intein appears to be a recent acquisition in this organism, because its codon usage and dG+dC content differ markedly from the norm. This observation provides further support for the horizontal intein transfer scenario suggested above. In comparison, codon usage of the DnaB intein of Synechocystis sp. PCC6803 is not markedly different from the norm, which might suggest an early origin of the DnaB intein in this organism. But we cannot rule out the possibility that Synechocystis sp. PCC6803 also acquired its DnaB intein recently but from an organism of similar codon usage.
Although the R. marinus DnaB intein appears to be of foreign origin, the exact source of this element is not known. The codon usage analysis might suggest that DnaB intein entered R. marinus more recently from an organism whose codon usage is similar to that of Synechocystis sp. PCC6803. Although the DnaB inteins of R. marinus and Synechocystis sp. PCC6803 are most likely related through horizontal intein transfer, the path of the suggested intein transfer is not known. It may involve a single intein transfer event between the two organisms or multiple steps through intermediary organisms. Alternatively, the two organisms may have acquired their DnaB inteins independently from other organisms. R. marinus grows optimally at temperatures around 75°C, which may present a barrier against DnaB intein transfer directly from organisms like Synechocystis sp. PCC6803, which grows at 25–39°C. There are cyanobacterial species that grow at temperatures above 50°C (35), but it is not known whether they also harbor the DnaB intein.
Acknowledgments
We thank Dr. Christoph Sensen for his assistance in using the GCG CodonPreference program. This work was supported by a grant from the Medical Research Council of Canada. X.-Q.L. is a Scholar of the Evolutionary Biology Program of the Canadian Institute for Advanced Research.
ABBREVIATION
- IPTG
isopropyl β-d-thiogalactoside
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF006675).
References
- 1.Perler F B, Davis E O, Dean G E, Gimble F S, Jack W E, Neff N, Noren C J, Thorner J, Belfort M. Nucleic Acids Res. 1994;22:1125–1127. doi: 10.1093/nar/22.7.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perler F B, Olsen G J, Adam E. Nucleic Acids Res. 1997;25:1087–1093. doi: 10.1093/nar/25.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kane P M, Yamashiro C T, Wolczyk D F, Neff N, Goebl M, Stevens T H. Science. 1990;250:651–657. doi: 10.1126/science.2146742. [DOI] [PubMed] [Google Scholar]
- 4.Gu H H, Xu J, Gallagher M, Dean G E. J Biol Chem. 1993;268:7372–7381. [PubMed] [Google Scholar]
- 5.Pietrokovski S. Protein Sci. 1994;3:2340–2350. doi: 10.1002/pro.5560031218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang S, Liu X-Q. J Biol Chem. 1997;272:11869–11873. doi: 10.1074/jbc.272.18.11869. [DOI] [PubMed] [Google Scholar]
- 7.Perler F B, Comb D G, Jack W E, Moran L S, Qiang B, Kucera R B, Benner J, Slatko B E, Nwankwo D O, Hempstead S K, Carlow C K S, Jannasch H. Proc Natl Acad Sci USA. 1992;89:5577–5581. doi: 10.1073/pnas.89.12.5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hodges R A, Perler F B, Noren C J, Jack W E. Nucleic Acids Res. 1992;20:6153–6157. doi: 10.1093/nar/20.23.6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bult C J, White W, Olsen G J, Zhouk L, Fleischmann R D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 10.Davis E O, Jenner P J, Brooks P C, Colston M J, Sedgwick S G. Cell. 1992;71:201–210. doi: 10.1016/0092-8674(92)90349-h. [DOI] [PubMed] [Google Scholar]
- 11.Davis E O, Thangaraj H S, Brooks P C, Colston M J. EMBO J. 1994;13:699–703. doi: 10.1002/j.1460-2075.1994.tb06309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fsihi H, Vincent V, Cole S T. Proc Natl Acad Sci USA. 1996;93:3410–3415. doi: 10.1073/pnas.93.8.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pietrokovski S. Trends Genet. 1996;12:287–288. doi: 10.1016/0168-9525(96)20005-8. [DOI] [PubMed] [Google Scholar]
- 14.Xu M Q, Southworth M W, Mersha F B, Hornstra L J, Perler F B. Cell. 1993;75:1371–1377. doi: 10.1016/0092-8674(93)90623-x. [DOI] [PubMed] [Google Scholar]
- 15.Wallace C J. Protein Sci. 1993;2:697–705. doi: 10.1002/pro.5560020501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clarke N D. Proc Natl Acad Sci USA. 1994;91:11084–11088. doi: 10.1073/pnas.91.23.11084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper A A, Stevens T H. Trends Biochem Sci. 1995;20:351–356. doi: 10.1016/s0968-0004(00)89075-1. [DOI] [PubMed] [Google Scholar]
- 18.Shao Y, Xu M Q, Paulus H. Biochemistry. 1996;35:3810–3815. doi: 10.1021/bi952592h. [DOI] [PubMed] [Google Scholar]
- 19.Xu M Q, Comb D G, Paulus H, Noren C J, Shao Y, Perler F B. EMBO J. 1994;13:5517–5522. doi: 10.1002/j.1460-2075.1994.tb06888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shao Y, Xu M Q, Paulus H. Biochemistry. 1995;34:10844–10850. doi: 10.1021/bi00034a017. [DOI] [PubMed] [Google Scholar]
- 21.Koonin E V. Trends Biochem Sci. 1995;20:141–142. doi: 10.1016/s0968-0004(00)88989-6. [DOI] [PubMed] [Google Scholar]
- 22.Shub D A, Goodrich-Blair H. Cell. 1992;71:183–186. doi: 10.1016/0092-8674(92)90345-d. [DOI] [PubMed] [Google Scholar]
- 23.Gimble F S, Thorner J. J Biol Chem. 1993;268:21844–21853. [PubMed] [Google Scholar]
- 24.Lambowitz A M, Belfort M. Annu Rev Biochem. 1993;62:587–622. doi: 10.1146/annurev.bi.62.070193.003103. [DOI] [PubMed] [Google Scholar]
- 25.Doolittle R F. Proc Natl Acad Sci USA. 1993;90:5379–5381. doi: 10.1073/pnas.90.12.5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belfort M, Reaban M E, Coetzee T, Dalgaard J Z. J Bacteriol. 1995;177:3897–3903. doi: 10.1128/jb.177.14.3897-3903.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gimble F S, Thorner J. Nature (London) 1992;357:301–306. doi: 10.1038/357301a0. [DOI] [PubMed] [Google Scholar]
- 28.Marians K J. Annu Rev Biochem. 1992;61:673–719. doi: 10.1146/annurev.bi.61.070192.003325. [DOI] [PubMed] [Google Scholar]
- 29.Reith M, Munholland J. Plant Mol Biol Rep. 1995;13:333–335. [Google Scholar]
- 30.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 31.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gribskov M, Devereux J, Burgess R R. Nucleic Acids Res. 1984;12:539–549. doi: 10.1093/nar/12.1part2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cooper A A, Chen Y J, Lindorfer M A, Stevens T H. EMBO J. 1993;12:2575–2583. doi: 10.1002/j.1460-2075.1993.tb05913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andresson O S, Fridjonsson O H. J Bacteriol. 1994;176:6165–6169. doi: 10.1128/jb.176.20.6165-6169.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stanier R Y, Kunisawa R, Mandel M, Cohen-Bazire G. Bacteriol Rev. 1971;35:171–205. doi: 10.1128/br.35.2.171-205.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]